39.3 Neutral Controls
Not all covariates used in regression adjustment are necessary for identification. Neutral controls do not help with causal identification but may affect estimation precision. Including them:
- Does not introduce bias, because they do not lie on back-door or collider paths.
- May reduce standard errors, by explaining additional variation in the outcome.
39.3.1 Good Predictive Controls
When a variable is correlated with the outcome \(Y\), but not a cause of the treatment \(X\), controlling for it is optional for identification but may increase precision.
39.3.1.1 \(Z\) predicts \(Y\), not \(X\)
# Clean workspace
rm(list = ls())
model <- dagitty("dag{
x -> y
z -> y
}")
coordinates(model) <- list(
x = c(x = 1, z = 2, y = 2),
y = c(x = 1, z = 2, y = 1)
)
ggdag(model) + theme_dag()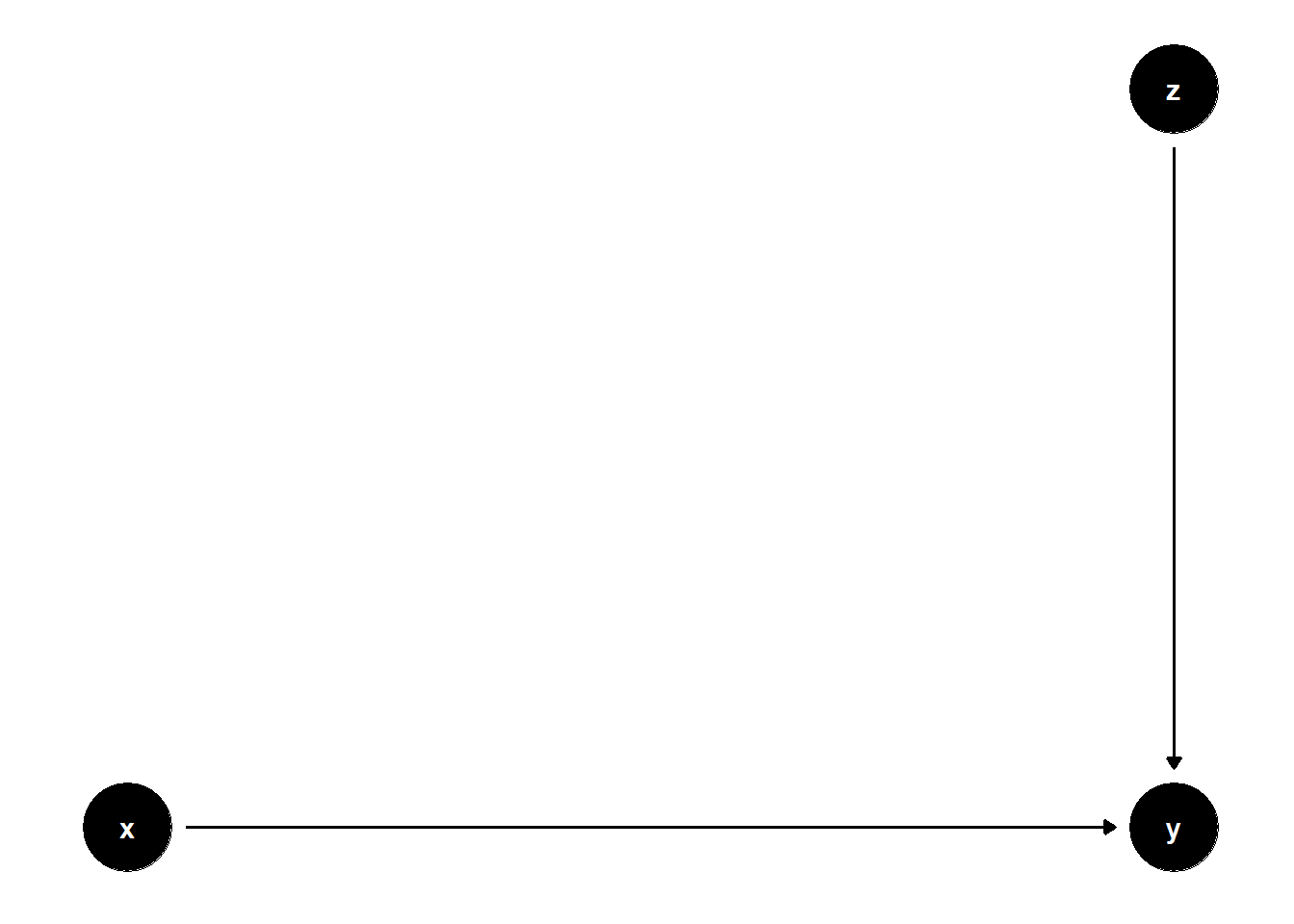
n <- 1e4
z <- rnorm(n)
