6.4 Application
6.4.1 Nonlinear Estimation Using Gauss-Newton Algorithm
This section demonstrates nonlinear parameter estimation using the Gauss-Newton algorithm and compares results with nls(). The model is given by:
\[ y_i = \frac{\theta_0 + \theta_1 x_i}{1 + \theta_2 \exp(0.4 x_i)} + \epsilon_i \]
where
- \(i = 1, \dots ,n\)
- \(\theta_0\), \(\theta_1\), \(\theta_2\) are the unknown parameters.
- \(\epsilon_i\) represents errors.
- Loading and Visualizing the Data
library(dplyr)
library(ggplot2)
library(investr)
# Load the dataset
my_data <- read.delim("images/S21hw1pr4.txt", header = FALSE, sep = "") %>%
dplyr::rename(x = V1, y = V2)# Plot data
ggplot(my_data, aes(x = x, y = y)) +
geom_point(color = "blue") +
labs(title = "Observed Data", x = "X", y = "Y") +
theme_minimal()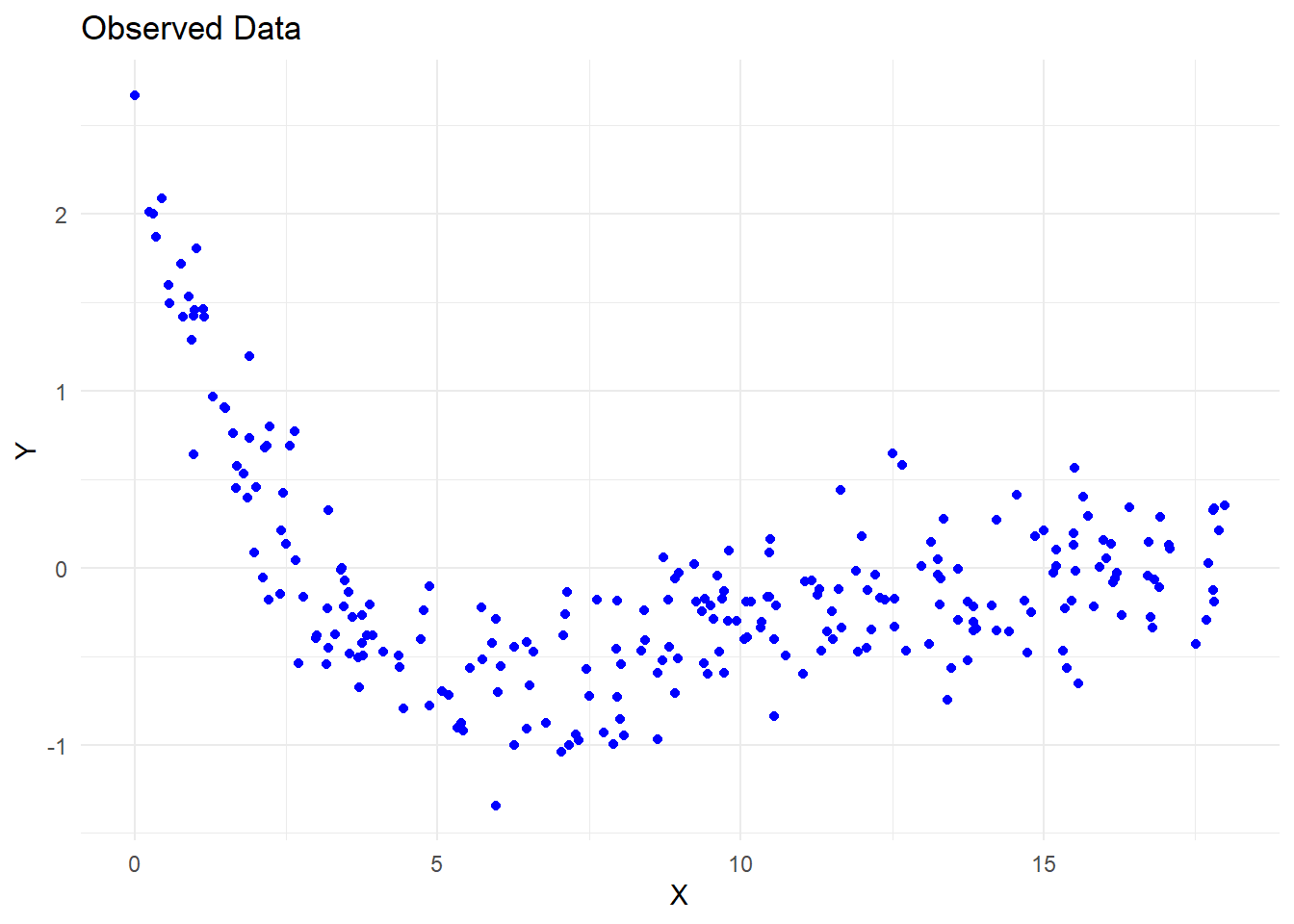
Figure 6.26: Observed Data
- Deriving Starting Values for Parameters
Since nonlinear optimization is sensitive to starting values, we estimate reasonable initial values based on model interpretation.
Finding the Maximum \(Y\) Value
When \(y = 2.6722\), the corresponding \(x = 0.0094\).
