44.4 Demonstration
###############################################################
# HPC Workflow Demo in R: Scaling Resources and Parallelism
###############################################################
# -- 1. Libraries --
library(parallel) # base parallel
library(doParallel) # foreach backend
library(foreach) # parallel loops
library(future) # futures
library(future.apply) # apply with futures
library(ggplot2) # plotting results
# Optional: install.packages("pryr") for memory usage (optional)
# library(pryr)
###############################################################
# -- 2. Generate or Load Data (Small and Large for Scaling) --
###############################################################
set.seed(42)
n_small <- 1e4 # Small dataset (test)
n_large <- 1e6 # Large dataset (scale-up demo)
generate_data <- function(n) {
data.frame(
id = 1:n,
x = rnorm(n, mean = 50, sd = 10),
y = rnorm(n, mean = 100, sd = 25)
)
}
data_small <- generate_data(n_small)
data_large <- generate_data(n_large)
###############################################################
# -- 3. Preprocessing Function --
###############################################################
clean_and_transform <- function(df) {
df$dist <- sqrt(df$x ^ 2 + df$y ^ 2)
return(df)
}
data_small_clean <- clean_and_transform(data_small)
data_large_clean <- clean_and_transform(data_large)
###############################################################
# -- 4. Heavy Computation (Simulated Workload) --
###############################################################
heavy_computation <- function(df, reps = 500) {
replicate(reps, sum(df$dist))
}
###############################################################
# -- 5. Resource Measurement Helper Function --
###############################################################
measure_resources <-
function(expr, approach_name, data_size, cores) {
start_time <- Sys.time()
# baseline_mem <- pryr::mem_used() # If pryr is available
result <- eval(expr)
end_time <- Sys.time()