x <- rnorm(n)
y <- x + 2 * z + rnorm(n)
jtools::export_summs(
lm(y ~ x),
lm(y ~ x + z),
model.names = c("Without Z", "With Predictive Z")
)| Without Z | With Predictive Z | |
|---|---|---|
| (Intercept) | -0.01 | -0.01 |
| (0.02) | (0.01) | |
| x | 1.01 *** | 1.01 *** |
| (0.02) | (0.01) | |
| z | 2.01 *** | |
| (0.01) | ||
| N | 10000 | 10000 |
| R2 | 0.17 | 0.83 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
The coefficient on \(X\) remains unbiased in both models, but standard errors are smaller in the model with \(Z\).
39.3.1.2 \(Z\) predicts a mediator \(M\)
rm(list = ls())
model <- dagitty("dag{
x -> y
x -> m
z -> m
m -> y
}")
coordinates(model) <- list(
x = c(x = 1, z = 2, m = 2, y = 3),
y = c(x = 1, z = 2, m = 1, y = 1)
)
ggdag(model) + theme_dag()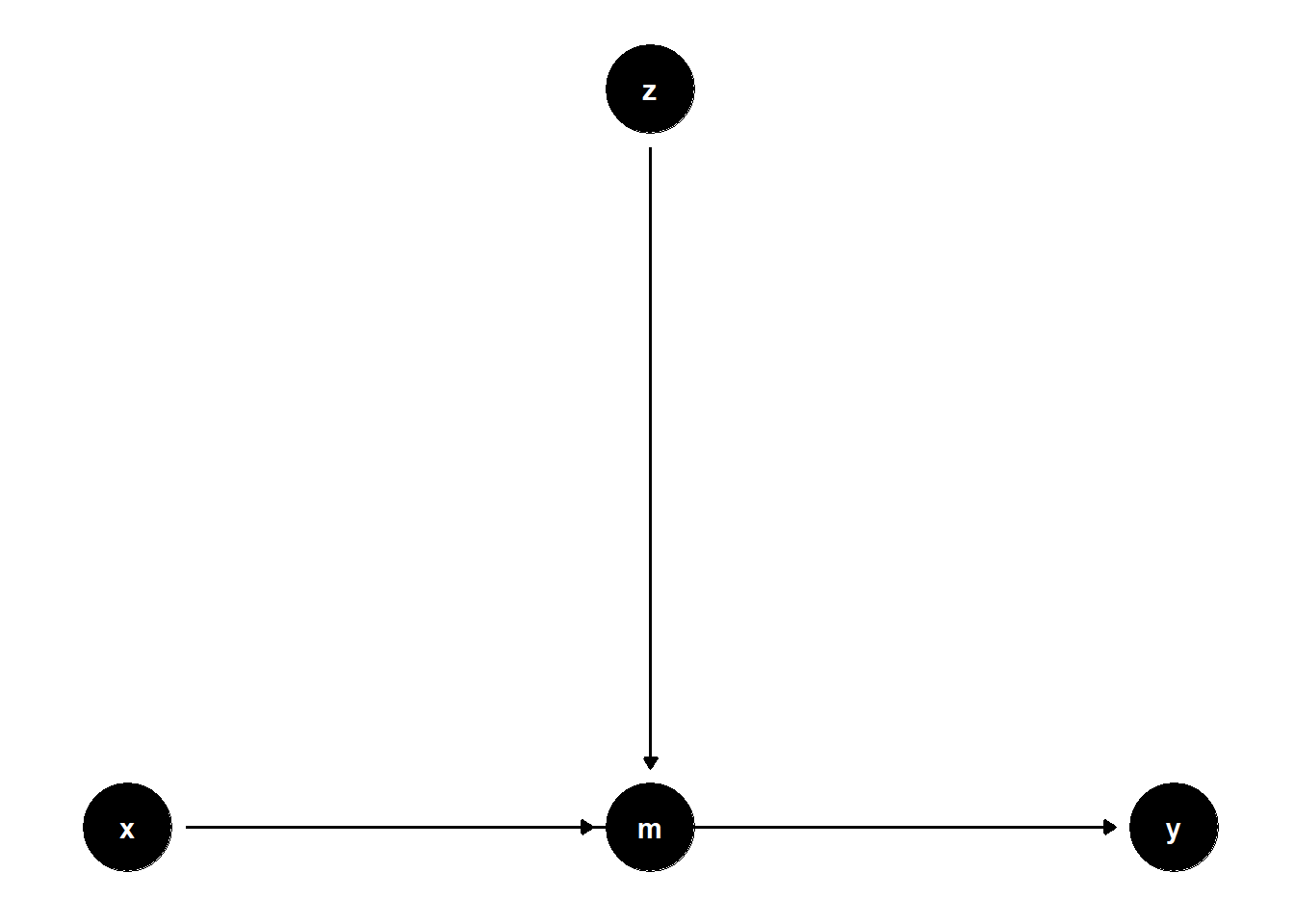
n <- 1e4
z <- rnorm(n)
x <- rnorm(n)
m <- 2 * z + rnorm(n)