From the model equation: \(\theta_0 + 0.0094 \theta_1 = 2.6722\)
Estimating \(\theta_2\) from the Median \(y\) Value
- The equation simplifies to: \(1 + \theta_2 \exp(0.4 x) = 2\)
# find mean y
mean(my_data$y)
#> [1] -0.0747864
# find y closest to its mean
my_data$y[which.min(abs(my_data$y - (mean(my_data$y))))]
#> [1] -0.0773
# find x closest to the mean y
my_data$x[which.min(abs(my_data$y - (mean(my_data$y))))]
#> [1] 11.0648- This yields the equation: \(83.58967 \theta_2 = 1\)
Finding the Value of \(\theta_0\) and \(\theta_1\)
# find value of x closet to 1
my_data$x[which.min(abs(my_data$x - 1))]
#> [1] 0.9895
# find index of x closest to 1
match(my_data$x[which.min(abs(my_data$x - 1))], my_data$x)
#> [1] 14
# find y value
my_data$y[match(my_data$x[which.min(abs(my_data$x - 1))], my_data$x)]
#> [1] 1.4577- This provides another equation: \(\theta_0 + \theta_1 \times 0.9895 - 2.164479 \theta_2 = 1.457\)
Solving for \(\theta_0, \theta_1, \theta_2\)
library(matlib)
# Define coefficient matrix
A = matrix(
c(0, 0.0094, 0, 0, 0, 83.58967, 1, 0.9895, -2.164479),
nrow = 3,
ncol = 3,
byrow = T
)
# Define constant vector
b <- c(2.6722, 1, 1.457)
# Display system of equations
showEqn(A, b)
#> 0*x1 + 0.0094*x2 + 0*x3 = 2.6722
#> 0*x1 + 0*x2 + 83.58967*x3 = 1
#> 1*x1 + 0.9895*x2 - 2.164479*x3 = 1.457
# Solve for parameters
theta_start <- Solve(A, b, fractions = FALSE)
#> x1 = -279.80879739
#> x2 = 284.27659574
#> x3 = 0.0119632
theta_start
#> [1] "x1 = -279.80879739" " x2 = 284.27659574"
#> [3] " x3 = 0.0119632"- Implementing the Gauss-Newton Algorithm
Using these estimates, we manually implement the Gauss-Newton optimization.
Defining the Model and Its Derivatives
# Starting values
theta_0_strt <- as.numeric(gsub(".*=\\s*", "", theta_start[1]))
theta_1_strt <- as.numeric(gsub(".*=\\s*", "", theta_start[2]))
theta_2_strt <- as.numeric(gsub(".*=\\s*", "", theta_start[3]))
# Model function
mod_4 <- function(theta_0, theta_1, theta_2, x) {
(theta_0 + theta_1 * x) / (1 + theta_2 * exp(0.4 * x))
}
# Define function expression
f_4 = expression((theta_0 + theta_1 * x) / (1 + theta_2 * exp(0.4 * x)))
# First derivatives
df_4.d_theta_0 <- D(f_4, 'theta_0')
df_4.d_theta_1 <- D(f_4, 'theta_1')
df_4.d_theta_2 <- D(f_4, 'theta_2')Iterative Gauss-Newton Optimization
# Initialize
theta_vec <- matrix(c(theta_0_strt, theta_1_strt, theta_2_strt))
delta <- matrix(NA, nrow = 3, ncol = 1)
i <- 1
# Evaluate function at initial estimates
f_theta <- as.matrix(eval(f_4, list(
x = my_data$x,
theta_0 = theta_vec[1, 1],
theta_1 = theta_vec[2, 1],
theta_2 = theta_vec[3, 1]
)))
repeat {
# Compute Jacobian matrix
F_theta_0 <- as.matrix(cbind(
eval(df_4.d_theta_0, list(
x = my_data$x,
theta_0 = theta_vec[1, i],
theta_1 = theta_vec[2, i],
