39.2 Good Controls
39.2.1 Omitted Variable Bias Correction
A variable \(Z\) is a good control when it blocks all back-door paths from the treatment \(X\) to the outcome \(Y\). This is the fundamental criterion from the back-door adjustment theorem in causal inference.
39.2.1.1 Simple Confounder
In this DAG, \(Z\) is a common cause of both \(X\) and \(Y\), i.e., a confounder.
rm(list = ls())
model <- dagitty("dag{
x -> y
z -> x
z -> y
}")
coordinates(model) <- list(
x = c(x = 1, z = 2, y = 3),
y = c(x = 1, z = 2, y = 1)
)
ggdag(model) + theme_dag()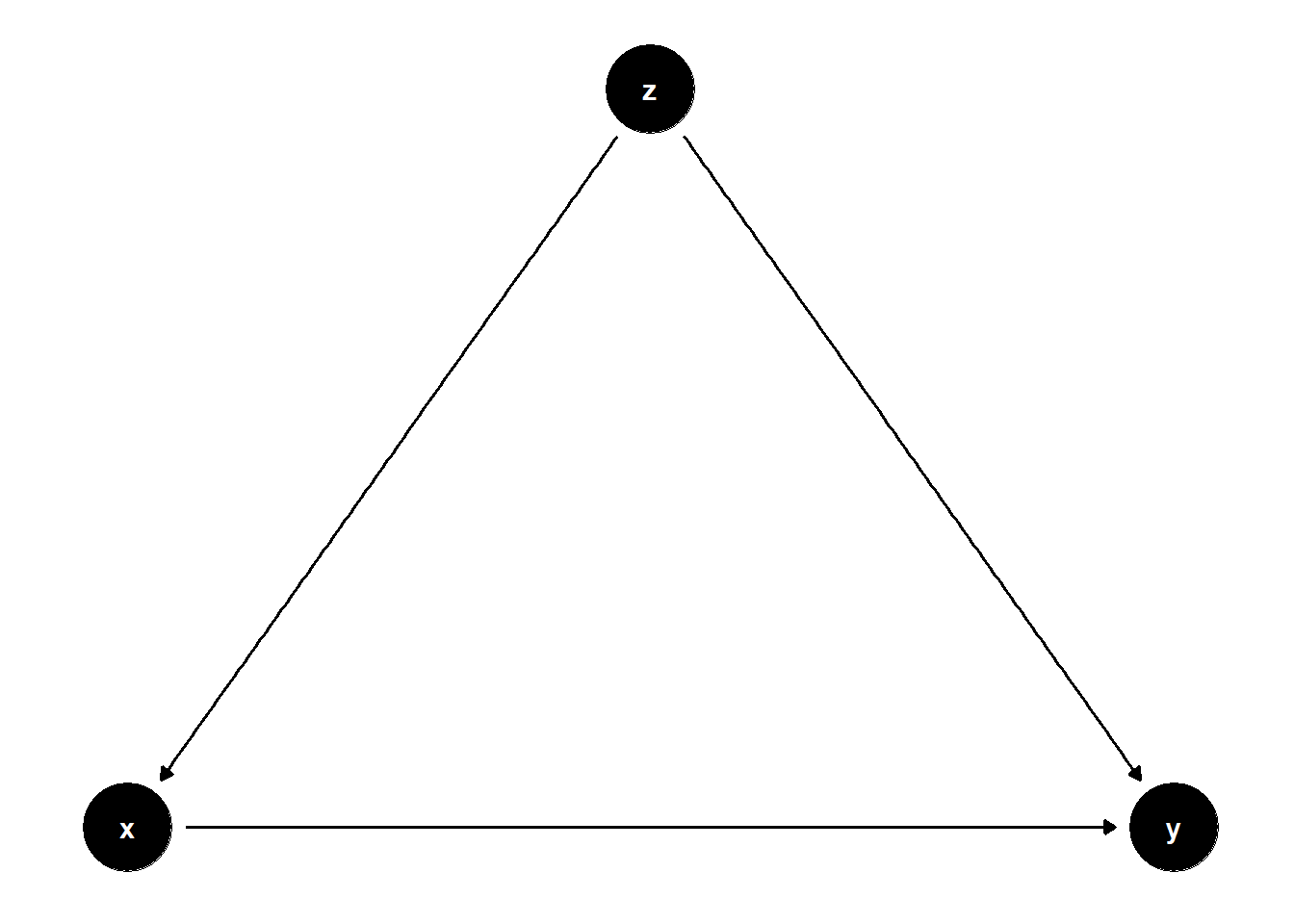
Controlling for \(Z\) removes the bias from the back-door path \(X \leftarrow Z \rightarrow Y\).
n <- 1e4
z <- rnorm(n)
causal_coef <- 2
beta2 <- 3
x <- z + rnorm(n)
y <- causal_coef * x + beta2 * z + rnorm(n)
jtools::export_summs(lm(y ~ x), lm(y ~ x + z))| Model 1 | Model 2 | |
|---|---|---|
| (Intercept) | -0.02 | 0.01 |
| (0.02) | (0.01) | |
| x | 3.49 *** | 2.00 *** |
| (0.02) | (0.01) | |
| z | 2.98 *** | |
| (0.01) | ||
| N | 10000 | 10000 |
| R2 | 0.82 | 0.97 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
