41.3 Missing Data
Missingness is information. First detect, then decide: delete, impute, or model it explicitly.
# Counts by variable
profile_missing(tx)
#> # A tibble: 13 × 3
#> feature num_missing pct_missing
#> <fct> <int> <dbl>
#> 1 city 0 0
#> 2 year 0 0
#> 3 month 0 0
#> 4 sales 568 0.0660
#> 5 volume 568 0.0660
#> 6 median 616 0.0716
#> 7 listings 1424 0.166
#> 8 inventory 1467 0.171
#> 9 date 0 0
#> 10 avg_price 568 0.0660
#> 11 absorption 1427 0.166
#> 12 quarter 0 0
#> 13 ym 0 0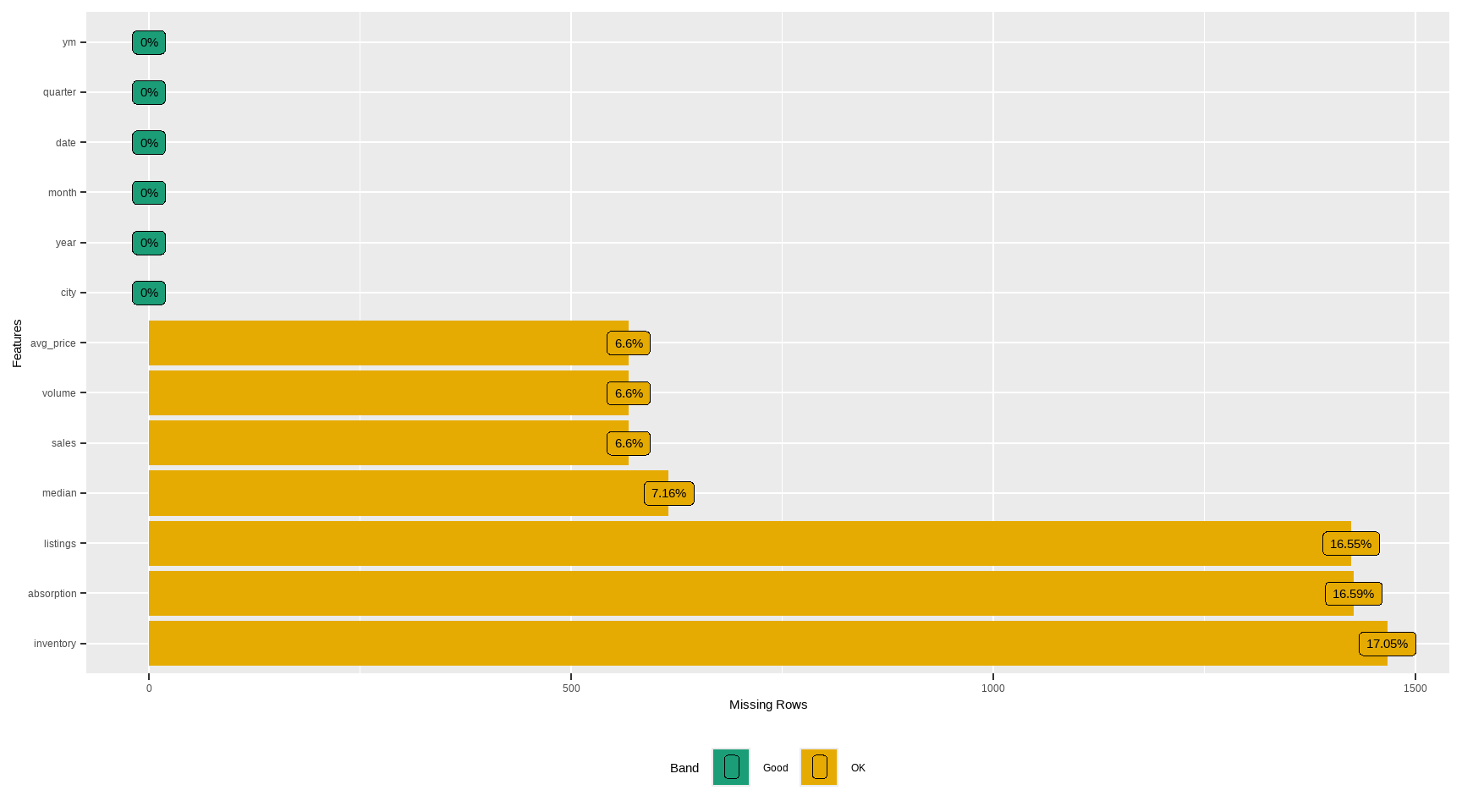
# Simple tidy summary (counts and proportions)
tx %>%
summarise(across(everything(),
~ sprintf(
"%d (%.1f%%)", sum(is.na(.)), 100 * mean(is.na(.))