theta_2 = theta_vec[3, i]
)),
eval(df_4.d_theta_1, list(
x = my_data$x,
theta_0 = theta_vec[1, i],
theta_1 = theta_vec[2, i],
theta_2 = theta_vec[3, i]
)),
eval(df_4.d_theta_2, list(
x = my_data$x,
theta_0 = theta_vec[1, i],
theta_1 = theta_vec[2, i],
theta_2 = theta_vec[3, i]
))
))
# Compute parameter updates
delta[, i] = (solve(t(F_theta_0)%*%F_theta_0))%*%t(F_theta_0)%*%(my_data$y-f_theta[,i])
# Update parameter estimates
theta_vec <- cbind(theta_vec, theta_vec[, i] + delta[, i])
theta_vec[, i + 1] = theta_vec[, i] + delta[, i]
# Increment iteration counter
i <- i + 1
# Compute new function values
f_theta <- cbind(f_theta, as.matrix(eval(f_4, list(
x = my_data$x,
theta_0 = theta_vec[1, i],
theta_1 = theta_vec[2, i],
theta_2 = theta_vec[3, i]
))))
delta = cbind(delta, matrix(NA, nrow = 3, ncol = 1))
# Convergence criteria based on SSE
if (abs(sum((my_data$y - f_theta[, i])^2) - sum((my_data$y - f_theta[, i - 1])^2)) /
sum((my_data$y - f_theta[, i - 1])^2) < 0.001) {
break
}
}
# Final parameter estimates
theta_vec[, ncol(theta_vec)]
#> [1] 3.6335135 -1.3055166 0.5043502- Checking Convergence and Variance
# Final objective function value (SSE)
sum((my_data$y - f_theta[, i])^2)
#> [1] 19.80165
sigma2 <- 1 / (nrow(my_data) - 3) *
(t(my_data$y - f_theta[, ncol(f_theta)]) %*%
(my_data$y - f_theta[, ncol(f_theta)])) # p = 3
# Asymptotic variance-covariance matrix
as.numeric(sigma2)*as.matrix(solve(crossprod(F_theta_0)))
#> [,1] [,2] [,3]
#> [1,] 0.11552571 -0.04817428 0.02685848
#> [2,] -0.04817428 0.02100861 -0.01158212
#> [3,] 0.02685848 -0.01158212 0.00703916- Validating with
nls()
nlin_4 <- nls(
y ~ mod_4(theta_0, theta_1, theta_2, x),
start = list(
theta_0 = as.numeric(gsub(".*=\\s*", "", theta_start[1])),
theta_1 = as.numeric(gsub(".*=\\s*", "", theta_start[2])),
theta_2 = as.numeric(gsub(".*=\\s*", "", theta_start[3]))
),
data = my_data
)
summary(nlin_4)
#>
#> Formula: y ~ mod_4(theta_0, theta_1, theta_2, x)
#>
#> Parameters:
#> Estimate Std. Error t value Pr(>|t|)
#> theta_0 3.63591 0.36528 9.954 < 2e-16 ***
#> theta_1 -1.30639 0.15561 -8.395 3.65e-15 ***
#> theta_2 0.50528 0.09215 5.483 1.03e-07 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 0.2831 on 247 degrees of freedom
#>
#> Number of iterations to convergence: 9
#> Achieved convergence tolerance: 2.307e-076.4.2 Logistic Growth Model
A classic logistic growth model follows the equation:
\[ P = \frac{K}{1 + \exp(P_0 + r t)} + \epsilon \]
where:
\(P\) = population at time \(t\)
\(K\) = carrying capacity (maximum population)
\(r\) = population growth rate
\(P_0\) = initial population log-ratio
However, R’s built-in SSlogis function uses a slightly different parameterization:
\[ P = \frac{asym}{1 + \exp\left(\frac{xmid - t}{scal}\right)} \]