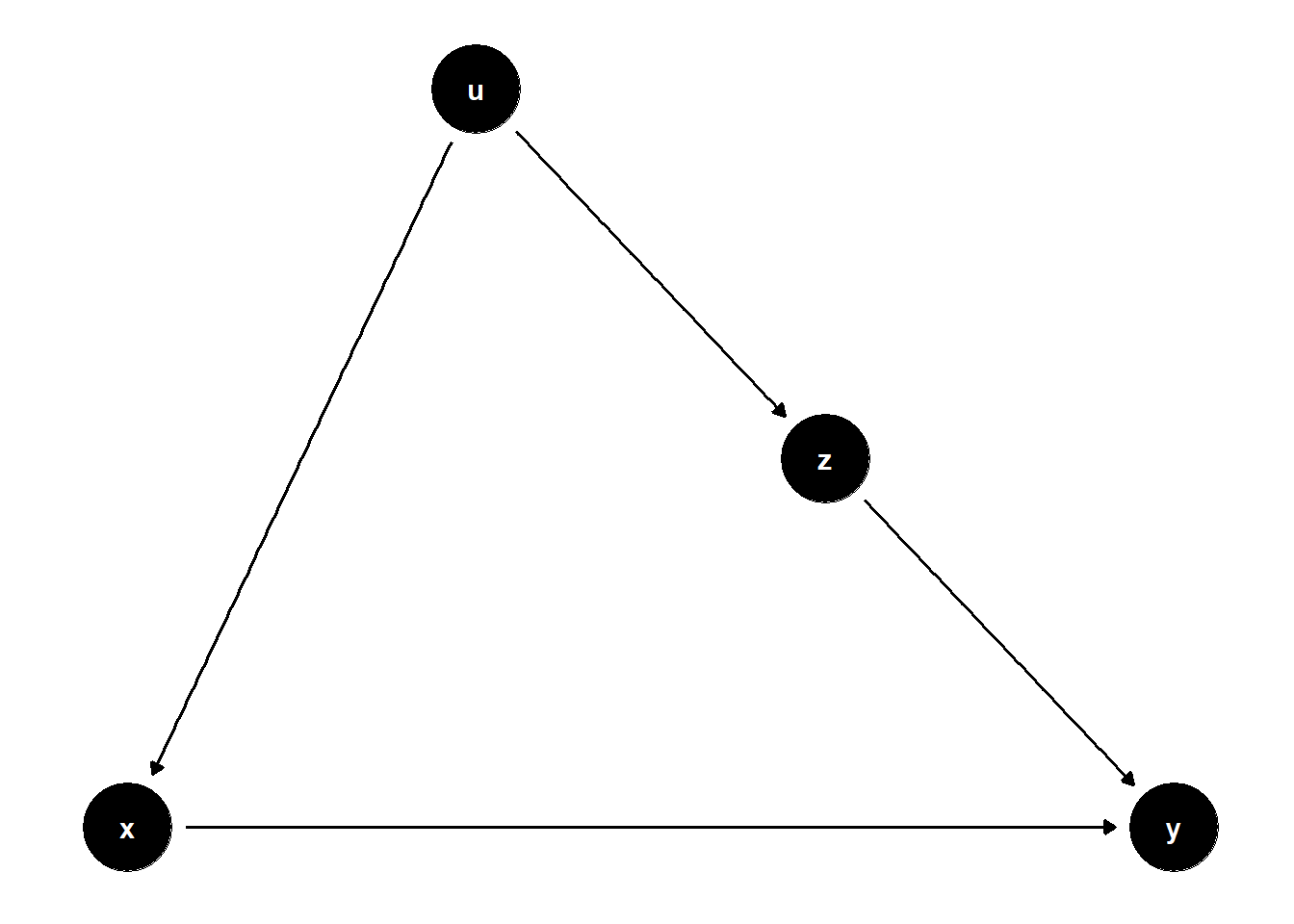
39.2.1.2 Confounding via a Latent Variable
In this structure, \(U\) is unobserved but causes both \(Z\) and \(Y\), and \(Z\) affects \(X\). Even though \(U\) is not observed, adjusting for \(Z\) helps block the back-door path from \(X\) to \(Y\) that goes through \(U\).
rm(list = ls())
model <- dagitty("dag{
x -> y
u -> z
z -> x
u -> y
}")
latents(model) <- "u"
coordinates(model) <- list(
x = c(x = 1, z = 2, u = 3, y = 4),
y = c(x = 1, z = 2, u = 3, y = 1)
)
ggdag(model) + theme_dag()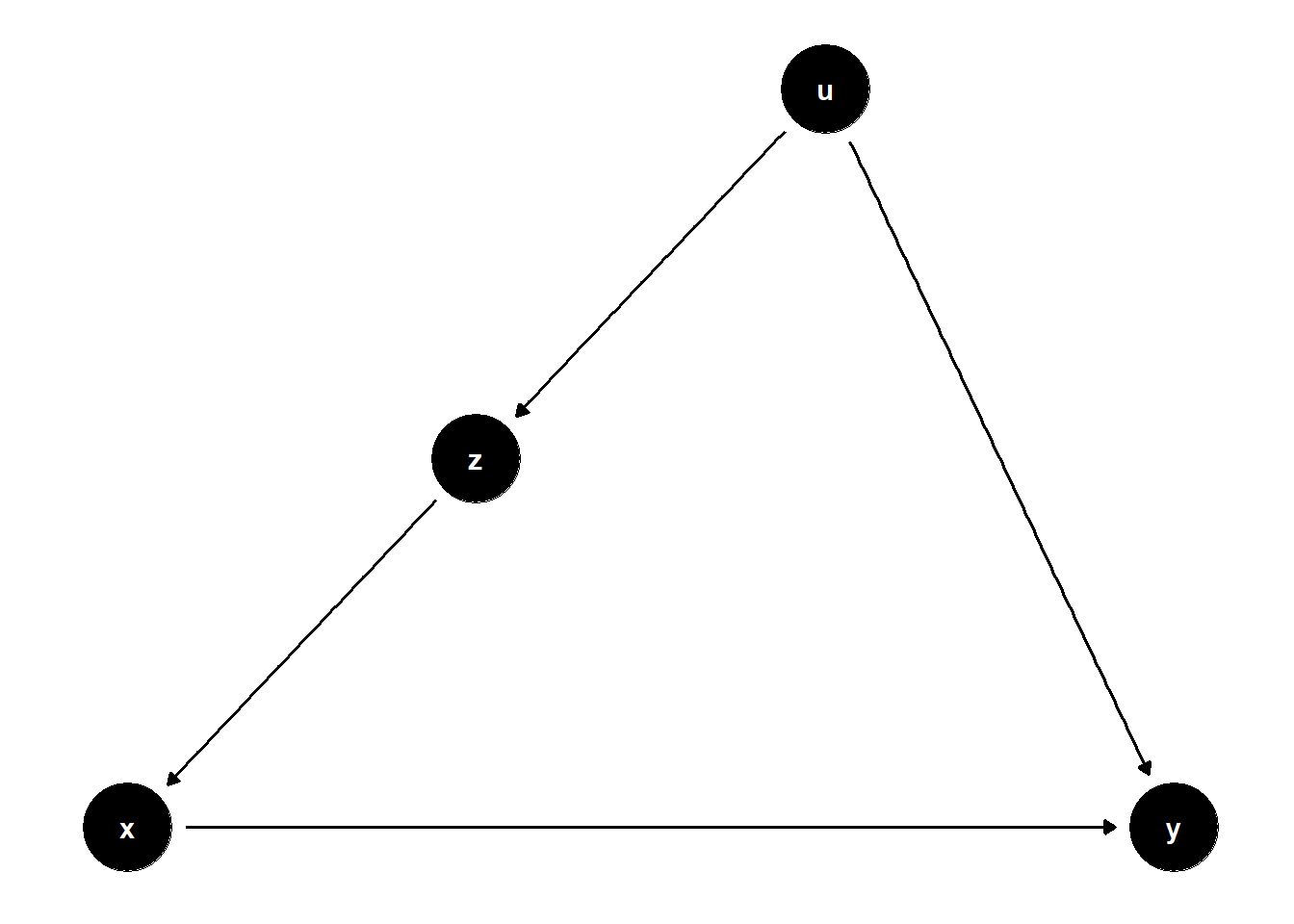
n <- 1e4
u <- rnorm(n)
z <- u + rnorm(n)
x <- z + rnorm(n)
y <- 2 * x + u + rnorm(n)
jtools::export_summs(lm(y ~ x), lm(y ~ x + z))| Model 1 | Model 2 | |
|---|---|---|
| (Intercept) | -0.02 | -0.01 |
| (0.01) | (0.01) | |
| x | 2.33 *** | 1.99 *** |
| (0.01) | (0.01) | |
| z | 0.51 *** | |
| (0.01) | ||
| N | 10000 | 10000 |
| R2 | 0.91 | 0.92 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
Even though $Z$ appears significant, its inclusion serves to reduce omitted variable bias rather than having a causal interpretation itself.
