2.5 Data Manipulation
# Load required packages
library(tidyverse)
library(lubridate)
# -----------------------------
# Data Structures in R
# -----------------------------
# Create vectors
x <- c(1, 4, 23, 4, 45)
n <- c(1, 3, 5)
g <- c("M", "M", "F")
# Create a data frame
df <- data.frame(n, g)
df # View the data frame
#> n g
#> 1 1 M
#> 2 3 M
#> 3 5 F
str(df) # Check its structure
#> 'data.frame': 3 obs. of 2 variables:
#> $ n: num 1 3 5
#> $ g: chr "M" "M" "F"
# Using tibble for cleaner outputs
df <- tibble(n, g)
df # View the tibble
#> # A tibble: 3 × 2
#> n g
#> <dbl> <chr>
#> 1 1 M
#> 2 3 M
#> 3 5 F
str(df)
#> tibble [3 × 2] (S3: tbl_df/tbl/data.frame)
#> $ n: num [1:3] 1 3 5
#> $ g: chr [1:3] "M" "M" "F"
# Create a list
lst <- list(x, n, g, df)
lst # Display the list
#> [[1]]
#> [1] 1 4 23 4 45
#>
#> [[2]]
#> [1] 1 3 5
#>
#> [[3]]
#> [1] "M" "M" "F"
#>
#> [[4]]
#> # A tibble: 3 × 2
#> n g
#> <dbl> <chr>
#> 1 1 M
#> 2 3 M
#> 3 5 F
# Name list elements
lst2 <- list(num = x, size = n, sex = g, data = df)
lst2 # Named list elements are easier to reference
#> $num
#> [1] 1 4 23 4 45
#>
#> $size
#> [1] 1 3 5
#>
#> $sex
#> [1] "M" "M" "F"
#>
#> $data
#> # A tibble: 3 × 2
#> n g
#> <dbl> <chr>
#> 1 1 M
#> 2 3 M
#> 3 5 F
# Another list example with numeric vectors
lst3 <- list(
x = c(1, 3, 5, 7),
y = c(2, 2, 2, 4, 5, 5, 5, 6),
z = c(22, 3, 3, 3, 5, 10)
)
lst3
#> $x
#> [1] 1 3 5 7
#>
#> $y
#> [1] 2 2 2 4 5 5 5 6
#>
#> $z
#> [1] 22 3 3 3 5 10
# Find means of list elements
# One at a time
mean(lst3$x)
#> [1] 4
mean(lst3$y)
#> [1] 3.875
mean(lst3$z)
#> [1] 7.666667
# Using lapply to calculate means
lapply(lst3, mean)
#> $x
#> [1] 4
#>
#> $y
#> [1] 3.875
#>
#> $z
#> [1] 7.666667
# Simplified output with sapply
sapply(lst3, mean)
#> x y z
#> 4.000000 3.875000 7.666667
# Tidyverse alternative: map() function
map(lst3, mean)
#> $x
#> [1] 4
#>
#> $y
#> [1] 3.875
#>
#> $z
#> [1] 7.666667
# Tidyverse with numeric output: map_dbl()
map_dbl(lst3, mean)
#> x y z
#> 4.000000 3.875000 7.666667
# -----------------------------
# Binding Data Frames
# -----------------------------
# Create tibbles for demonstration
dat01 <- tibble(x = 1:5, y = 5:1)