where:
\(asym\) = carrying capacity (\(K\))
\(xmid\) = the \(x\)-value at the inflection point of the curve
\(scal\) = scaling parameter
This gives the parameter relationships:
\(K = asym\)
\(r = -1 / scal\)
\(P_0 = -r \cdot xmid\)
# Simulated time-series data
time <- c(1, 2, 3, 5, 10, 15, 20, 25, 30, 35)
population <- c(2.8, 4.2, 3.5, 6.3, 15.7, 21.3, 23.7, 25.1, 25.8, 25.9)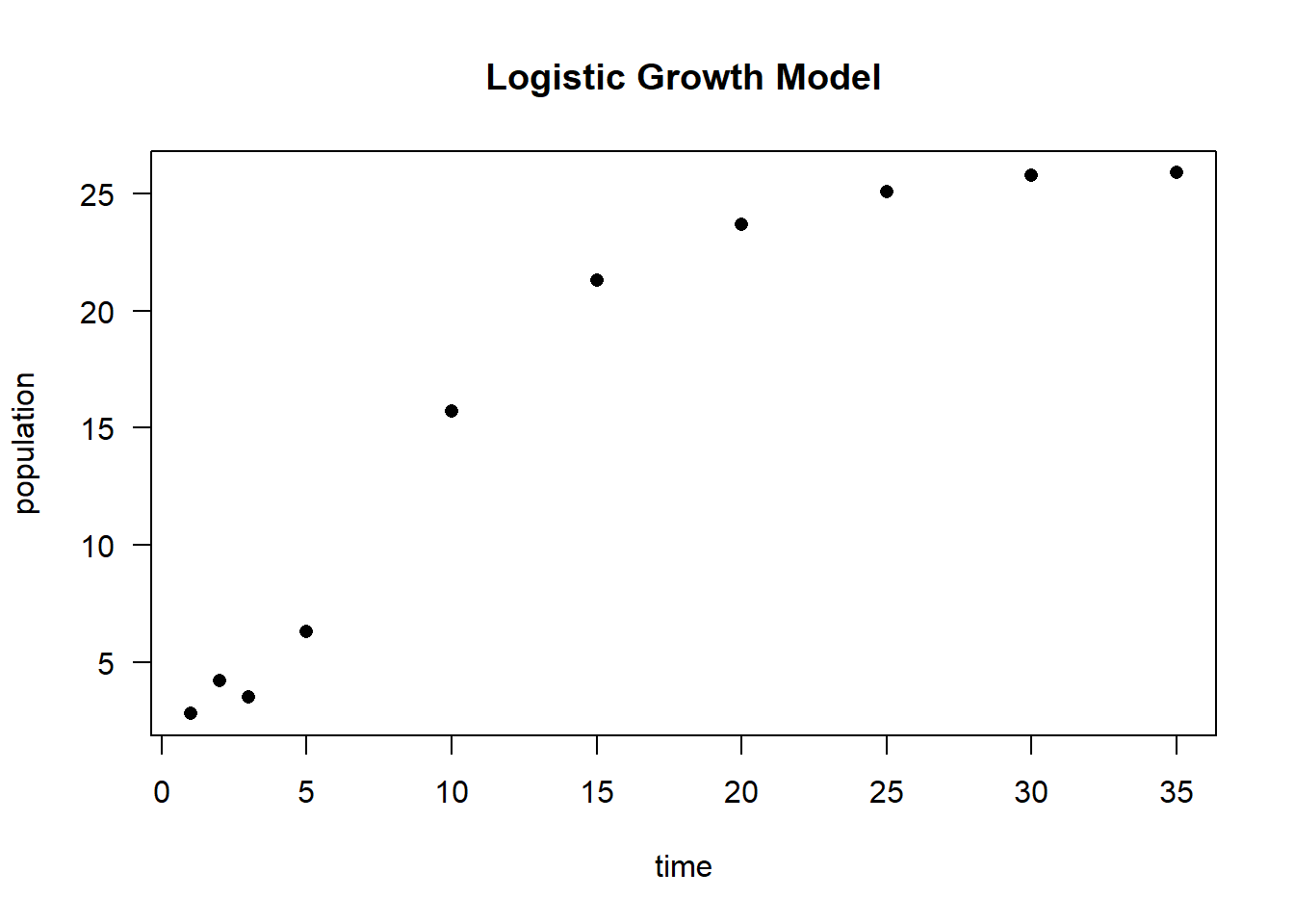
Figure 6.27: Logistic Growth Model
# Fit the logistic growth model using programmed starting values
logisticModelSS <- nls(population ~ SSlogis(time, Asym, xmid, scal))
# Model summary
summary(logisticModelSS)
#>
#> Formula: population ~ SSlogis(time, Asym, xmid, scal)
#>
#> Parameters:
#> Estimate Std. Error t value Pr(>|t|)
#> Asym 25.5029 0.3666 69.56 3.34e-11 ***
#> xmid 8.7347 0.3007 29.05 1.48e-08 ***
#> scal 3.6353 0.2186 16.63 6.96e-07 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 0.6528 on 7 degrees of freedom
#>
#> Number of iterations to convergence: 1
#> Achieved convergence tolerance: 1.906e-06
# Extract parameter estimates
coef(logisticModelSS)
#> Asym xmid scal
#> 25.502890 8.734698 3.635333To fit the model using an alternative parameterization (\(K, r, P_0\)), we convert the estimated coefficients:
# Convert parameter estimates to alternative logistic model parameters
Ks <- as.numeric(coef(logisticModelSS)[1]) # Carrying capacity (K)
rs <- -1 / as.numeric(coef(logisticModelSS)[3]) # Growth rate (r)
Pos <- -rs * as.numeric(coef(logisticModelSS)[2]) # P_0
# Fit the logistic model with the alternative parameterization
logisticModel <- nls(
population ~ K / (1 + exp(Po + r * time)),
start = list(Po = Pos, r = rs, K = Ks)
)
# Model summary
summary(logisticModel)
#>
#> Formula: population ~ K/(1 + exp(Po + r * time))
#>
#> Parameters:
#> Estimate Std. Error t value Pr(>|t|)
#> Po 2.40272 0.12702 18.92 2.87e-07 ***
#> r -0.27508 0.01654 -16.63 6.96e-07 ***
#> K 25.50289 0.36665 69.56 3.34e-11 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 0.6528 on 7 degrees of freedom
#>
#> Number of iterations to convergence: 0
#> Achieved convergence tolerance: 1.91e-06Visualizing the Logistic Model Fit
# Plot original data
plot(time,
population,
las = 1,
pch = 16,
main = "Logistic Growth Model Fit")
# Overlay the fitted logistic curve
lines(time,
predict(logisticModel),
col = "red",
lwd = 2)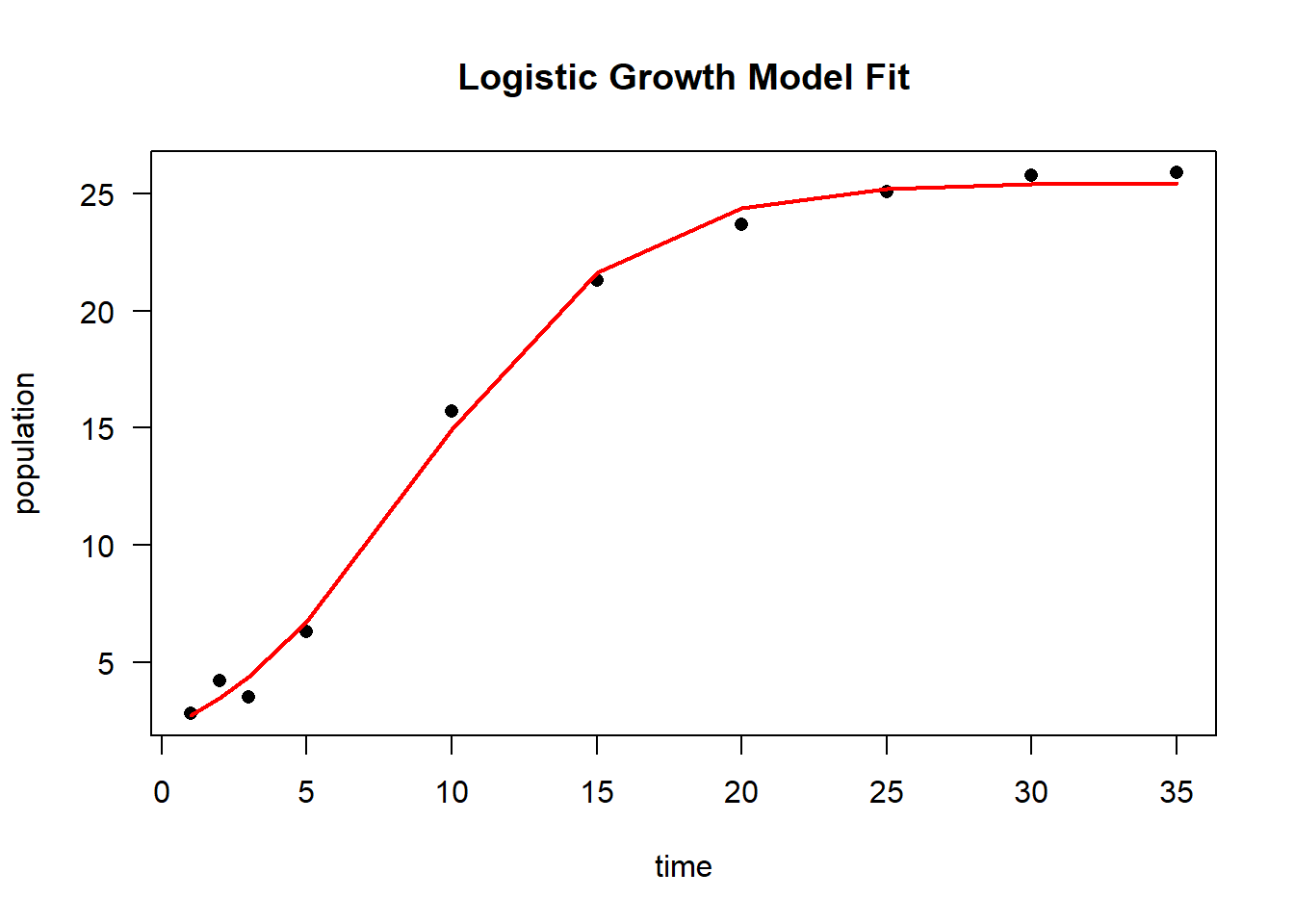
Figure 6.28: Logistic Growth Model Fit
6.4.3 Nonlinear Plateau Model
This example is based on (Schabenberger and Pierce 2001) and demonstrates the use of a plateau model to estimate the relationship between soil nitrate (\(NO_3\)) concentration and relative yield percent (RYP) at two different depths (30 cm and 60 cm).