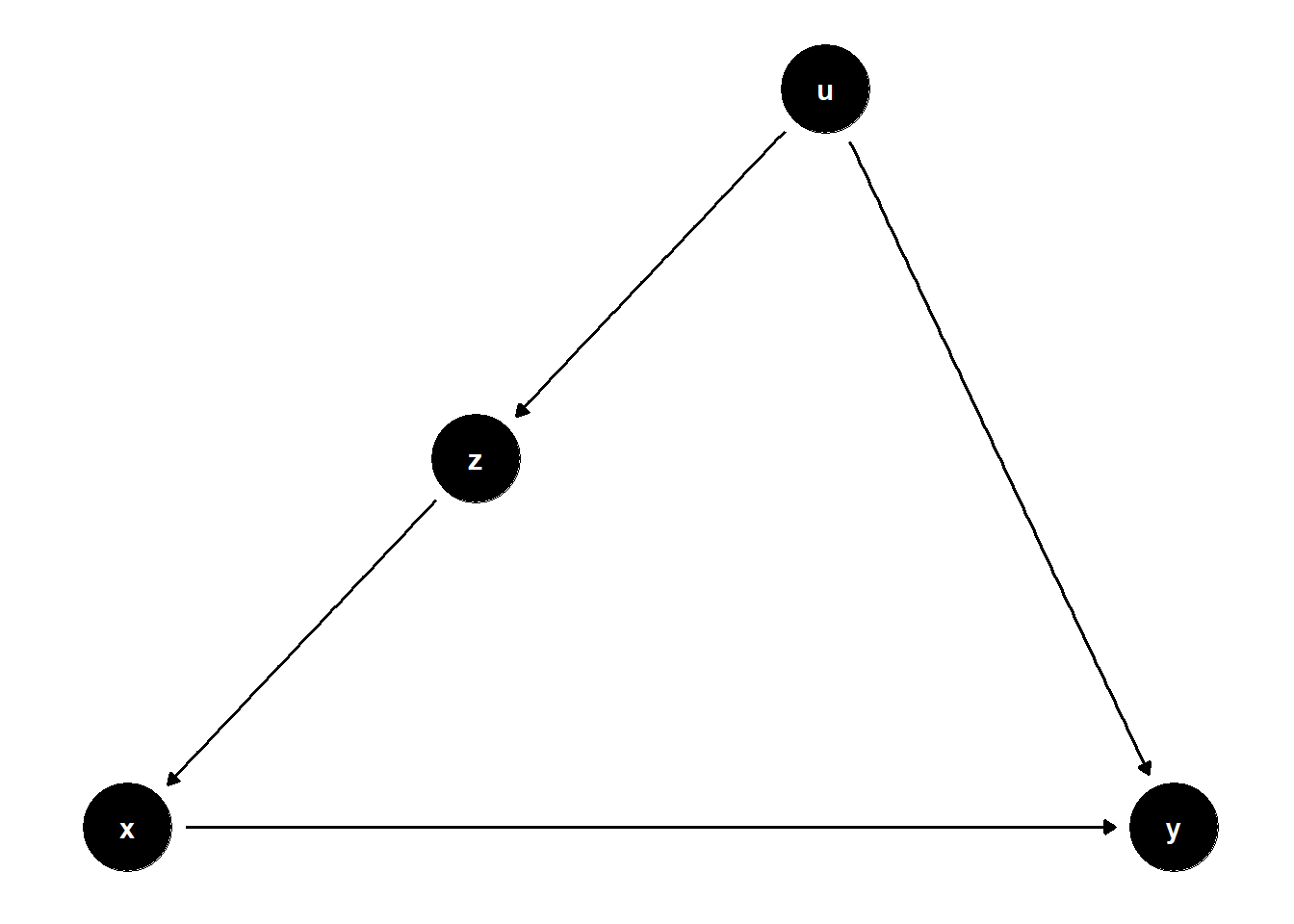
39.2.1.3 \(Z\) is caused by \(U\), but also causes \(Y\)
This DAG illustrates a subtle case where \(Z\) is on a non-causal path from \(X\) to \(Y\) and helps block bias through a shared cause \(U\).
rm(list = ls())
model <- dagitty("dag{
x -> y
u -> z
u -> x
z -> y
}")
latents(model) <- "u"
coordinates(model) <- list(
x = c(x = 1, z = 3, u = 2, y = 4),
y = c(x = 1, z = 2, u = 3, y = 1)
)
ggdag(model) + theme_dag()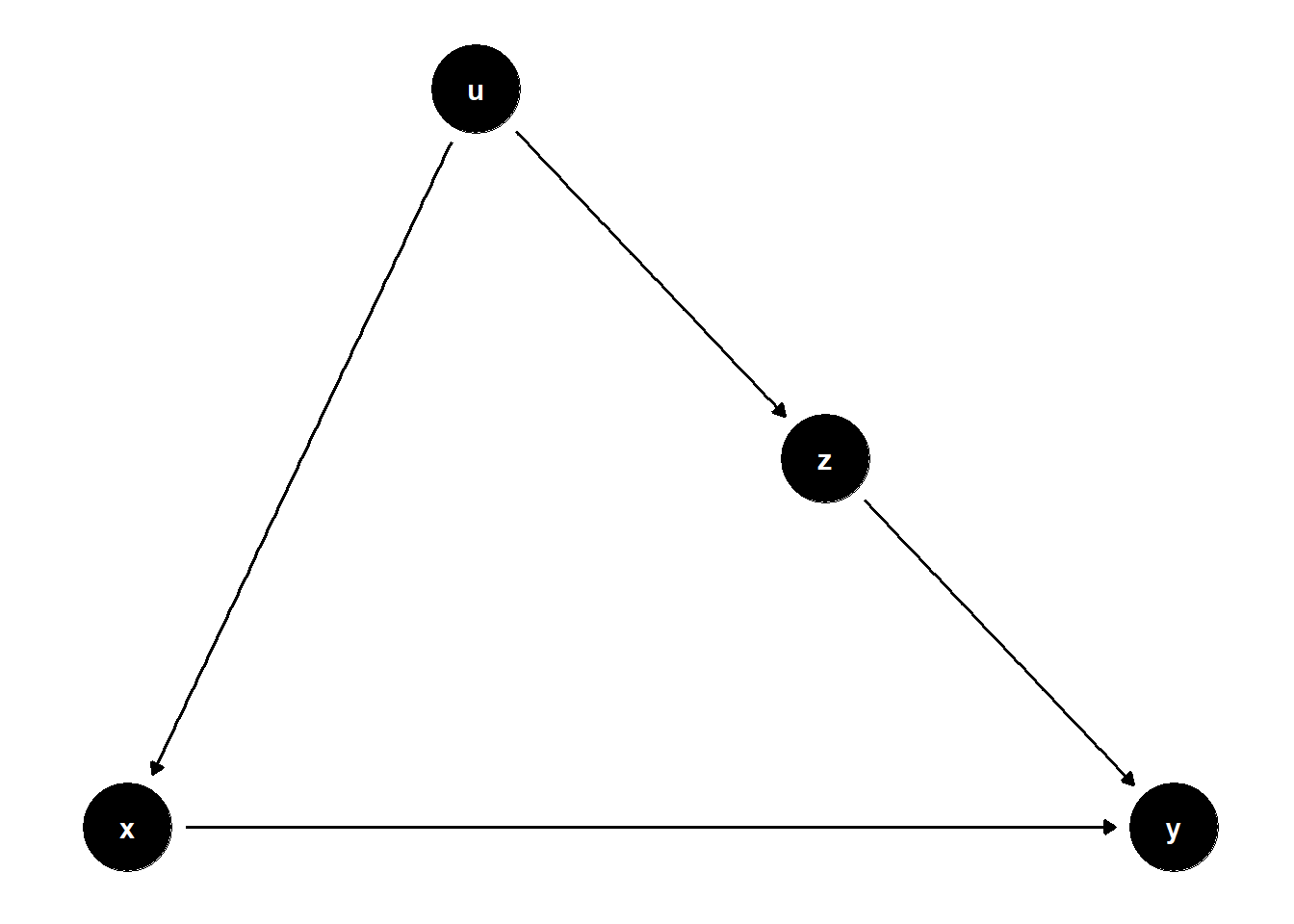
n <- 1e4
u <- rnorm(n)
z <- u + rnorm(n)
x <- u + rnorm(n)
y <- 2 * x + z + rnorm(n)
jtools::export_summs(lm(y ~ x), lm(y ~ x + z))| Model 1 | Model 2 | |
|---|---|---|
| (Intercept) | 0.02 | 0.01 |
| (0.02) | (0.01) | |
| x | 2.49 *** | 1.98 *** |
| (0.01) | (0.01) | |
| z | 1.02 *** | |
| (0.01) | ||
| N | 10000 | 10000 |
| R2 | 0.83 | 0.93 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
Again, we cannot interpret the coefficient on \(Z\) causally, but including \(Z\) helps reduce omitted variable bias from the unobserved confounder \(U\).