dat02 <- tibble(x = 10:16, y = x / 2)
dat03 <- tibble(z = runif(5)) # 5 random numbers from (0, 1)
# Row binding
bind_rows(dat01, dat02, dat01)
#> # A tibble: 17 × 2
#> x y
#> <int> <dbl>
#> 1 1 5
#> 2 2 4
#> 3 3 3
#> 4 4 2
#> 5 5 1
#> 6 10 5
#> 7 11 5.5
#> 8 12 6
#> 9 13 6.5
#> 10 14 7
#> 11 15 7.5
#> 12 16 8
#> 13 1 5
#> 14 2 4
#> 15 3 3
#> 16 4 2
#> 17 5 1
# Add a new identifier column with .id
bind_rows(dat01, dat02, .id = "id")
#> # A tibble: 12 × 3
#> id x y
#> <chr> <int> <dbl>
#> 1 1 1 5
#> 2 1 2 4
#> 3 1 3 3
#> 4 1 4 2
#> 5 1 5 1
#> 6 2 10 5
#> 7 2 11 5.5
#> 8 2 12 6
#> 9 2 13 6.5
#> 10 2 14 7
#> 11 2 15 7.5
#> 12 2 16 8
# Use named inputs for better identification
bind_rows("dat01" = dat01, "dat02" = dat02, .id = "id")
#> # A tibble: 12 × 3
#> id x y
#> <chr> <int> <dbl>
#> 1 dat01 1 5
#> 2 dat01 2 4
#> 3 dat01 3 3
#> 4 dat01 4 2
#> 5 dat01 5 1
#> 6 dat02 10 5
#> 7 dat02 11 5.5
#> 8 dat02 12 6
#> 9 dat02 13 6.5
#> 10 dat02 14 7
#> 11 dat02 15 7.5
#> 12 dat02 16 8
# Bind a list of data frames
list01 <- list("dat01" = dat01, "dat02" = dat02)
bind_rows(list01, .id = "source")
#> # A tibble: 12 × 3
#> source x y
#> <chr> <int> <dbl>
#> 1 dat01 1 5
#> 2 dat01 2 4
#> 3 dat01 3 3
#> 4 dat01 4 2
#> 5 dat01 5 1
#> 6 dat02 10 5
#> 7 dat02 11 5.5
#> 8 dat02 12 6
#> 9 dat02 13 6.5
#> 10 dat02 14 7
#> 11 dat02 15 7.5
#> 12 dat02 16 8
# Column binding
bind_cols(dat01, dat03)
#> # A tibble: 5 × 3
#> x y z
#> <int> <int> <dbl>
#> 1 1 5 0.960
#> 2 2 4 0.357
#> 3 3 3 0.883
#> 4 4 2 0.123
#> 5 5 1 0.628
# -----------------------------
# String Manipulation
# -----------------------------
names <- c("Ford, MS", "Jones, PhD", "Martin, Phd", "Huck, MA, MLS")
# Remove everything after the first comma
str_remove(names, pattern = ", [[:print:]]+")
#> [1] "Ford" "Jones" "Martin" "Huck"
# Explanation: [[:print:]]+ matches one or more printable characters
# -----------------------------
# Reshaping Data
# -----------------------------
# Wide format data
wide <- data.frame(
name = c("Clay", "Garrett", "Addison"),
test1 = c(78, 93, 90),
test2 = c(87, 91, 97),
test3 = c(88, 99, 91)
)
# Long format data
long <- data.frame(
name = rep(c("Clay", "Garrett", "Addison"), each = 3),