y <- x + 2 * m + rnorm(n)
jtools::export_summs(
lm(y ~ x),
lm(y ~ x + z),
model.names = c("Without Z", "With Predictive Z")
)| Without Z | With Predictive Z | |
|---|---|---|
| (Intercept) | 0.02 | 0.02 |
| (0.05) | (0.02) | |
| x | 1.04 *** | 1.00 *** |
| (0.05) | (0.02) | |
| z | 3.98 *** | |
| (0.02) | ||
| N | 10000 | 10000 |
| R2 | 0.05 | 0.77 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
Even though \(Z\) is not on any causal path from \(X\) to \(Y\), controlling for it may reduce residual variance in \(Y\), hence increasing precision.
39.3.2 Good Selection Bias
In more complex selection structures, adjusting for selection variables can improve identification, but only in the presence of additional post-selection information.
39.3.2.1 \(W\) is a collider; \(Z\) helps condition on selection
rm(list = ls())
model <- dagitty("dag{
x -> y
x -> z
z -> w
u -> w
u -> y
}")
latents(model) <- "u"
coordinates(model) <- list(
x = c(x = 1, z = 2, w = 3, u = 3, y = 5),
y = c(x = 3, z = 2, w = 1, u = 4, y = 3)
)
ggdag(model) + theme_dag()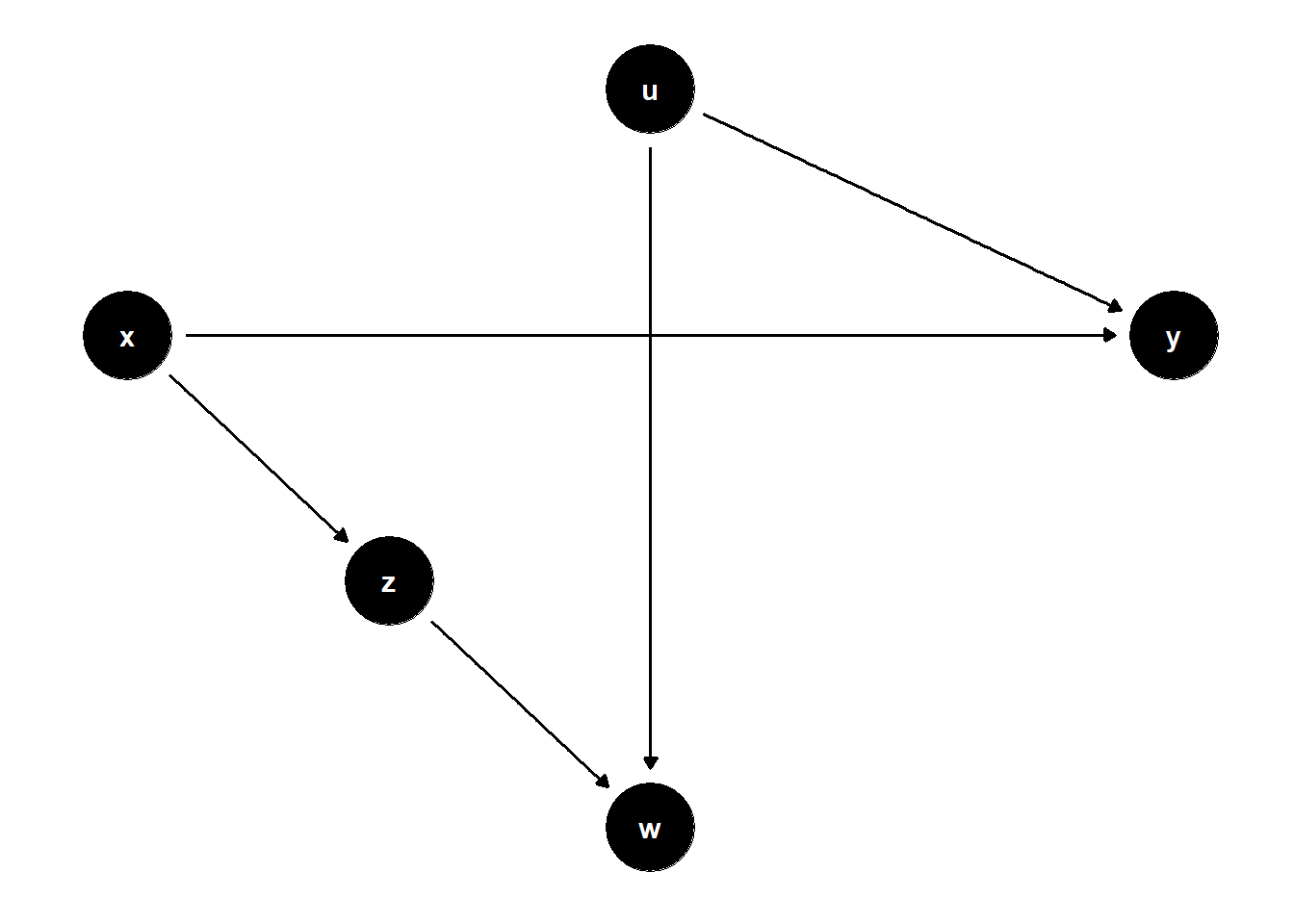
n <- 1e4
x <- rnorm(n)
u <- rnorm(n)
z <- x + rnorm(n)
w <- z + u + rnorm(n)
y <- x - 2 * u + rnorm(n)
jtools::export_summs(
lm(y ~ x),
lm(y ~ x + w),
lm(y ~ x + z + w),
model.names = c("Unadjusted", "Control W", "Control W + Z")
)| Unadjusted | Control W | Control W + Z | |
|---|---|---|---|
| (Intercept) | -0.00 | 0.00 | -0.02 |
| (0.02) | (0.02) | (0.02) | |
| x | 1.00 *** | 1.69 *** | 1.01 *** |
| (0.02) | (0.02) | (0.02) | |
| w | -0.67 *** | -1.00 *** | |
| (0.01) | (0.01) | ||
| z | 1.01 *** | ||
| (0.02) | |||
| N | 10000 | 10000 | 10000 |
| R2 | 0.18 | 0.40 | 0.51 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | |||
Unadjusted model is unbiased.