# Load data
dat <- read.table("images/dat.txt", header = TRUE)
# Plot NO3 concentration vs. relative yield percent, colored by depth
library(ggplot2)
dat.plot <- ggplot(dat) +
geom_point(aes(x = no3, y = ryp, color = as.factor(depth))) +
labs(color = 'Depth (cm)') +
xlab('Soil NO3 Concentration') +
ylab('Relative Yield Percent') +
theme_minimal()
# Display plot
dat.plot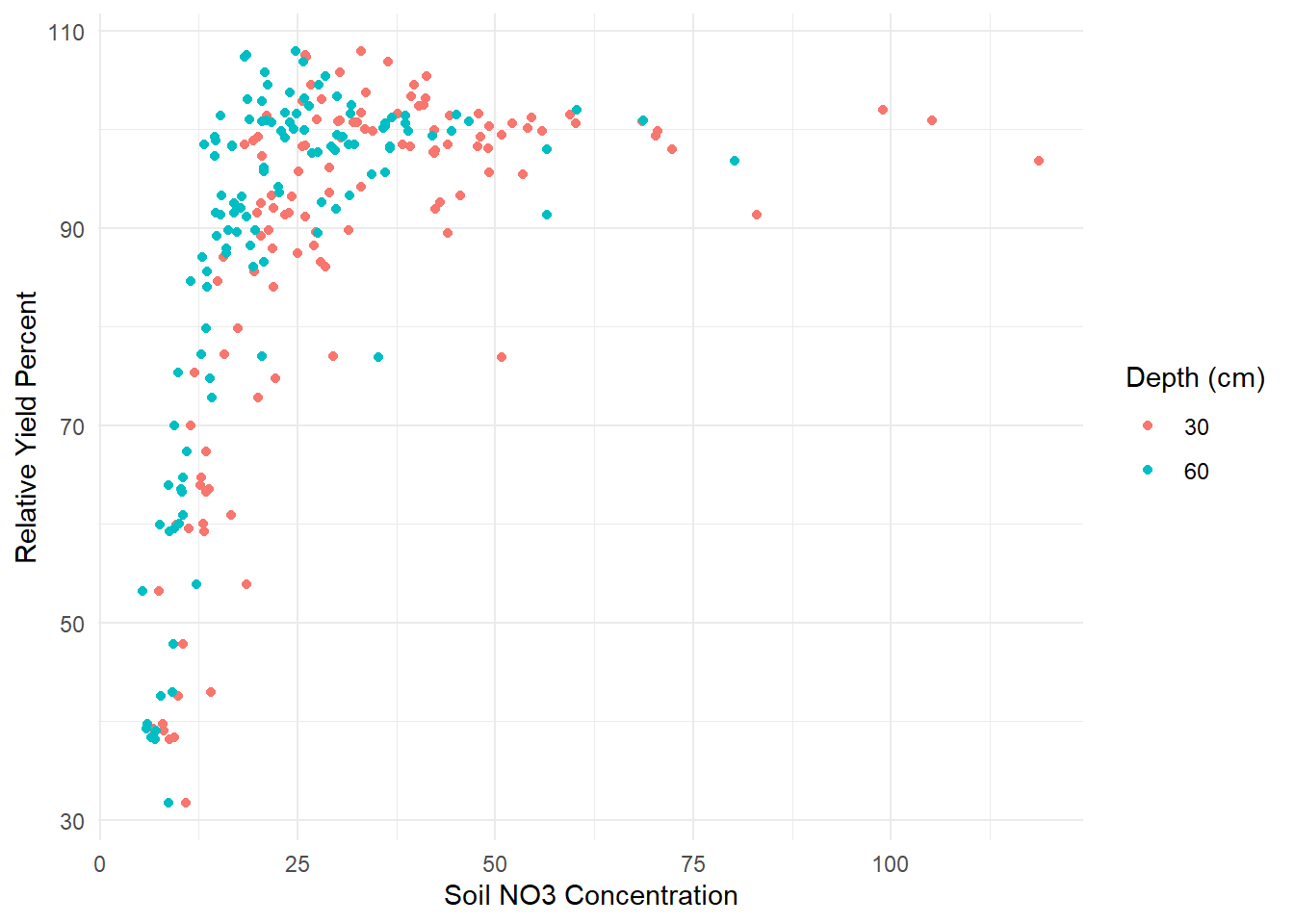
Figure 6.29: Nonlinear Plateau Plot
The suggested nonlinear plateau model is given by:
\[ E(Y_{ij}) = (\beta_{0j} + \beta_{1j}N_{ij})I_{N_{ij}\le \alpha_j} + (\beta_{0j} + \beta_{1j}\alpha_j)I_{N_{ij} > \alpha_j} \]
where:
\(N_{ij}\) represents the soil nitrate (\(NO_3\)) concentration for observation \(i\) at depth \(j\).
\(i\) indexes individual observations.
\(j = 1, 2\) corresponds to depths 30 cm and 60 cm.
This model assumes a linear increase up to a threshold (\(\alpha_j\)), beyond which the response levels off (plateaus).
Defining the Plateau Model as a Function
# Define the nonlinear plateau model function
nonlinModel <- function(predictor, b0, b1, alpha) {
ifelse(predictor <= alpha,
b0 + b1 * predictor, # Linear growth below threshold
b0 + b1 * alpha) # Plateau beyond threshold
}Creating a Self-Starting Function for nls
Since the model is piecewise linear, we can estimate starting values using:
A linear regression on the first half of sorted predictor values to estimate \(b_0\) and \(b_1\).
The last predictor value used in the regression as the plateau threshold (\(\alpha\))
# Define initialization function for self-starting plateau model
nonlinModelInit <- function(mCall, LHS, data) {
# Sort data by increasing predictor value
xy <- sortedXyData(mCall[['predictor']], LHS, data)
n <- nrow(xy)
# Fit a simple linear model using the first half of the sorted data
lmFit <- lm(xy[1:(n / 2), 'y'] ~ xy[1:(n / 2), 'x'])
# Extract initial estimates
b0 <- coef(lmFit)[1] # Intercept
b1 <- coef(lmFit)[2] # Slope
alpha <- xy[(n / 2), 'x'] # Last x-value in the fitted linear range
# Return initial parameter estimates
value <- c(b0, b1, alpha)
names(value) <- mCall[c('b0', 'b1', 'alpha')]
value
}Combining Model and Self-Start Function
# Define a self-starting nonlinear model for nls
SS_nonlinModel <- selfStart(nonlinModel,
nonlinModelInit,
c('b0', 'b1', 'alpha'))The nls function is used to estimate parameters separately for each soil depth (30 cm and 60 cm).
# Fit the model for depth = 30 cm
sep30_nls <- nls(ryp ~ SS_nonlinModel(predictor = no3, b0, b1, alpha),
data = dat[dat$depth == 30,])
# Fit the model for depth = 60 cm
sep60_nls <- nls(ryp ~ SS_nonlinModel(predictor = no3, b0, b1, alpha),
data = dat[dat$depth == 60,])We generate separate plots for 30 cm and 60 cm depths, showing both confidence and prediction intervals.