test = rep(1:3, 3),
score = c(78, 87, 88, 93, 91, 99, 90, 97, 91)
)
# Summary statistics
aggregate(score ~ name, data = long, mean) # Mean score per student
#> name score
#> 1 Addison 92.66667
#> 2 Clay 84.33333
#> 3 Garrett 94.33333
aggregate(score ~ test, data = long, mean) # Mean score per test
#> test score
#> 1 1 87.00000
#> 2 2 91.66667
#> 3 3 92.66667# Line plot of scores over tests
ggplot(long,
aes(
x = factor(test),
y = score,
color = name,
group = name
)) +
geom_point() +
geom_line() +
xlab("Test") +
ggtitle("Test Scores by Student")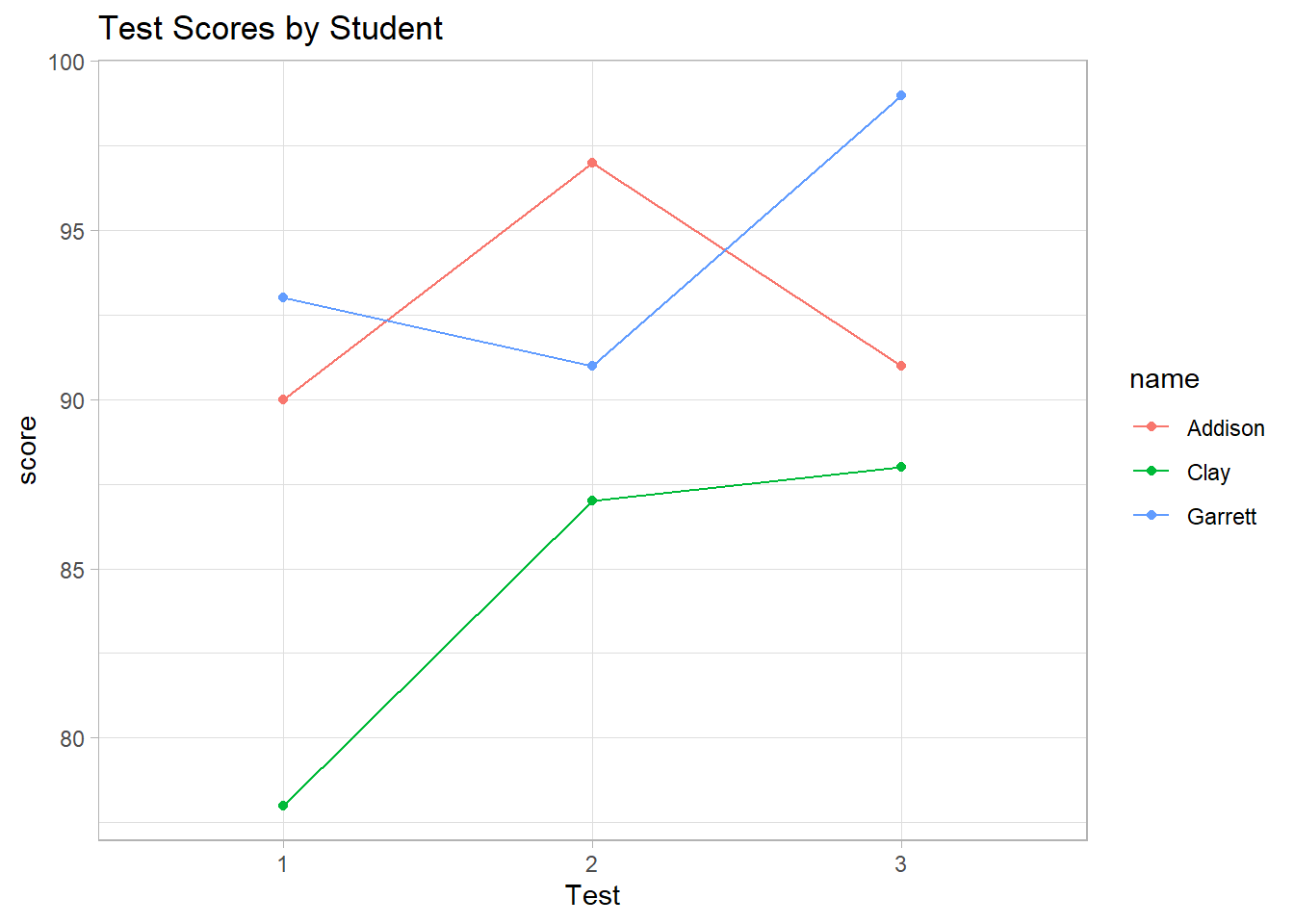
Figure 2.22: Test Scores by Student
# Reshape wide to long
pivot_longer(wide, test1:test3, names_to = "test", values_to = "score")
#> # A tibble: 9 × 3
#> name test score
#> <chr> <chr> <dbl>
#> 1 Clay test1 78
#> 2 Clay test2 87
#> 3 Clay test3 88
#> 4 Garrett test1 93
#> 5 Garrett test2 91
#> 6 Garrett test3 99
#> 7 Addison test1 90
#> 8 Addison test2 97
#> 9 Addison test3 91
# Use names_prefix to clean column names
pivot_longer(
wide,
-name,
names_to = "test",
values_to = "score",
names_prefix = "test"
)
#> # A tibble: 9 × 3
#> name test score
#> <chr> <chr> <dbl>
#> 1 Clay 1 78
#> 2 Clay 2 87
#> 3 Clay 3 88
#> 4 Garrett 1 93
#> 5 Garrett 2 91
#> 6 Garrett 3 99
#> 7 Addison 1 90
#> 8 Addison 2 97
#> 9 Addison 3 91
# Reshape long to wide with explicit id_cols argument
pivot_wider(
long,
id_cols = name,
names_from = test,
values_from = score
)
#> # A tibble: 3 × 4
#> name `1` `2` `3`
#> <chr> <dbl> <dbl> <dbl>
#> 1 Clay 78 87 88
#> 2 Garrett 93 91 99
#> 3 Addison 90 97 91
# Add a prefix to the resulting columns
pivot_wider(
long,
id_cols = name,
names_from = test,
values_from = score,
names_prefix = "test"
)
#> # A tibble: 3 × 4
#> name test1 test2 test3
#> <chr> <dbl> <dbl> <dbl>
#> 1 Clay 78 87 88
#> 2 Garrett 93 91 99
#> 3 Addison 90 97 91The verbs of data manipulation