39.2.1.4 Summary of Omitted Variable Correction
# Model 1: Z is a confounder
model1 <- dagitty("dag{
x -> y
z -> x
z -> y
}")
coordinates(model1) <-
list(x = c(x = 1, z = 2, y = 3), y = c(x = 1, z = 2, y = 1))
# Model 2: Z is on path from U to X
model2 <- dagitty("dag{
x -> y
u -> z
z -> x
u -> y
}")
latents(model2) <- "u"
coordinates(model2) <-
list(x = c(
x = 1,
z = 2,
u = 3,
y = 4
),
y = c(
x = 1,
z = 2,
u = 3,
y = 1
))
# Model 3: Z influenced by U, affects Y
model3 <- dagitty("dag{
x -> y
u -> z
u -> x
z -> y
}")
latents(model3) <- "u"
coordinates(model3) <-
list(x = c(
x = 1,
z = 3,
u = 2,
y = 4
),
y = c(
x = 1,
z = 2,
u = 3,
y = 1
))
par(mfrow = c(1, 3))
ggdag(model1) + theme_dag()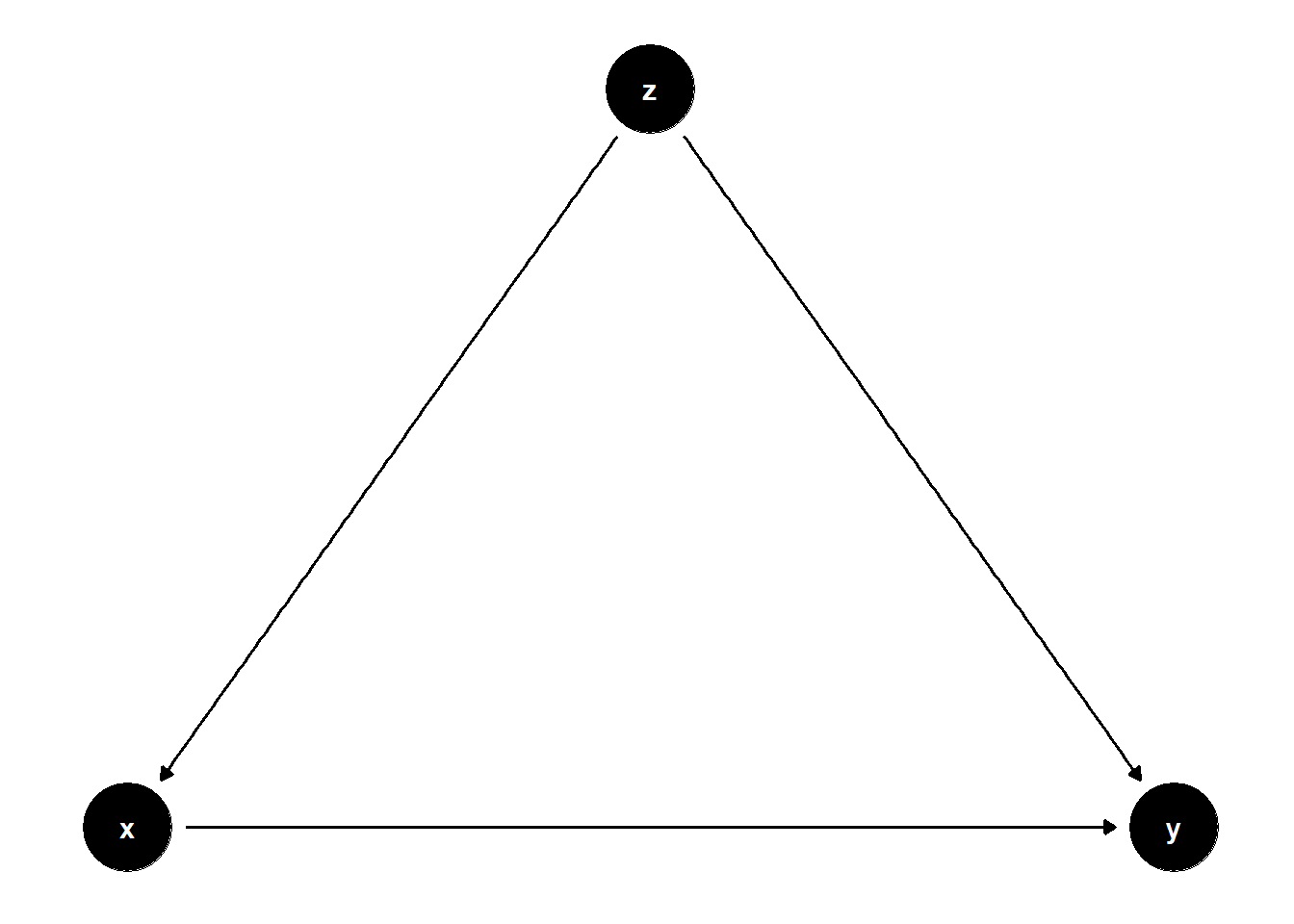
39.2.2 Omitted Variable Bias in Mediation Correction
When a variable \(Z\) is a confounder of both the treatment \(X\) and a mediator \(M\), controlling for \(Z\) helps isolate the indirect and direct effects more accurately.
