39.3 Neutral Controls
39.3.1 Good Predictive Controls
Good for precision
# cleans workspace
rm(list = ls())
# DAG
## specify edges
model <- dagitty("dag{x->y; z->y}")
## coordinates for plotting
coordinates(model) <- list(
x = c(x=1, z=2, y=2),
y = c(x=1, z=2, y=1))
## ggplot
ggdag(model) + theme_dag()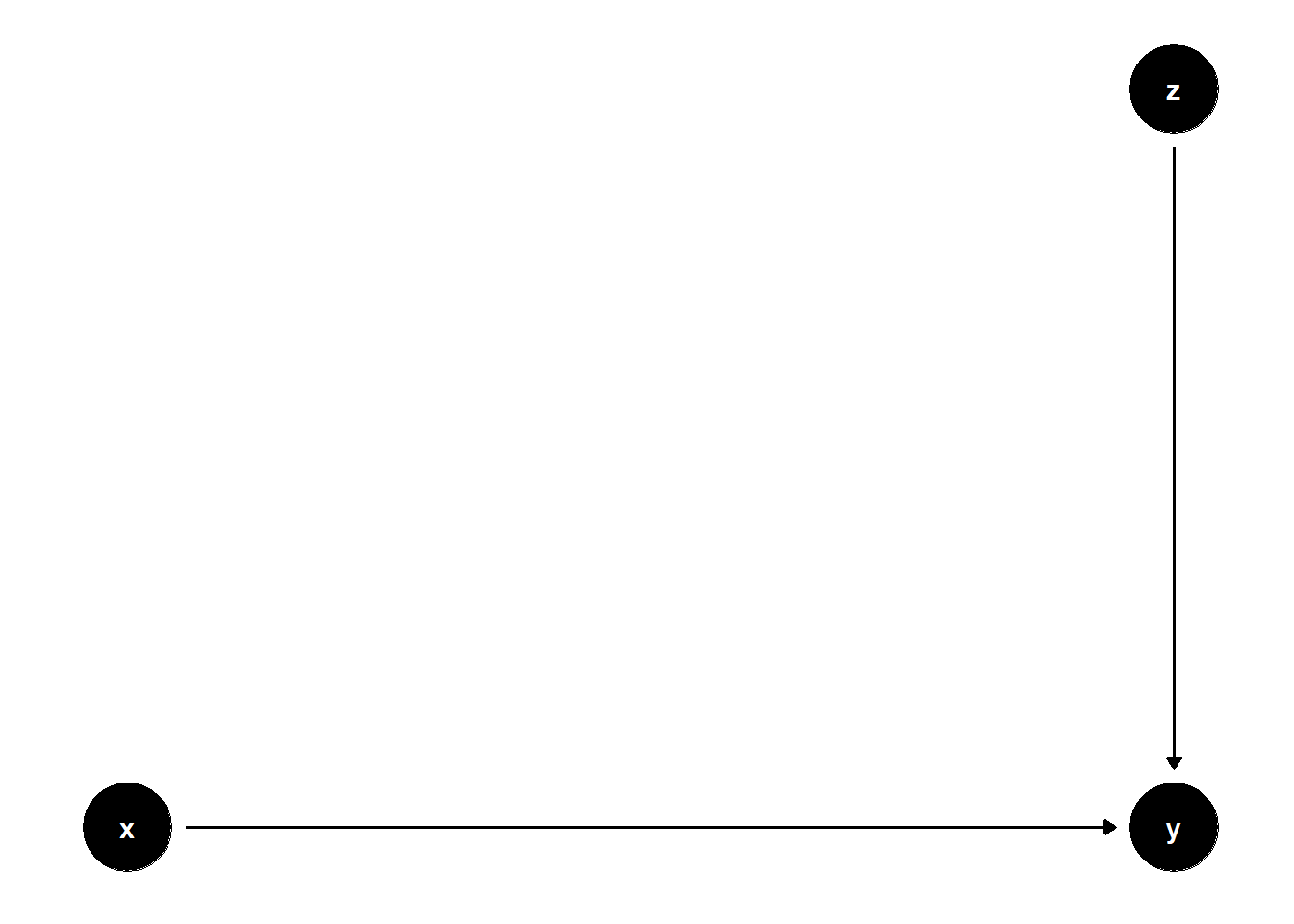
Controlling for \(Z\) does not help or hurt identification, but it can increase precision (i.e., reducing SE)
n <- 1e4
z <- rnorm(n)
x <- rnorm(n)
y <- x + 2 * z + rnorm(n)
jtools::export_summs(lm(y ~ x), lm(y ~ x + z))| Model 1 | Model 2 | |
|---|---|---|