time_elapsed <-
as.numeric(difftime(end_time, start_time, units = "secs"))
# used_mem <- as.numeric(pryr::mem_used() - baseline_mem) / 1e6 # MB
# If pryr isn't available, approximate:
used_mem <- as.numeric(object.size(result)) / 1e6 # MB
return(
data.frame(
Approach = approach_name,
Data_Size = data_size,
Cores = cores,
Time_Sec = time_elapsed,
Memory_MB = round(used_mem, 2)
)
)
}
###############################################################
# -- 6. Define and Run Parallel Approaches --
###############################################################
# Collect results in a dataframe
results <- data.frame()
### 6.1 Single Core (Baseline)
res_single_small <- measure_resources(
expr = quote(heavy_computation(data_small_clean)),
approach_name = "SingleCore",
data_size = nrow(data_small_clean),
cores = 1
)
res_single_large <- measure_resources(
expr = quote(heavy_computation(data_large_clean)),
approach_name = "SingleCore",
data_size = nrow(data_large_clean),
cores = 1
)
results <- rbind(results, res_single_small, res_single_large)
# clusterExport(cl, varlist = c("heavy_computation"))
### 6.2 Base Parallel (Cross-Platform): parLapply (Cluster Based)
cores_to_test <- c(2, 4)
for (cores in cores_to_test) {
cl <- makeCluster(cores)
# Export both the heavy function and the datasets
clusterExport(cl,
varlist = c(
"heavy_computation",
"data_small_clean",
"data_large_clean"
))
res_par_small <- measure_resources(
expr = quote({
parLapply(
cl = cl,
X = 1:cores,
fun = function(i)
heavy_computation(data_small_clean)
)
}),
approach_name = "parLapply",
data_size = nrow(data_small_clean),
cores = cores
)
res_par_large <- measure_resources(
expr = quote({
parLapply(
cl = cl,
X = 1:cores,
fun = function(i)
heavy_computation(data_large_clean)
)
}),
approach_name = "parLapply",
data_size = nrow(data_large_clean),
cores = cores
)
stopCluster(cl)
results <- rbind(results, res_par_small, res_par_large)
}
### 6.3 foreach + doParallel
for (cores in cores_to_test) {