39.2.2.1 Observed Confounder of Mediator and Treatment
rm(list = ls())
model <- dagitty("dag{
x -> y
z -> x
x -> m
z -> m
m -> y
}")
coordinates(model) <-
list(x = c(
x = 1,
z = 2,
m = 3,
y = 4
),
y = c(
x = 1,
z = 2,
m = 1,
y = 1
))
ggdag(model) + theme_dag()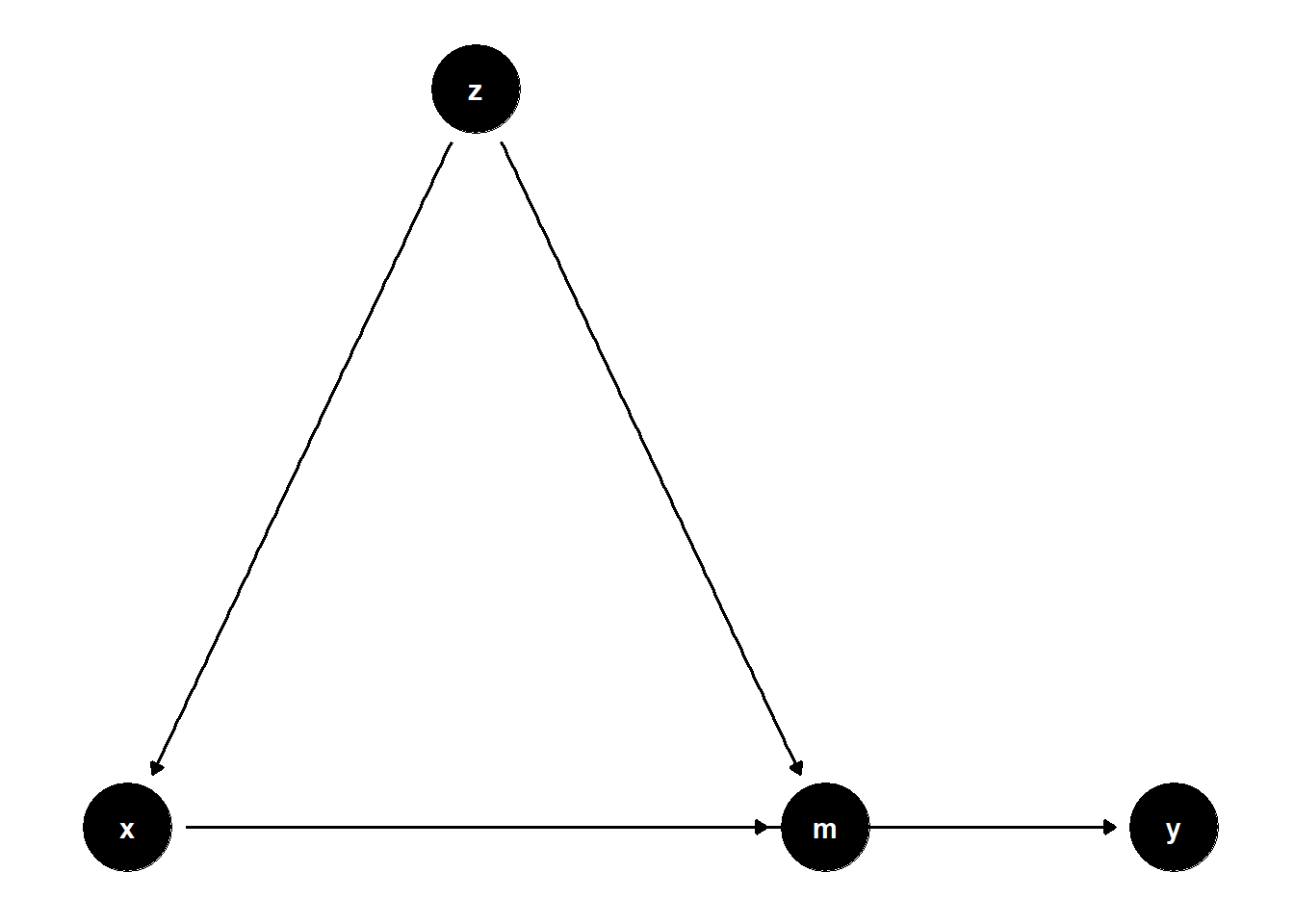
n <- 1e4
z <- rnorm(n)
x <- z + rnorm(n)
m <- 2 * x + z + rnorm(n)
y <- m + rnorm(n)
jtools::export_summs(lm(y ~ x), lm(y ~ x + z))| Model 1 | Model 2 | |
|---|---|---|
| (Intercept) | -0.01 | 0.01 |
| (0.02) | (0.01) | |
| x | 2.50 *** | 1.99 *** |
| (0.01) | (0.01) | |
| z | 1.01 *** | |
| (0.02) | ||
| N | 10000 | 10000 |
| R2 | 0.84 | 0.87 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
39.2.2.2 Latent Common Cause of Mediator and Treatment
rm(list = ls())
model <- dagitty("dag{
x -> y
u -> z
z -> x
x -> m
u -> m