select: selecting (or not selecting) columns based on their names (eg: select columns Q1 through Q25)slice: selecting (or not selecting) rows based on their position (eg: select rows 1:10)mutate: add or derive new columns (or variables) based on existing columns (eg: create a new column that expresses measurement in cm based on existing measure in inches)rename: rename variables or change column names (eg: change “GraduationRate100” to “grad100”)filter: selecting rows based on a condition (eg: all rows where gender = Male)arrange: ordering rows based on variable(s) numeric or alphabetical order (eg: sort in descending order of Income)sample: take random samples of data (eg: sample 80% of data to create a “training” set)summarize: condense or aggregate multiple values into single summary values (eg: calculate median income by age group)group_by: convert a tbl into a grouped tbl so that operations are performed “by group”; allows us to summarize data or apply verbs to data by groups (eg, by gender or treatment)- the pipe:
%>%Use Ctrl + Shift + M (Win) or Cmd + Shift + M (Mac) to enter in RStudio
The pipe takes the output of a function and “pipes” into the first argument of the next function.
new pipe is
|>It should be identical to the old one, except for certain special cases.
:=(Walrus operator): similar to=, but for cases where you want to use thegluepackage (i.e., dynamic changes in the variable name in the left-hand side)
Writing function in R
Tunneling
{{ (called curly-curly) allows you to tunnel data-variables through arg-variables (i.e., function arguments)
library(tidyverse)
# -----------------------------
# Writing Functions with {{ }}
# -----------------------------
# Define a custom function using {{ }}
get_mean <- function(data, group_var, var_to_mean) {
data %>%
group_by({{group_var}}) %>%
summarize(mean = mean({{var_to_mean}}, na.rm = TRUE))
}
# Apply the function
data("mtcars")
mtcars %>%
get_mean(group_var = cyl, var_to_mean = mpg)
#> # A tibble: 3 × 2
#> cyl mean
#> <dbl> <dbl>
#> 1 4 26.7
#> 2 6 19.7
#> 3 8 15.1
# Dynamically name the resulting variable
get_mean <- function(data, group_var, var_to_mean, prefix = "mean_of") {
data %>%
group_by({{group_var}}) %>%
summarize("{prefix}_{{var_to_mean}}" := mean({{var_to_mean}}, na.rm = TRUE))
}
# Apply the modified function
mtcars %>%
get_mean(group_var = cyl, var_to_mean = mpg)
#> # A tibble: 3 × 2
#> cyl mean_of_mpg
#> <dbl> <dbl>
#> 1 4 26.7
#> 2 6 19.7
#> 3 8 15.1