| (Intercept) | 0.01 | 0.01 |
| (0.02) | (0.01) | |
| x | 1.00 *** | 1.01 *** |
| (0.02) | (0.01) | |
| z | 2.00 *** | |
| (0.01) | ||
| N | 10000 | 10000 |
| R2 | 0.17 | 0.83 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
Similar coefficients, but smaller SE when controlling for \(Z\)
Another variation is
# cleans workspace
rm(list = ls())
# DAG
## specify edges
model <- dagitty("dag{x->y; x->m; z->m; m->y}")
## coordinates for plotting
coordinates(model) <- list(
x = c(x=1, z=2, m=2, y=3),
y = c(x=1, z=2, m=1, y=1))
## ggplot
ggdag(model) + theme_dag()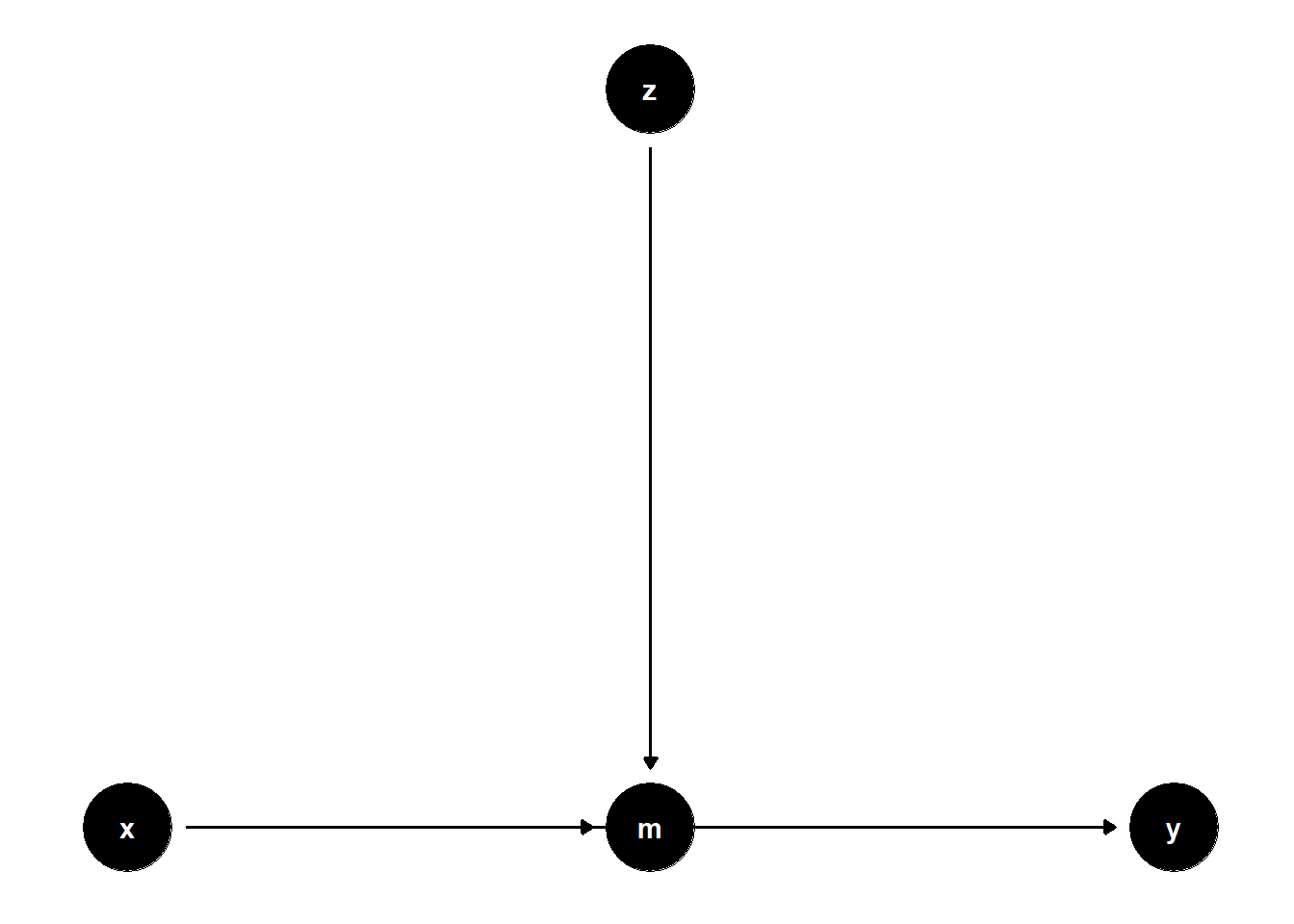