cl <- makeCluster(cores)
registerDoParallel(cl)
clusterExport(cl,
varlist = c(
"heavy_computation",
"data_small_clean",
"data_large_clean"
))
registerDoParallel(cl)
# Small dataset
res_foreach_small <- measure_resources(
expr = quote({
foreach(i = 1:cores, .combine = c) %dopar% {
heavy_computation(data_small_clean)
}
}),
approach_name = "foreach_doParallel",
data_size = nrow(data_small_clean),
cores = cores
)
# Large dataset
res_foreach_large <- measure_resources(
expr = quote({
foreach(i = 1:cores, .combine = c) %dopar% {
heavy_computation(data_large_clean)
}
}),
approach_name = "foreach_doParallel",
data_size = nrow(data_large_clean),
cores = cores
)
stopCluster(cl)
results <- rbind(results, res_foreach_small, res_foreach_large)
}
### 6.4 future + future.apply
for (cores in cores_to_test) {
plan(multicore, workers = cores) # multicore only works on Unix; use multisession on Windows
# Small dataset
res_future_small <- measure_resources(
expr = quote({
future_lapply(1:cores, function(i)
heavy_computation(data_small_clean))
}),
approach_name = "future_lapply",
data_size = nrow(data_small_clean),
cores = cores
)
# Large dataset
res_future_large <- measure_resources(
expr = quote({
future_lapply(1:cores, function(i)
heavy_computation(data_large_clean))
}),
approach_name = "future_lapply",
data_size = nrow(data_large_clean),
cores = cores
)
results <- rbind(results, res_future_small, res_future_large)
}
# Reset plan to sequential
plan(sequential)
###############################################################
# -- 7. Summarize and Plot Results --
###############################################################
# Print table
print(results)
#> Approach Data_Size Cores Time_Sec Memory_MB
#> 1 SingleCore 10000 1 0.01586890 0.00
#> 2 SingleCore 1000000 1 1.34883380 0.00
#> 3 parLapply 10000 2 0.01907897 0.01
#> 4 parLapply 1000000 2 1.33863306 0.01
#> 5 parLapply 10000 4 0.01780701 0.02
#> 6 parLapply 1000000 4 1.32461405 0.02
#> 7 foreach_doParallel 10000 2 0.05314898 0.01