Controlling only for \(W\) is biased due to collider path \(X \to Z \to W \leftarrow U \to Y\).
Adding \(Z\) restores identification by blocking that path.
39.3.3 Bad Predictive Controls
Not all predictive variables are useful — some may reduce precision by soaking up degrees of freedom or increasing multicollinearity.
39.3.3.1 \(Z\) predicts \(X\), not \(Y\)
rm(list = ls())
model <- dagitty("dag{
x -> y
z -> x
}")
coordinates(model) <- list(
x = c(x = 1, z = 1, y = 2),
y = c(x = 1, z = 2, y = 1)
)
ggdag(model) + theme_dag()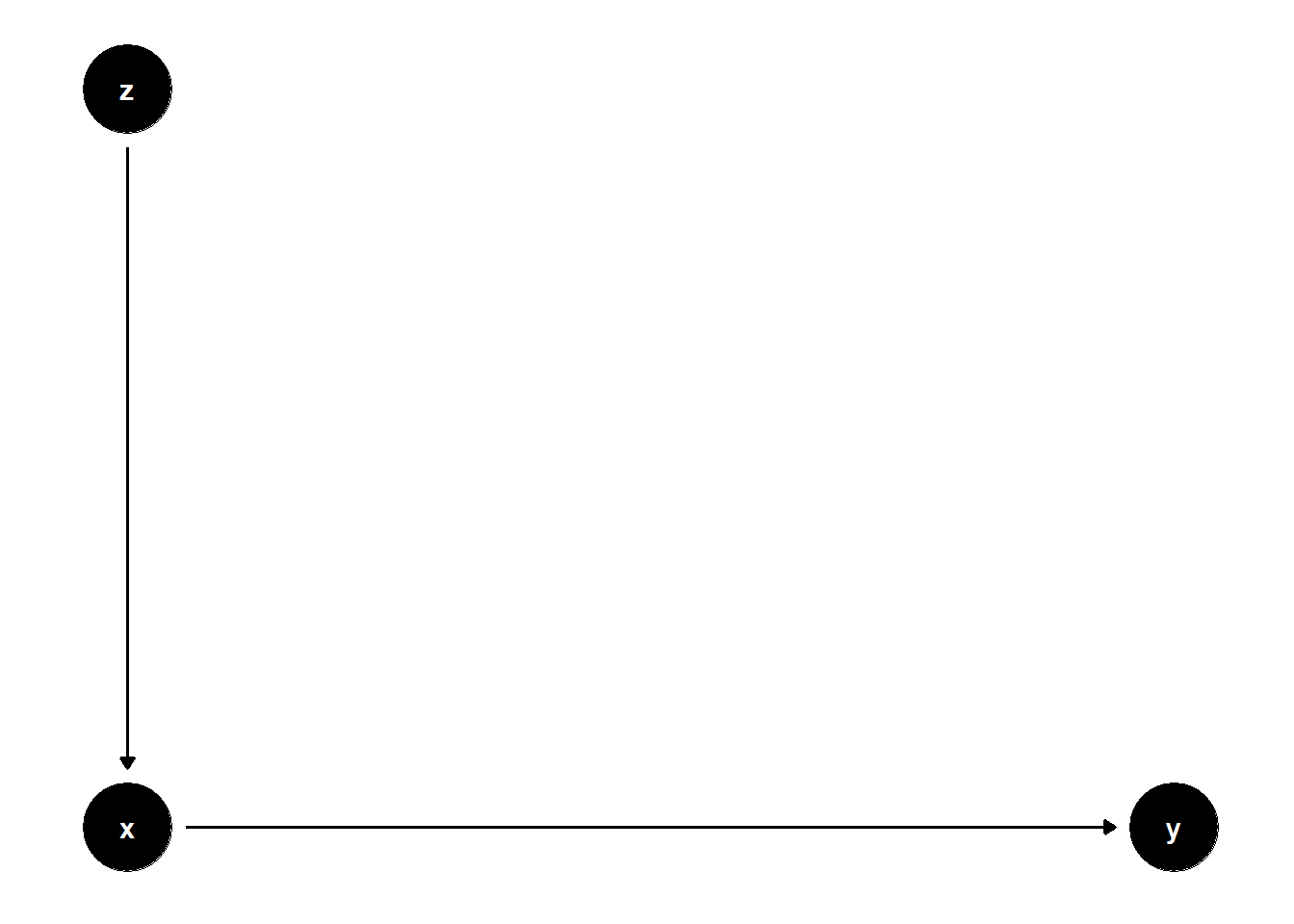
n <- 1e4
z <- rnorm(n)
x <- 2 * z + rnorm(n)
y <- x + rnorm(n)
jtools::export_summs(
lm(y ~ x),
lm(y ~ x + z),
model.names = c("Without Z", "With Z (predicts X)")
)| Without Z | With Z (predicts X) | |
|---|---|---|
| (Intercept) | 0.01 | 0.01 |
| (0.01) | (0.01) | |
| x | 1.00 *** | 1.00 *** |