n <- 1e4
z <- rnorm(n)
x <- rnorm(n)
m <- 2 * z + rnorm(n)
y <- x + 2 * m + rnorm(n)
jtools::export_summs(lm(y ~ x), lm(y ~ x + z))| Model 1 | Model 2 | |
|---|---|---|
| (Intercept) | -0.00 | -0.00 |
| (0.05) | (0.02) | |
| x | 0.97 *** | 0.99 *** |
| (0.05) | (0.02) | |
| z | 4.02 *** | |
| (0.02) | ||
| N | 10000 | 10000 |
| R2 | 0.04 | 0.77 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
Controlling for \(Z\) can reduce SE
39.3.2 Good Selection Bias
# cleans workspace
rm(list = ls())
# DAG
## specify edges
model <- dagitty("dag{x->y; x->z; z->w; u->w;u->y}")
# set u as latent
latents(model) <- "u"
## coordinates for plotting
coordinates(model) <- list(
x = c(x=1, z=2, w=3, u=3, y=5),
y = c(x=3, z=2, w=1, u=4, y=3))
## ggplot
ggdag(model) + theme_dag()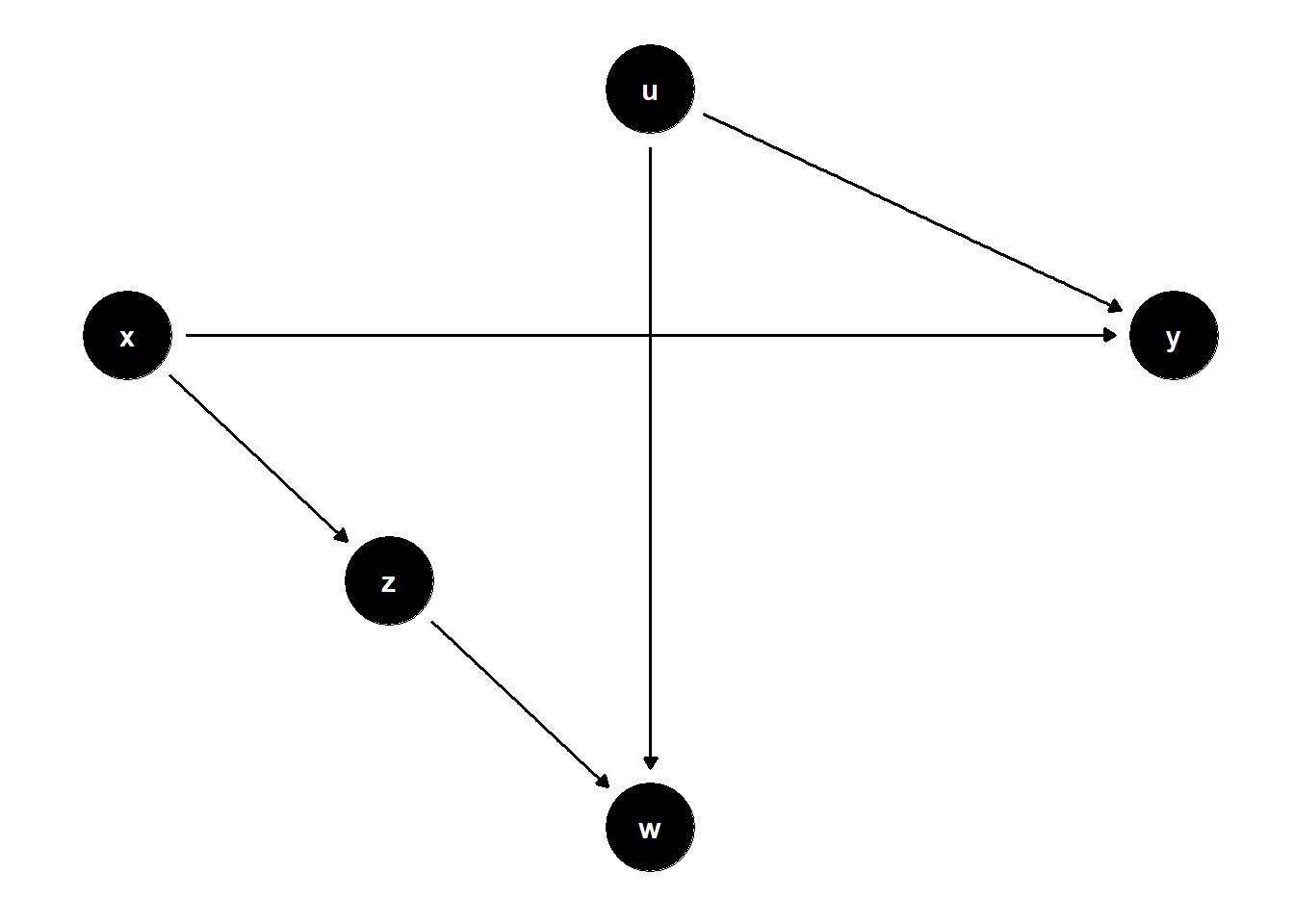
- Unadjusted estimate is unbiased
- Controlling for Z can increase SE
- Controlling for Z while having on W can help identify X
n <- 1e4
x <- rnorm(n)
u <- rnorm(n)
z <- x + rnorm(n)
w <- z + u + rnorm(n)
y <- x - 2*u + rnorm(n)
jtools::export_summs(lm(y ~ x), lm(y ~ x + w), lm(y ~ x + z + w))| Model 1 | Model 2 | Model 3 | |
|---|---|---|---|
| (Intercept) | 0.01 | 0.01 | 0.03 |