# Set plotting layout
par(mfrow = c(1, 2))
# Plot model fit for 30 cm depth
investr::plotFit(
sep30_nls,
interval = "both",
pch = 19,
shade = TRUE,
col.conf = "skyblue4",
col.pred = "lightskyblue2",
data = dat[dat$depth == 30,],
main = "Results at 30 cm Depth",
ylab = "Relative Yield Percent",
xlab = "Soil NO3 Concentration",
xlim = c(0, 120)
)
# Plot model fit for 60 cm depth
investr::plotFit(
sep60_nls,
interval = "both",
pch = 19,
shade = TRUE,
col.conf = "lightpink4",
col.pred = "lightpink2",
data = dat[dat$depth == 60,],
main = "Results at 60 cm Depth",
ylab = "Relative Yield Percent",
xlab = "Soil NO3 Concentration",
xlim = c(0, 120)
)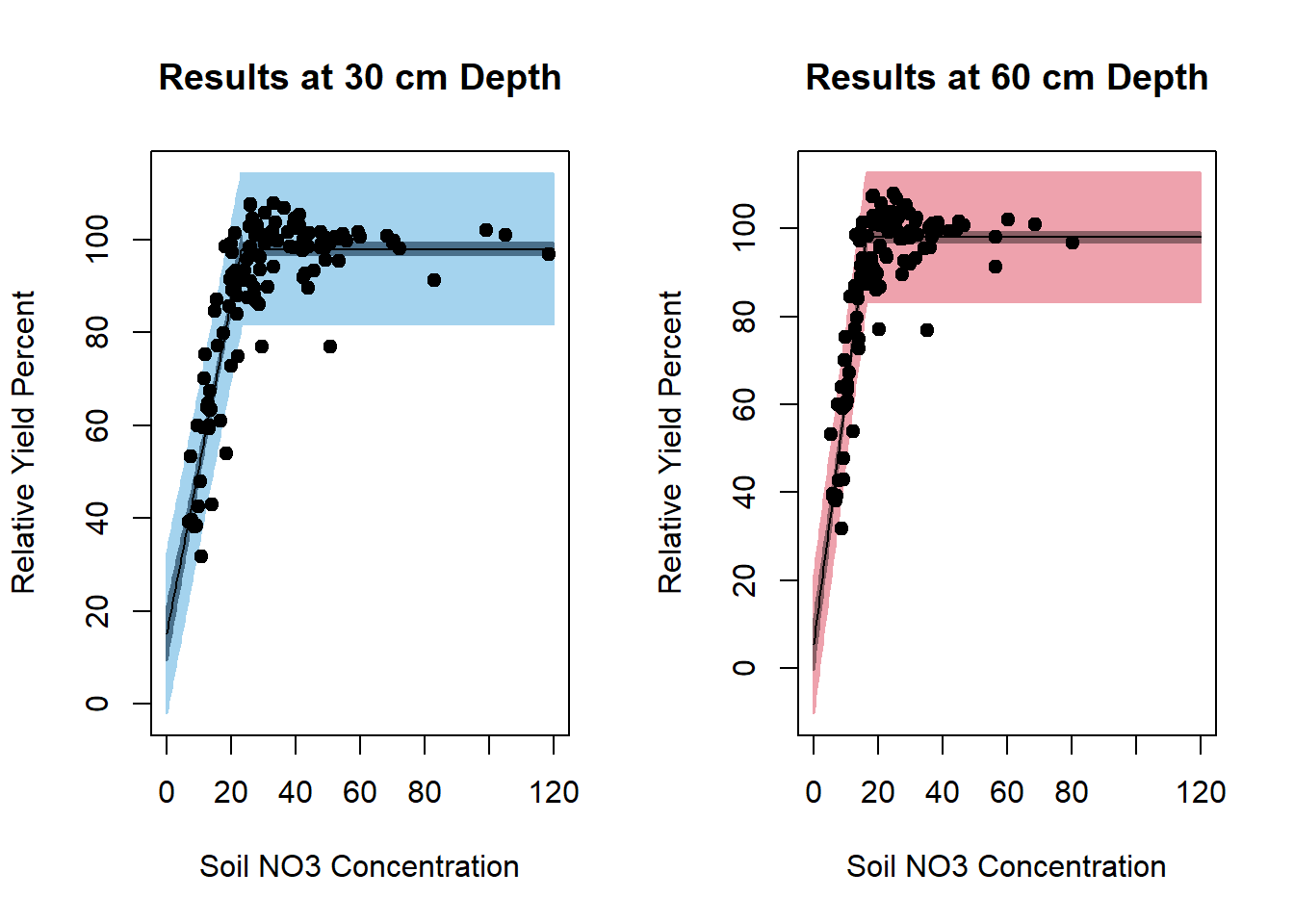
Figure 6.30: Fitted Curves for Relative Yield Response by Soil NO3 Concentration at 30 cm and 60 cm Depths
summary(sep30_nls)
#>
#> Formula: ryp ~ SS_nonlinModel(predictor = no3, b0, b1, alpha)
#>
#> Parameters:
#> Estimate Std. Error t value Pr(>|t|)
#> b0 15.1943 2.9781 5.102 6.89e-07 ***
#> b1 3.5760 0.1853 19.297 < 2e-16 ***
#> alpha 23.1324 0.5098 45.373 < 2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 8.258 on 237 degrees of freedom
#>
#> Number of iterations to convergence: 6
#> Achieved convergence tolerance: 1.166e-08
summary(sep60_nls)
#>
#> Formula: ryp ~ SS_nonlinModel(predictor = no3, b0, b1, alpha)
#>
#> Parameters:
#> Estimate Std. Error t value Pr(>|t|)
#> b0 5.4519 2.9785 1.83 0.0684 .
#> b1 5.6820 0.2529 22.46 <2e-16 ***
#> alpha 16.2863 0.2818 57.80 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 7.427 on 237 degrees of freedom
#>
#> Number of iterations to convergence: 5
#> Achieved convergence tolerance: 2.196e-08Modeling Soil Depths Together and Comparing Models
Instead of fitting separate models for different soil depths, we first fit a combined model where all observations share a common slope, intercept, and plateau. We then test whether modeling the two depths separately provides a significantly better fit.