),
.names = "{.col}")) %>%
pivot_longer(everything(), names_to = "variable", values_to = "n_pct_na") %>%
arrange(desc(n_pct_na)) %>%
head(12)
#> # A tibble: 12 × 2
#> variable n_pct_na
#> <chr> <chr>
#> 1 median 616 (7.2%)
#> 2 sales 568 (6.6%)
#> 3 volume 568 (6.6%)
#> 4 avg_price 568 (6.6%)
#> 5 inventory 1467 (17.1%)
#> 6 absorption 1427 (16.6%)
#> 7 listings 1424 (16.6%)
#> 8 city 0 (0.0%)
#> 9 year 0 (0.0%)
#> 10 month 0 (0.0%)
#> 11 date 0 (0.0%)
#> 12 quarter 0 (0.0%)# Example: median imputation for a few numerics (for demonstration only)
# (In modeling, prefer imputations *within* resampling using recipes/caret/tidymodels.)
tx_imputed <- tx %>%
mutate(
sales = ifelse(is.na(sales), median(sales, na.rm = TRUE), sales),
listings = ifelse(is.na(listings), median(listings, na.rm = TRUE), listings),
inventory = ifelse(is.na(inventory), median(inventory, na.rm = TRUE), inventory),
median = ifelse(is.na(median), median(median, na.rm = TRUE), median)
)
skimr::skim(tx_imputed %>% select(sales, listings, inventory, median))| Name | tx_imputed %>% select(sal… |
| Number of rows | 8602 |
| Number of columns | 4 |
| _______________________ | |
| Column type frequency: | |
| numeric | 4 |
| ________________________ | |
| Group variables | None |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| sales | 0 | 1 | 524.44 | 1077.59 | 6 | 90.0 | 169.0 | 432.00 | 8945.0 | ▇▁▁▁▁ |
| listings | 0 | 1 | 2896.76 | 5499.11 | 0 | 756.0 | 1283.0 | 2527.75 | 43107.0 | ▇▁▁▁▁ |
| inventory | 0 | 1 | 7.01 | 4.22 | 0 | 5.2 | 6.2 | 7.57 | 55.9 | ▇▁▁▁▁ |
| median | 0 | 1 | 127821.26 | 36014.21 | 50000 | 101725.0 | 123800.0 | 147900.00 | 304200.0 | ▃▇▃▁▁ |
# dlookr helpers (diagnose and visualize NA concentration)
diagnose(tx)
#> # A tibble: 13 × 6
#> variables types missing_count missing_percent unique_count unique_rate
#> <chr> <chr> <int> <dbl> <int> <dbl>
#> 1 city factor 0 0 46 0.00535
#> 2 year integer 0 0 16 0.00186
#> 3 month integer 0 0 12 0.00140
#> 4 sales numeric 568 6.60 1712 0.199
#> 5 volume numeric 568 6.60 7495 0.871
#> 6 median numeric 616 7.16 1538 0.179
#> 7 listings numeric 1424 16.6 3703 0.430
#> 8 inventory numeric 1467 17.1 296 0.0344
#> 9 date Date 0 0 187 0.0217
#> 10 avg_price numeric 568 6.60 7917 0.920
#> 11 absorption numeric 1427 16.6 6880 0.800
#> 12 quarter factor 0 0 4 0.000465
#> 13 ym factor 0 0 187 0.0217
plot_na_pareto(tx)
Best practice: Impute inside a modeling pipeline (e.g., recipes::step_impute_median()), and consider adding “was missing” flags to retain signal from missingness patterns.