m -> y
}")
latents(model) <- "u"
coordinates(model) <-
list(x = c(
x = 1,
z = 2,
u = 3,
m = 4,
y = 5
),
y = c(
x = 1,
z = 2,
u = 3,
m = 1,
y = 1
))
ggdag(model) + theme_dag()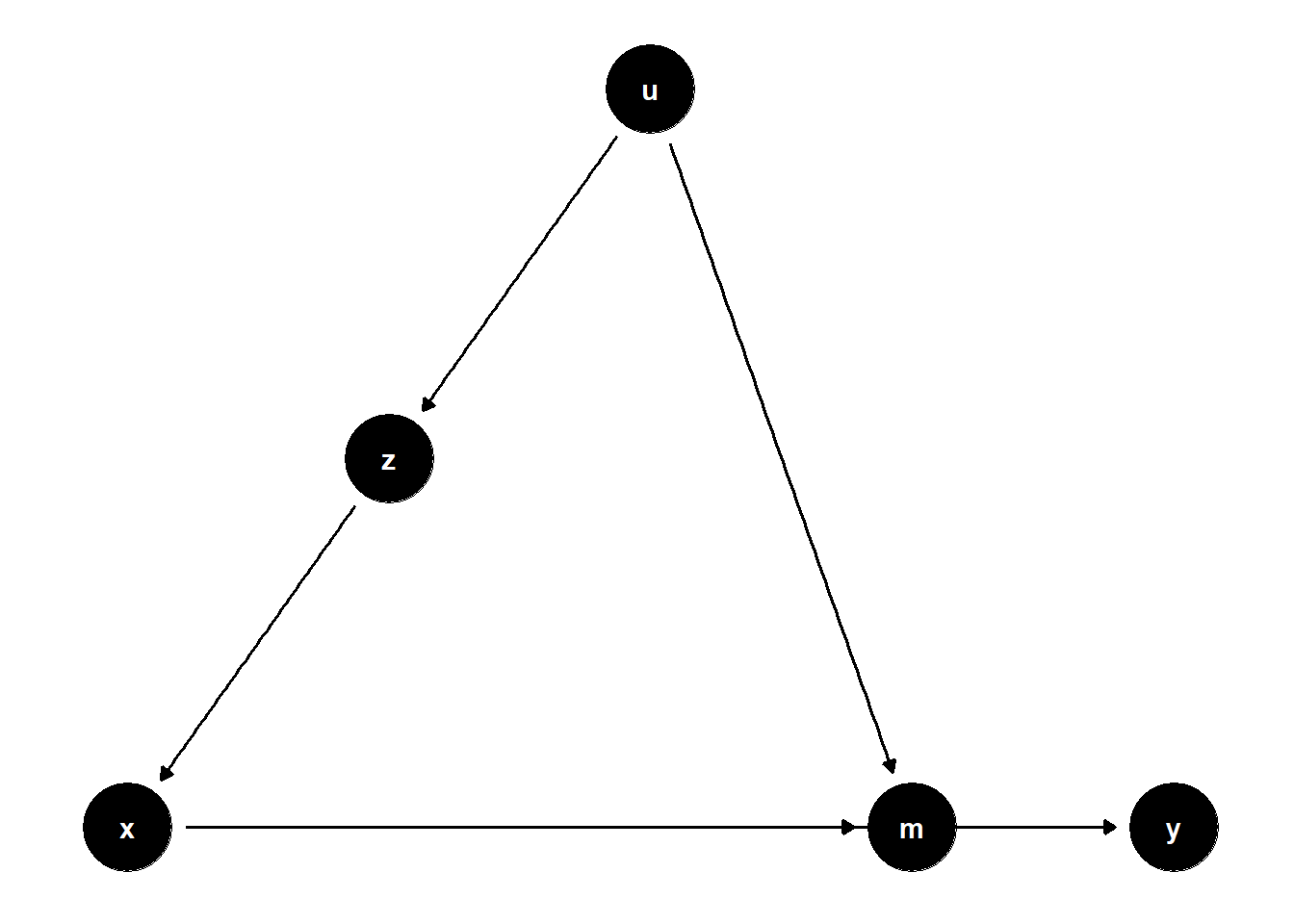
n <- 1e4
u <- rnorm(n)
z <- u + rnorm(n)
x <- z + rnorm(n)
m <- 2 * x + u + rnorm(n)
y <- m + rnorm(n)
jtools::export_summs(lm(y ~ x), lm(y ~ x + z))| Model 1 | Model 2 | |
|---|---|---|
| (Intercept) | 0.01 | 0.01 |
| (0.02) | (0.02) | |
| x | 2.34 *** | 2.01 *** |
| (0.01) | (0.02) | |
| z | 0.50 *** | |
| (0.02) | ||
| N | 10000 | 10000 |
| R2 | 0.86 | 0.87 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
39.2.2.3 Z Affects Mediator, U Affects Both X and Z
rm(list = ls())
model <- dagitty("dag{
x -> y
u -> z
z -> m
x -> m
u -> x
m -> y
}")
latents(model) <- "u"
coordinates(model) <-
list(x = c(
x = 1,