- Fitting a Reduced (Combined) Model
The reduced model assumes that all soil depths follow the same nonlinear relationship.
# Fit the combined model (common parameters across all depths)
red_nls <- nls(
ryp ~ SS_nonlinModel(predictor = no3, b0, b1, alpha),
data = dat
)
# Display model summary
summary(red_nls)
#>
#> Formula: ryp ~ SS_nonlinModel(predictor = no3, b0, b1, alpha)
#>
#> Parameters:
#> Estimate Std. Error t value Pr(>|t|)
#> b0 8.7901 2.7688 3.175 0.0016 **
#> b1 4.8995 0.2207 22.203 <2e-16 ***
#> alpha 18.0333 0.3242 55.630 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 9.13 on 477 degrees of freedom
#>
#> Number of iterations to convergence: 7
#> Achieved convergence tolerance: 8.568e-09# Visualizing the combined model fit
par(mfrow = c(1, 1))
plotFit(
red_nls,
interval = "both",
pch = 19,
shade = TRUE,
col.conf = "lightblue4",
col.pred = "lightblue2",
data = dat,
main = 'Results for Combined Model',
ylab = 'Relative Yield Percent',
xlab = 'Soil NO3 Concentration'
)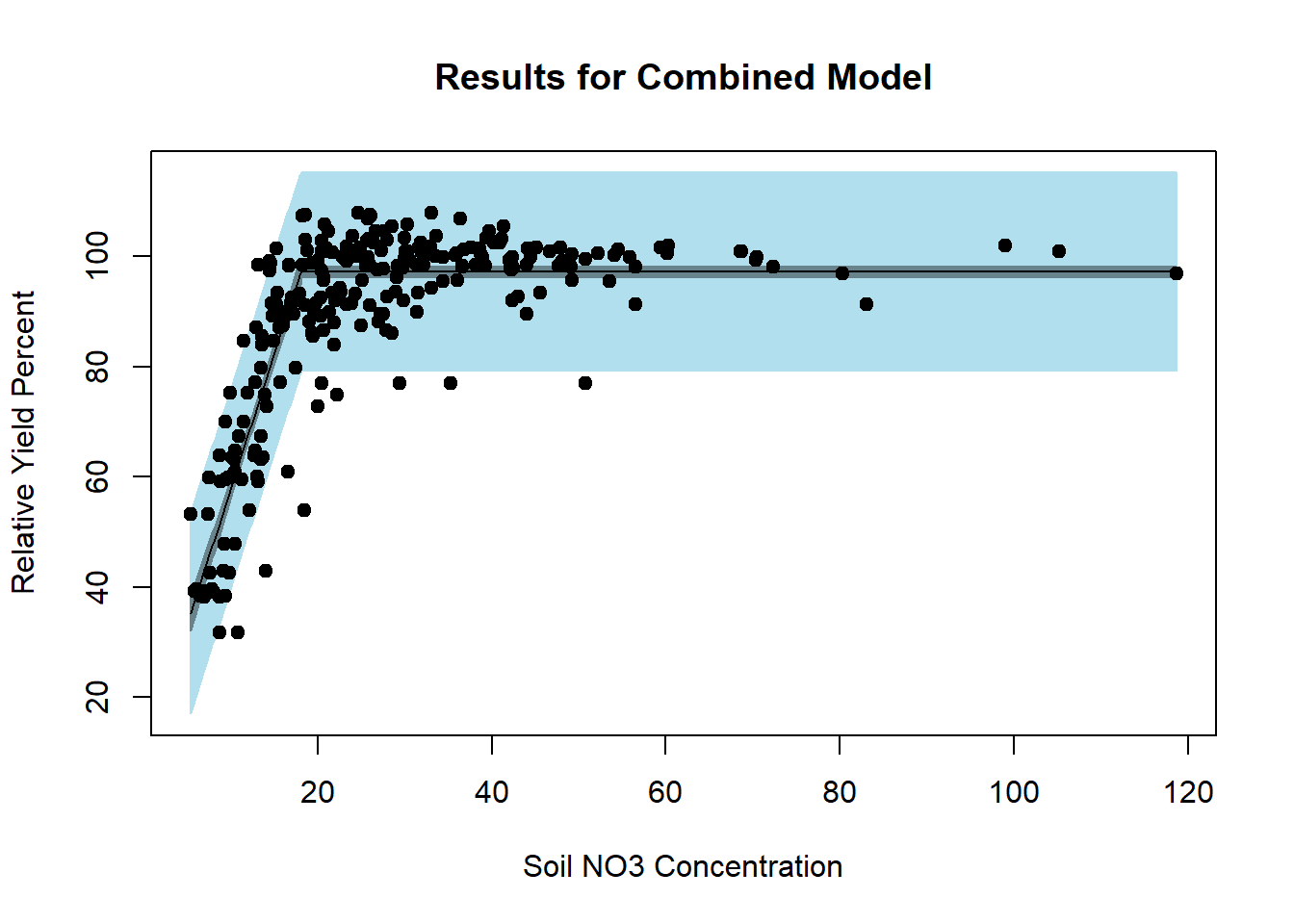
Figure 6.31: Results for Combined Model
- Examining Residuals for the Combined Model
Checking residuals helps diagnose potential lack of fit.