| (0.00) | (0.01) | |
| z | -0.00 | |
| (0.02) | ||
| N | 10000 | 10000 |
| R2 | 0.84 | 0.84 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
\(Z\) adds no explanatory power for \(Y\), and thus increases SE for the estimate of \(X\)’s effect.
39.3.3.2 \(Z\) is a child of \(X\)
rm(list = ls())
model <- dagitty("dag{
x -> y
x -> z
}")
coordinates(model) <- list(
x = c(x = 1, z = 1, y = 2),
y = c(x = 1, z = 2, y = 1)
)
ggdag(model) + theme_dag()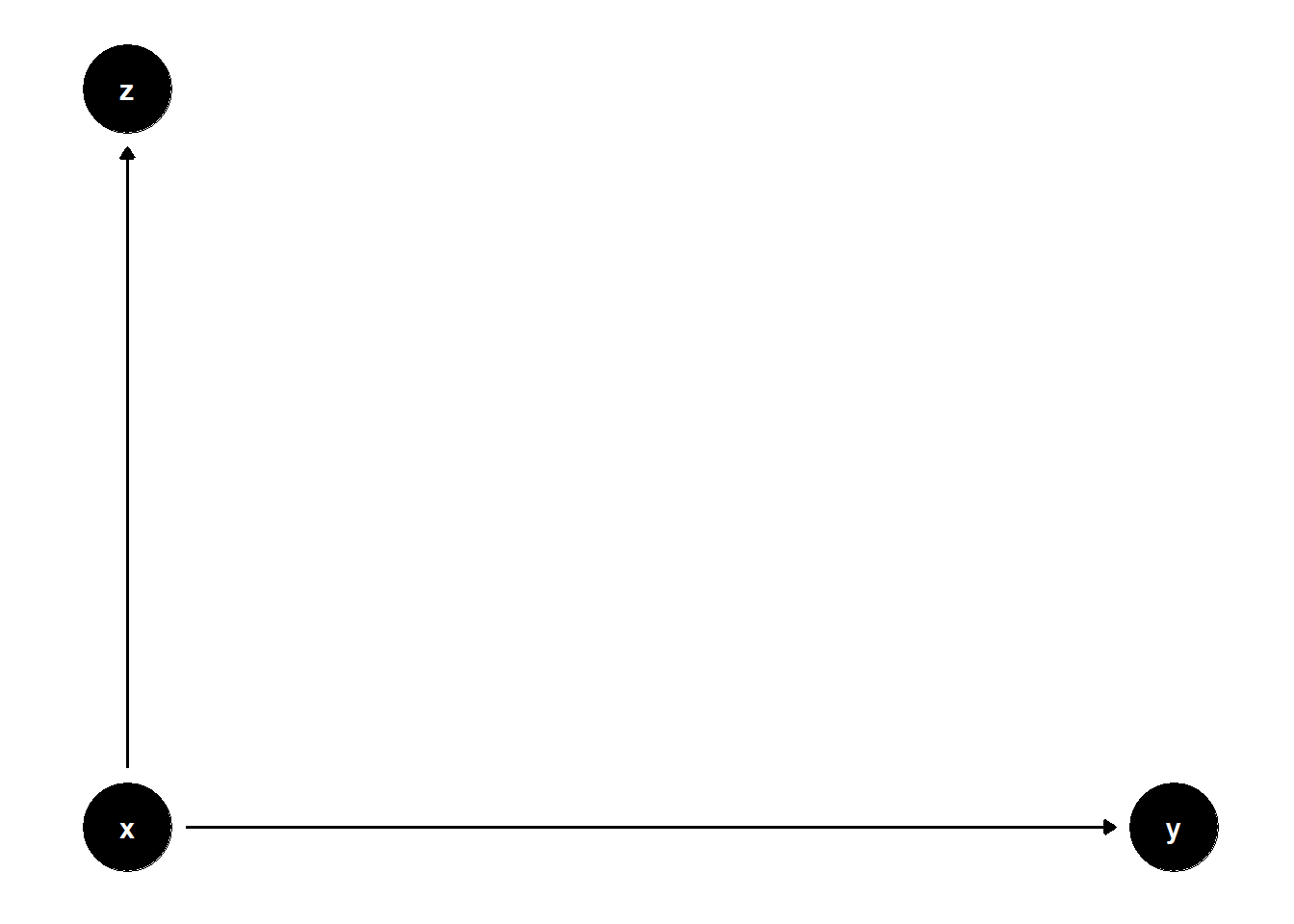
n <- 1e4
x <- rnorm(n)
z <- 2 * x + rnorm(n)
y <- x + rnorm(n)
jtools::export_summs(
lm(y ~ x),
lm(y ~ x + z),
model.names = c("Without Z", "With Child Z")
)| Without Z | With Child Z | |
|---|---|---|
| (Intercept) | 0.00 | 0.00 |
| (0.01) | (0.01) | |
| x | 1.02 *** | 1.06 *** |
| (0.01) | (0.02) | |
| z | -0.02 * | |
| (0.01) | ||
| N | 10000 | 10000 |