z = 3,
u = 2,
m = 4,
y = 5
),
y = c(
x = 1,
z = 2,
u = 3,
m = 1,
y = 1
))
ggdag(model) + theme_dag()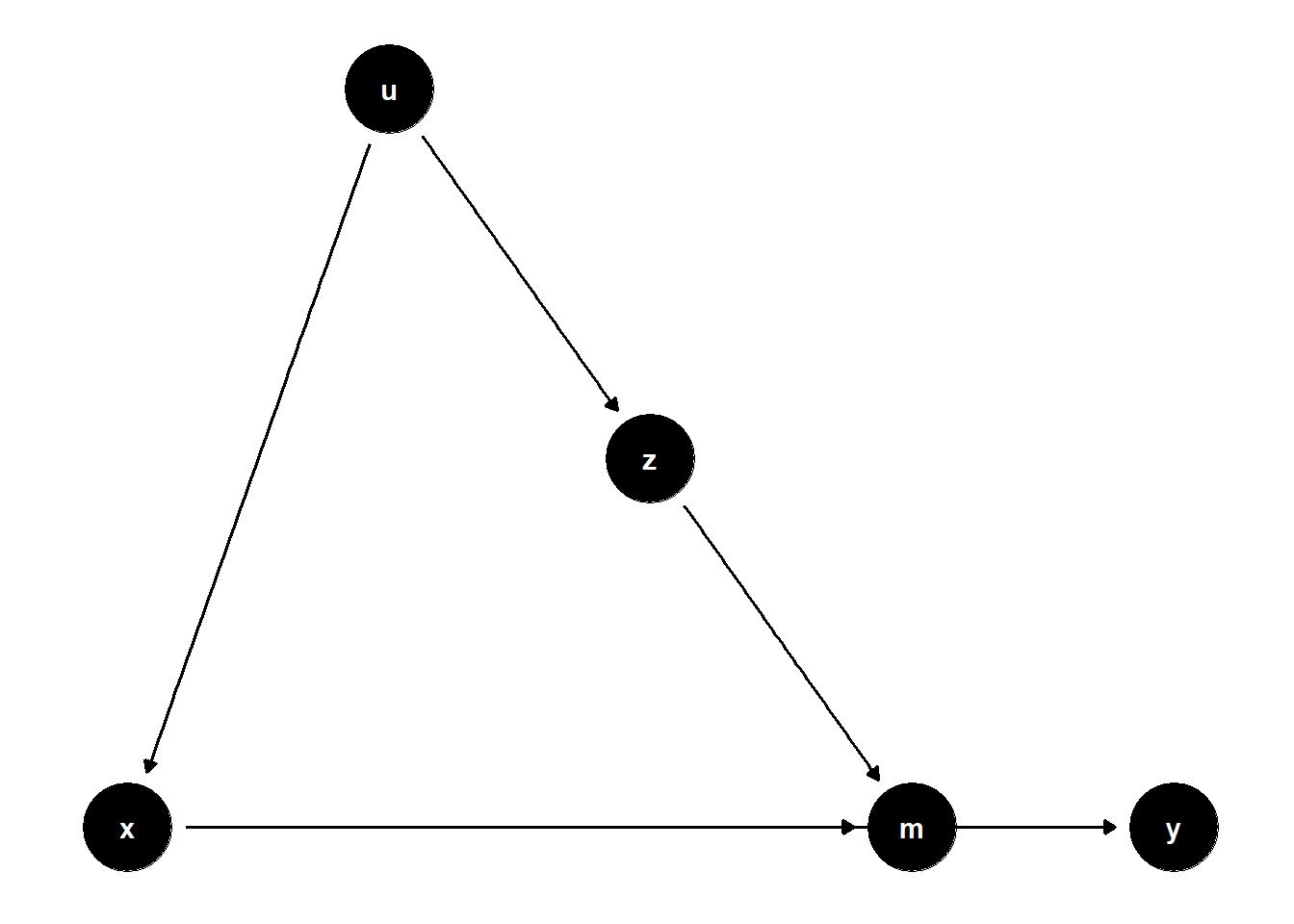
n <- 1e4
u <- rnorm(n)
z <- u + rnorm(n)
x <- u + rnorm(n)
m <- 2 * x + z + rnorm(n)
y <- m + rnorm(n)
jtools::export_summs(lm(y ~ x), lm(y ~ x + z))| Model 1 | Model 2 | |
|---|---|---|
| (Intercept) | 0.00 | 0.00 |
| (0.02) | (0.01) | |
| x | 2.49 *** | 1.99 *** |
| (0.01) | (0.01) | |
| z | 1.00 *** | |
| (0.01) | ||
| N | 10000 | 10000 |
| R2 | 0.78 | 0.87 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
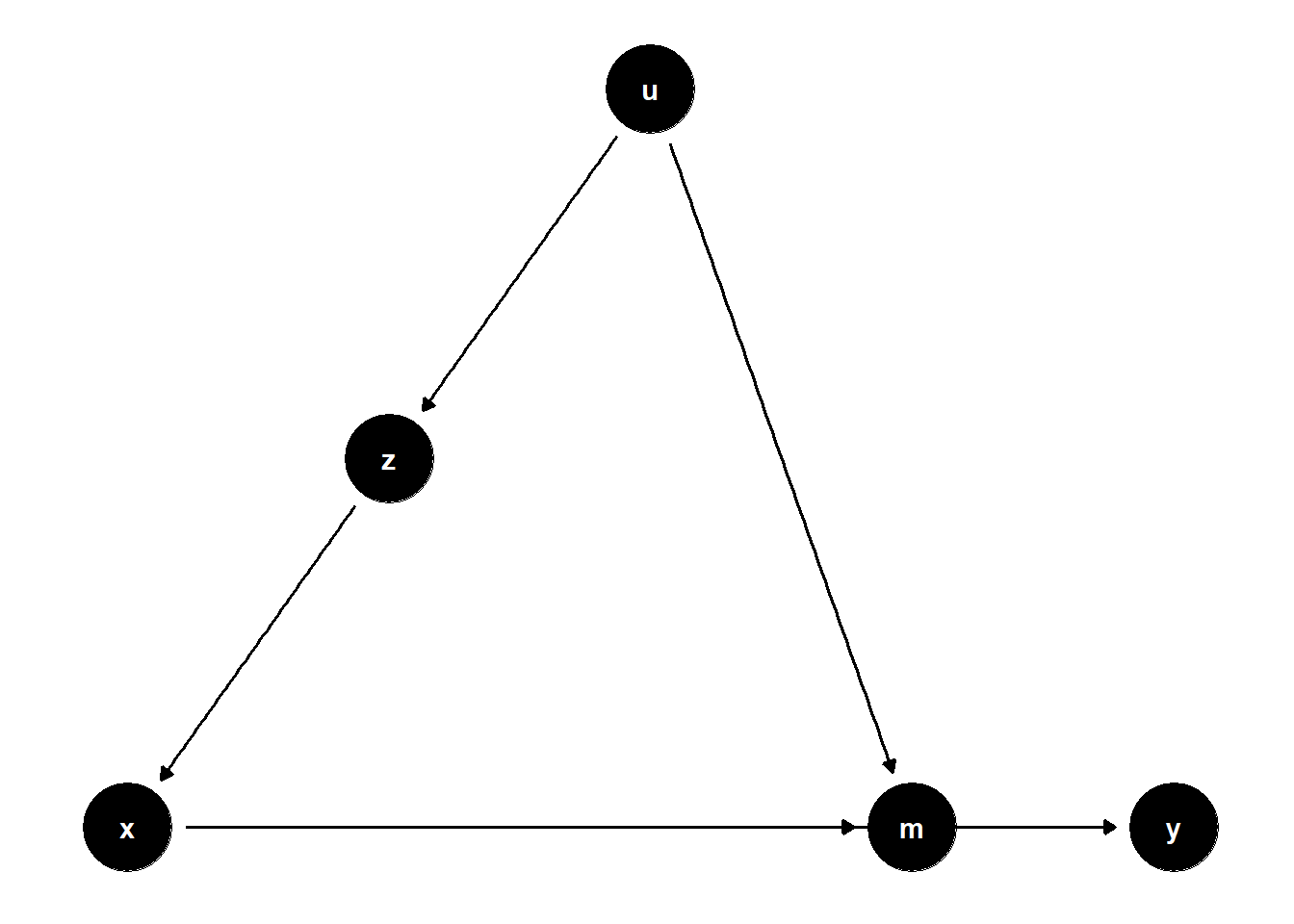
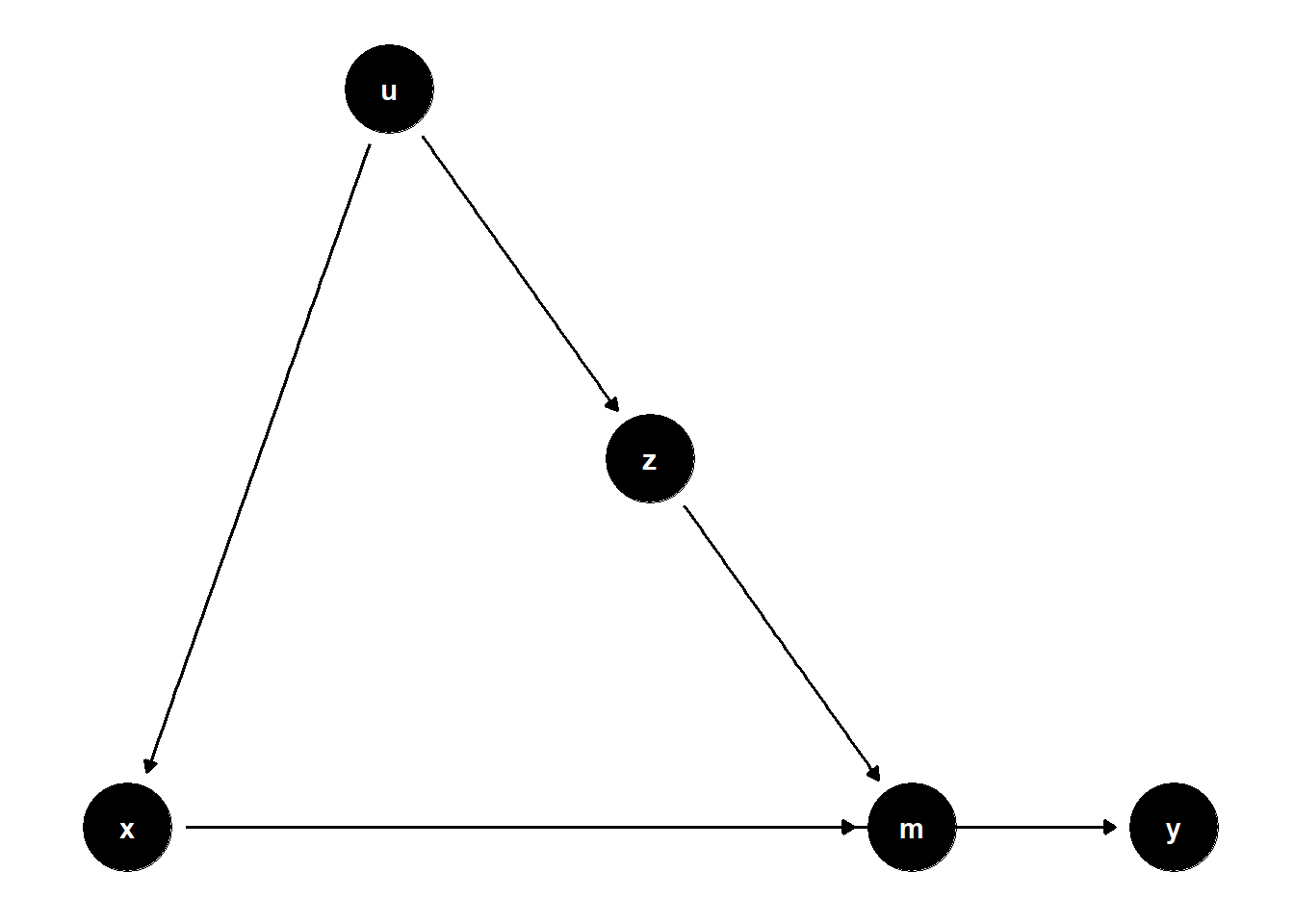
39.2.2.4 Summary of Mediation Correction
# Model 4
model4 <- dagitty("dag{
x -> y
z -> x
x -> m
z -> m
m -> y
}")
coordinates(model4) <-
list(x = c(
x = 1,
z = 2,
m = 3,
y = 4
),
y = c(
x = 1,