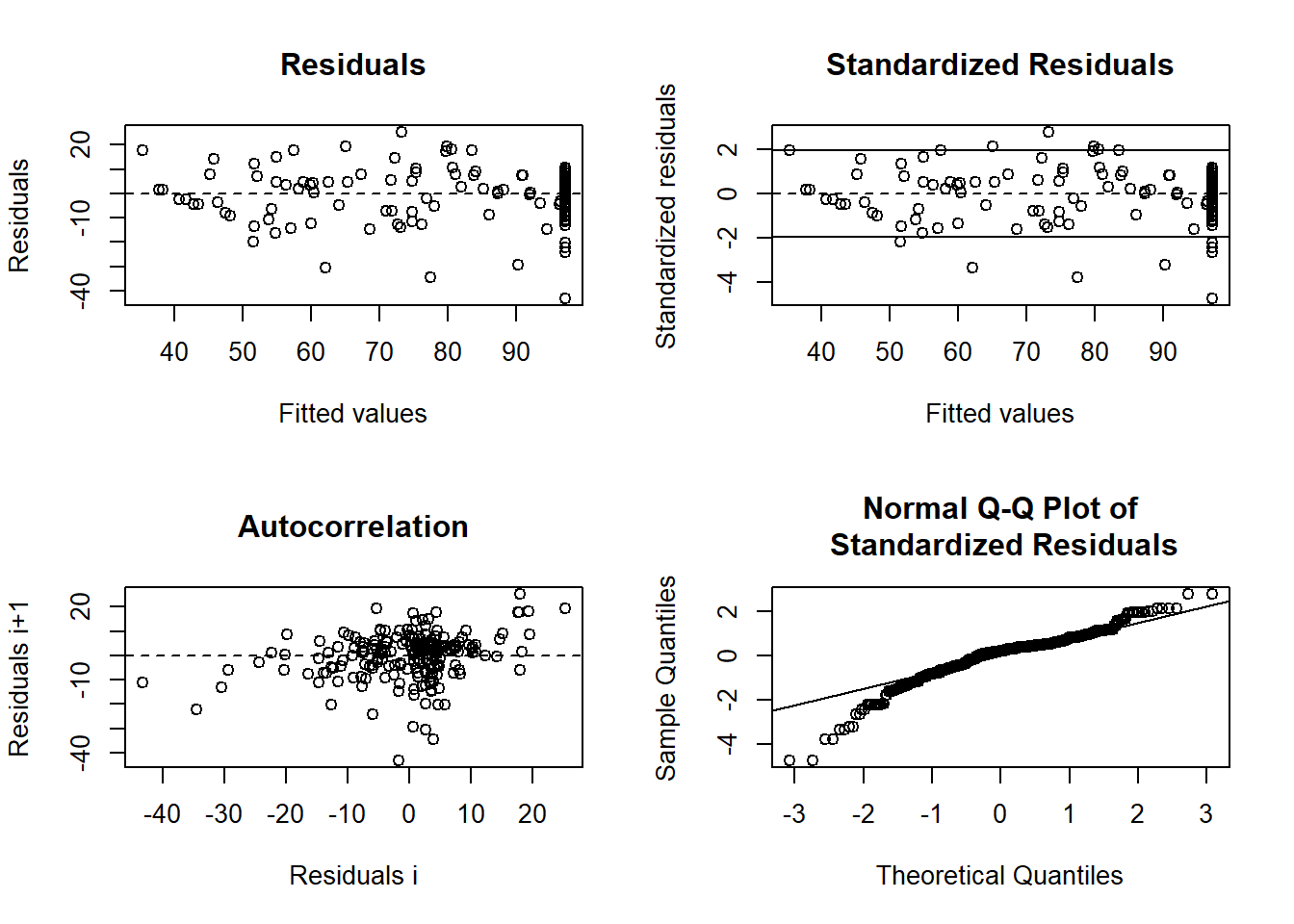
Figure 6.32: Residual Plot Nonlinear Example
If there is a pattern in the residuals (e.g., systematic deviations based on soil depth), this suggests that a separate model for each depth may be necessary.
- Testing Whether Depths Require Separate Models
To formally test whether soil depth significantly affects the model parameters, we introduce a parameterization where depth-specific parameters are increments from a baseline model (30 cm depth):
\[ \begin{aligned} \beta_{02} &= \beta_{01} + d_0 \\ \beta_{12} &= \beta_{11} + d_1 \\ \alpha_{2} &= \alpha_{1} + d_a \end{aligned} \]
where:
\(\beta_{01}, \beta_{11}, \alpha_1\) are parameters for 30 cm depth.
\(d_0, d_1, d_a\) represent depth-specific differences for 60 cm depth.
If \(d_0, d_1, d_a\) are significantly different from 0, the two depths should be modeled separately.
- Defining the Full (Depth-Specific) Model
nonlinModelF <- function(predictor, soildep, b01, b11, a1, d0, d1, da) {
# Define parameters for 60 cm depth as increments from 30 cm parameters
b02 <- b01 + d0
b12 <- b11 + d1
a2 <- a1 + da
# Compute model output for 30 cm depth
y1 <- ifelse(
predictor <= a1,
b01 + b11 * predictor,
b01 + b11 * a1
)
# Compute model output for 60 cm depth
y2 <- ifelse(
predictor <= a2,
b02 + b12 * predictor,
b02 + b12 * a2
)
# Assign correct model output based on depth
y <- y1 * (soildep == 30) + y2 * (soildep == 60)
return(y)
}- Fitting the Full (Depth-Specific) Model
The starting values are taken from the separately fitted models for each depth.
Soil_full <- nls(
ryp ~ nonlinModelF(
predictor = no3,
soildep = depth,
b01,
b11,
a1,
d0,
d1,
da
),
data = dat,
start = list(
b01 = 15.2, # Intercept for 30 cm depth
b11 = 3.58, # Slope for 30 cm depth
a1 = 23.13, # Plateau cutoff for 30 cm depth
d0 = -9.74, # Intercept difference (60 cm - 30 cm)
d1 = 2.11, # Slope difference (60 cm - 30 cm)
da = -6.85 # Plateau cutoff difference (60 cm - 30 cm)
)
)
# Display model summary
summary(Soil_full)
#>
#> Formula: ryp ~ nonlinModelF(predictor = no3, soildep = depth, b01, b11,
#> a1, d0, d1, da)
#>
#> Parameters:
#> Estimate Std. Error t value Pr(>|t|)
#> b01 15.1943 2.8322 5.365 1.27e-07 ***
#> b11 3.5760 0.1762 20.291 < 2e-16 ***
#> a1 23.1324 0.4848 47.711 < 2e-16 ***
#> d0 -9.7424 4.2357 -2.300 0.0219 *
#> d1 2.1060 0.3203 6.575 1.29e-10 ***
#> da -6.8461 0.5691 -12.030 < 2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 7.854 on 474 degrees of freedom
#>
#> Number of iterations to convergence: 1
#> Achieved convergence tolerance: 3.742e-06- Model Comparison: Does Depth Matter?
If \(d_0, d_1, d_a\) are significantly different from 0, the depths should be modeled separately.
The p-values for these parameters indicate whether depth-specific modeling is necessary.