#> 8 foreach_doParallel 1000000 2 1.33675098 0.01
#> 9 foreach_doParallel 10000 4 0.06010795 0.02
#> 10 foreach_doParallel 1000000 4 1.34139991 0.02
#> 11 future_lapply 10000 2 0.09521294 0.01
#> 12 future_lapply 1000000 2 2.73076987 0.01
#> 13 future_lapply 10000 4 0.10914207 0.02
#> 14 future_lapply 1000000 4 5.50541496 0.02
# Save to CSV
# write.csv(results, "HPC_parallel_results.csv", row.names = FALSE)
# Plot Time vs. Data Size / Cores
ggplot(results, aes(x = as.factor(Cores), y = Time_Sec, fill = Approach)) +
geom_bar(stat = "identity", position = "dodge") +
facet_wrap( ~ Data_Size, scales = "free", labeller = label_both) +
labs(title = "Runtime by Cores, Approach, and Data Size",
x = "Number of Cores",
y = "Time (Seconds)") +
theme_minimal()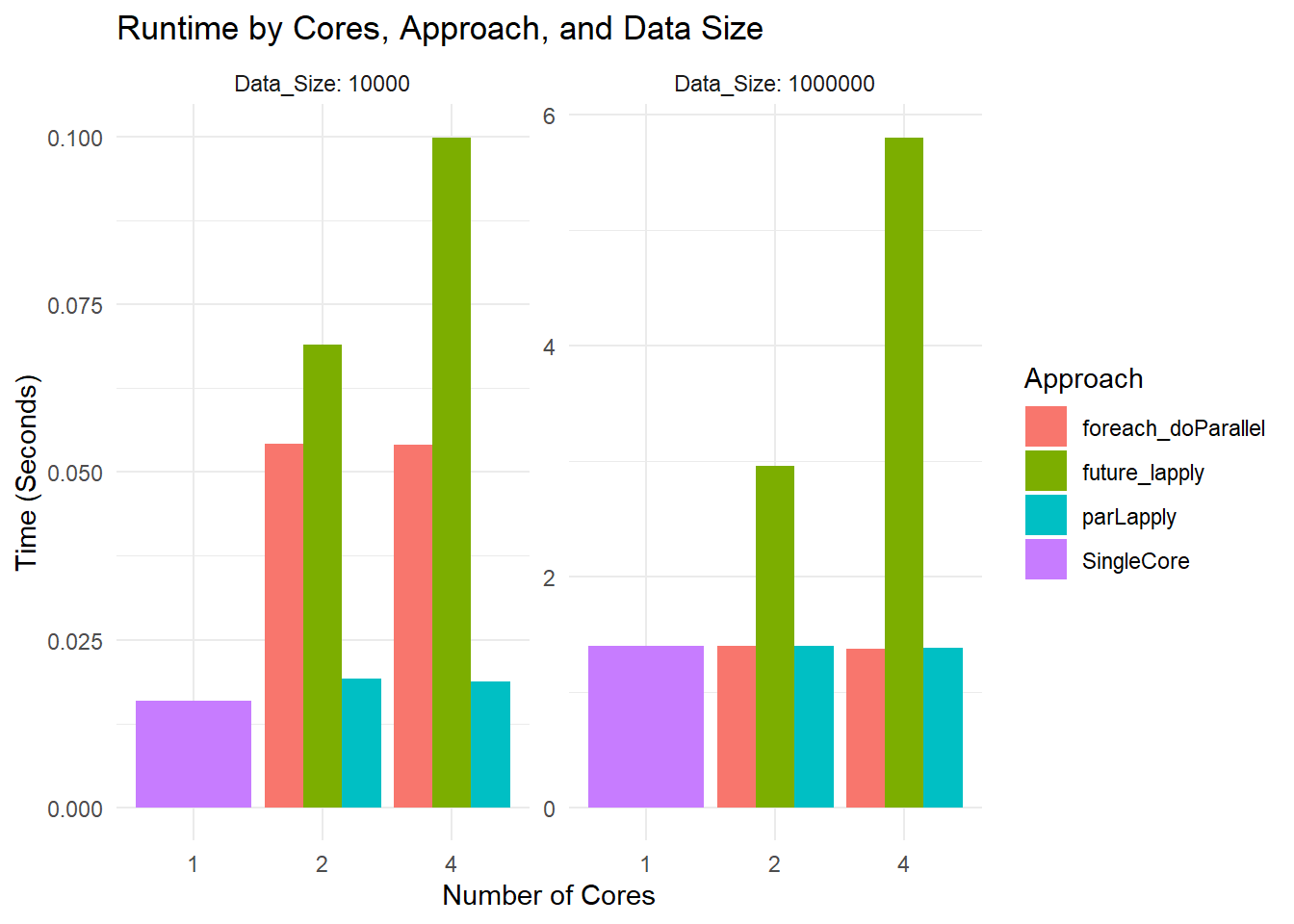
# Plot Memory Usage
ggplot(results, aes(x = as.factor(Cores), y = Memory_MB, fill = Approach)) +
geom_bar(stat = "identity", position = "dodge") +
facet_wrap(~ Data_Size, scales = "free", labeller = label_both) +
labs(title = "Memory Usage by Cores, Approach, and Data Size",
x = "Number of Cores",
y = "Memory (MB)") +
theme_minimal()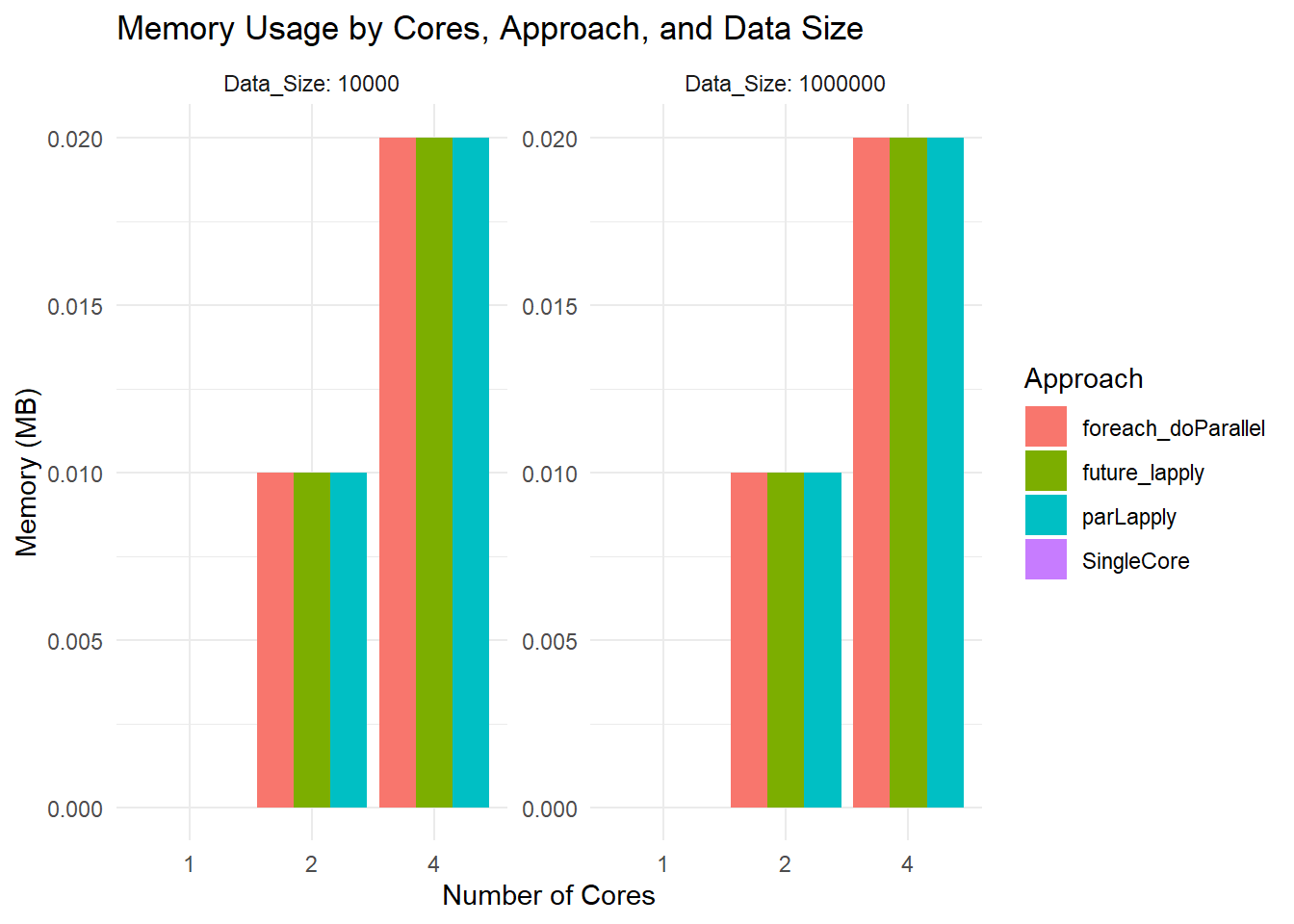
Runtime Plot
Small Dataset: All methods are so fast that you are mostly seeing overhead differences. The differences in bar heights reflect how each parallel framework handles overhead.
Large Dataset: You see more separation. For instance,
parLapplywith 2 cores might be slower than 1 core due to overhead. At 4 cores, it might improve or not, depending on how the overhead and actual workload balance out.
Memory Usage Plot
- All bars hover around 0.02 MB or so because you are measuring the size of the returned result, not the in-memory size of the large dataset. The function
object.size(result)is not capturing total RAM usage by the entire parallel job.
- All bars hover around 0.02 MB or so because you are measuring the size of the returned result, not the in-memory size of the large dataset. The function