z = 2,
m = 1,
y = 1
))
# Model 5
model5 <- dagitty("dag{
x -> y
u -> z
z -> x
x -> m
u -> m
m -> y
}")
latents(model5) <- "u"
coordinates(model5) <-
list(x = c(
x = 1,
z = 2,
u = 3,
m = 4,
y = 5
),
y = c(
x = 1,
z = 2,
u = 3,
m = 1,
y = 1
))
# Model 6
model6 <- dagitty("dag{
x -> y
u -> z
z -> m
x -> m
u -> x
m -> y
}")
latents(model6) <- "u"
coordinates(model6) <-
list(x = c(
x = 1,
z = 3,
u = 2,
m = 4,
y = 5
),
y = c(
x = 1,
z = 2,
u = 3,
m = 1,
y = 1
))
par(mfrow = c(1, 3))
ggdag(model4) + theme_dag()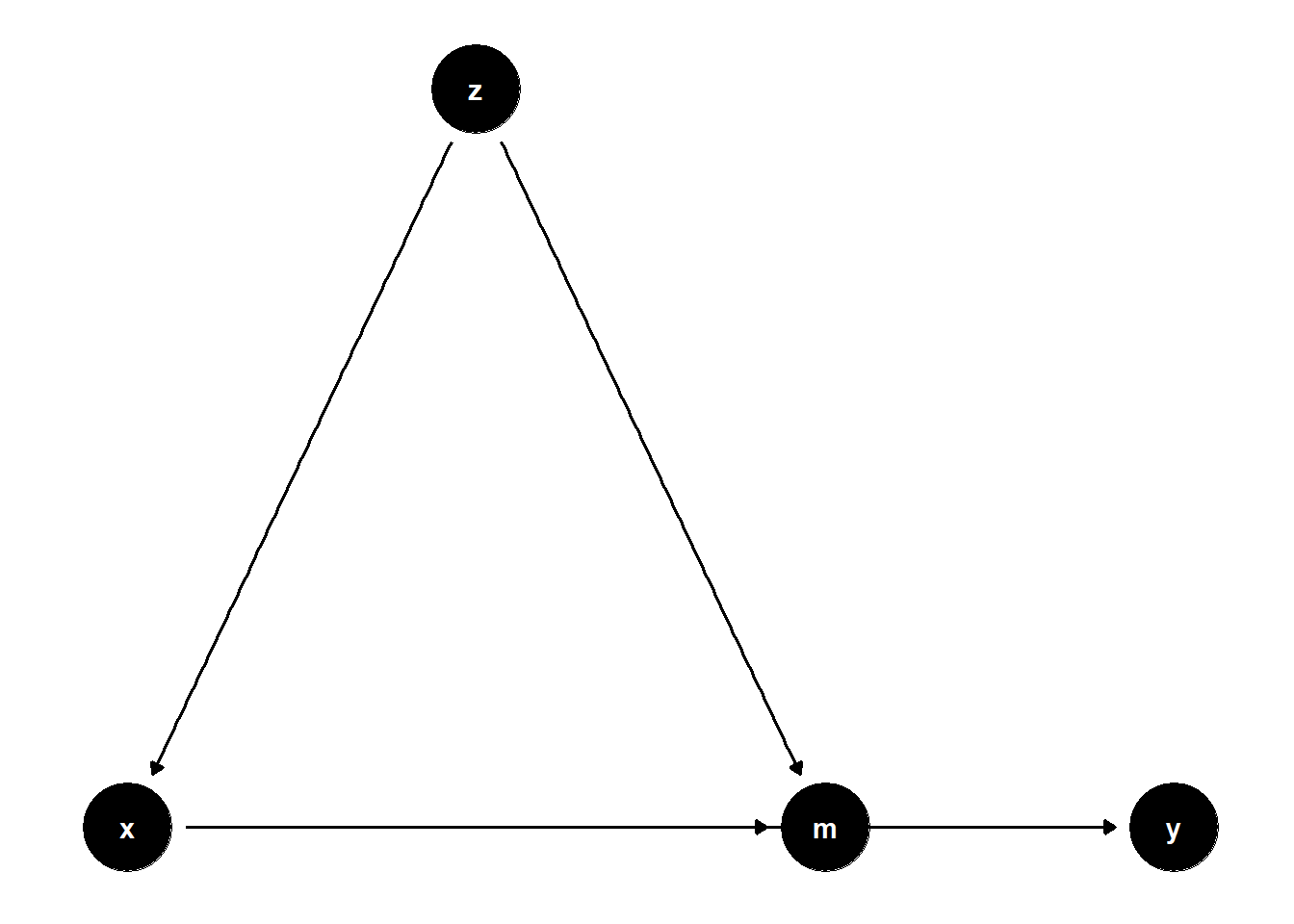
While \(Z\) may be statistically significant, this does not imply a causal effect unless \(Z\) is directly on the causal path from \(X\) to \(Y\). In many valid control scenarios, \(Z\) simply serves to isolate the causal effect of \(X\), not to be interpreted as a cause itself.