| R2 | 0.49 | 0.49 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
Here, \(Z\) is a post-treatment variable. Though it does not bias the estimate of \(X\), it adds noise and increases the SE.
39.3.4 Bad Selection Bias
Controlling for some post-treatment variables can hurt precision without affecting bias.
rm(list = ls())
model <- dagitty("dag{
x -> y
x -> z
}")
coordinates(model) <- list(
x = c(x = 1, z = 2, y = 2),
y = c(x = 1, z = 2, y = 1)
)
ggdag(model) + theme_dag()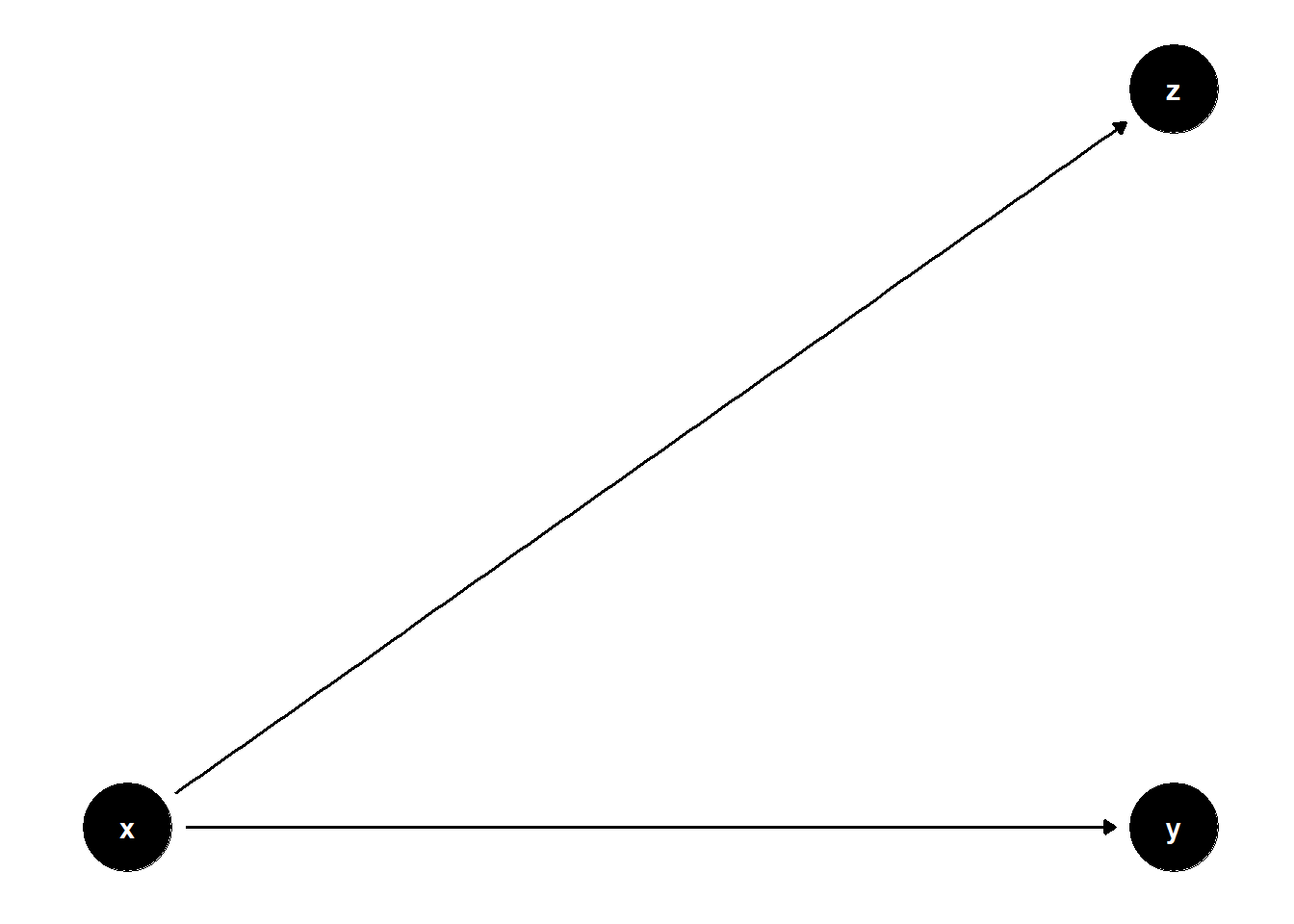
n <- 1e4
x <- rnorm(n)
z <- 2 * x + rnorm(n)
y <- x + rnorm(n)
jtools::export_summs(
lm(y ~ x),
lm(y ~ x + z),
model.names = c("Without Z", "With Post-treatment Z")
)| Without Z | With Post-treatment Z | |
|---|---|---|
| (Intercept) | -0.00 | -0.00 |
| (0.01) | (0.01) | |
| x | 1.00 *** | 0.99 *** |
| (0.01) | (0.02) | |
| z | 0.01 | |
| (0.01) | ||
| N | 10000 | 10000 |
| R2 | 0.50 | 0.50 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
Although \(Z\) lies on a causal path from \(X\) to \(Z\) (and not from \(Z\) to \(Y\)), including \(Z\) adds redundant information and may inflate standard errors. In this case, it’s not a “bad control” in the M-bias sense, but still suboptimal.
39.3.5 Summary Table: Predictive vs. Causal Utility of Controls
| Control Type | Bias Impact | SE Impact | Causal Use? |
|---|---|---|---|
| Predictive of \(Y\), not \(X\) | None | Improves SE | Optional |
| Predictive of \(X\), not \(Y\) | None | Hurts SE | Avoid if possible |
| On causal path (\(X \to Z\)) | None (if not a mediator) | SE or undercontrol | Avoid unless estimating direct effect |
| Post-treatment collider | May induce bias | Hurts SE | Avoid |
| Aids blocking collider paths | May help | Mixed | Conditional |