| (0.02) | (0.02) | (0.02) | |
| x | 0.99 *** | 1.65 *** | 0.99 *** |
| (0.02) | (0.02) | (0.02) | |
| w | -0.67 *** | -1.01 *** | |
| (0.01) | (0.01) | ||
| z | 1.02 *** | ||
| (0.02) | |||
| N | 10000 | 10000 | 10000 |
| R2 | 0.16 | 0.39 | 0.50 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | |||
39.3.3 Bad Predictive Controls
# cleans workspace
rm(list = ls())
# DAG
## specify edges
model <- dagitty("dag{x->y; z->x}")
## coordinates for plotting
coordinates(model) <- list(
x = c(x=1, z=1, y=2),
y = c(x=1, z=2, y=1))
## ggplot
ggdag(model) + theme_dag()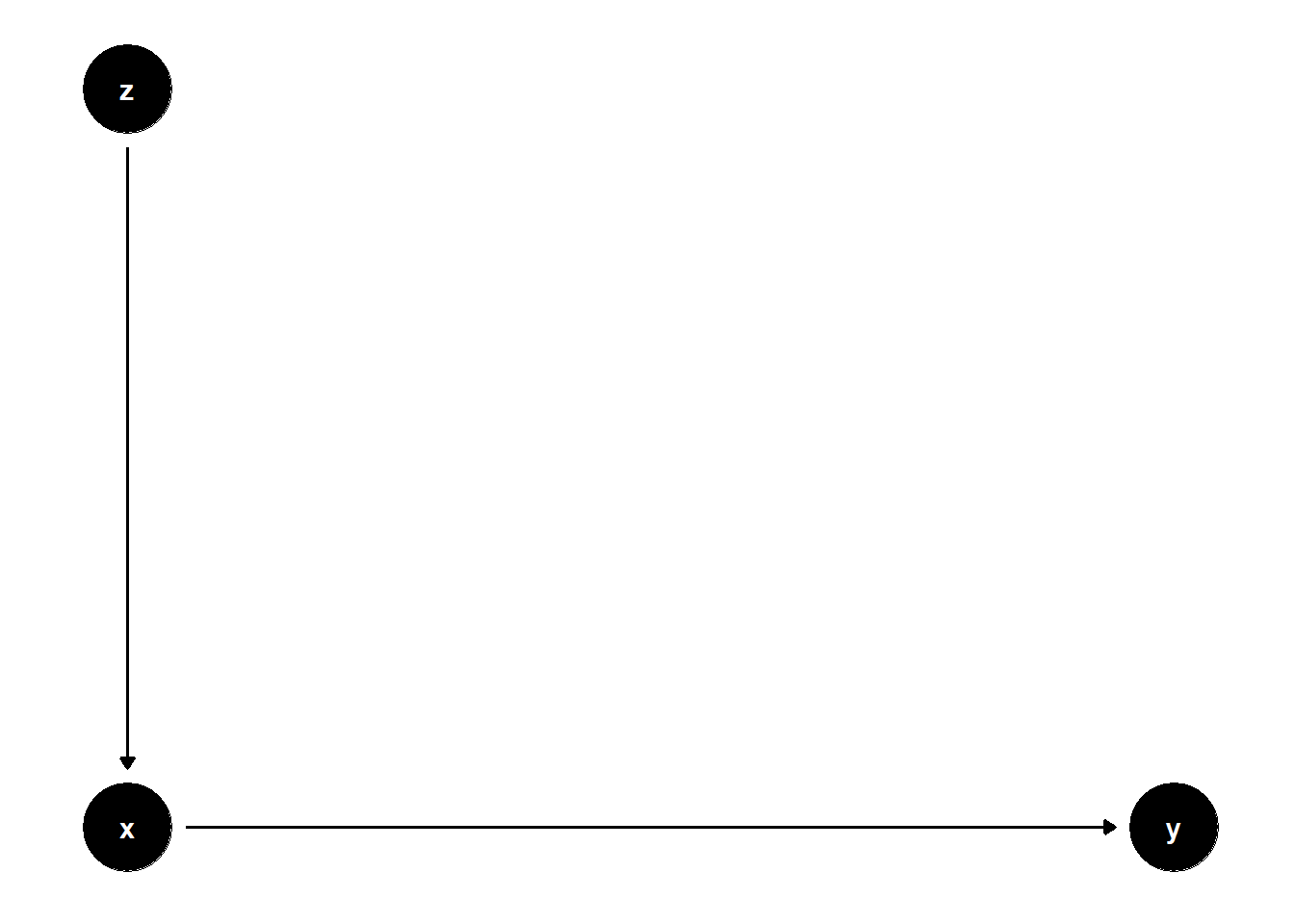
n <- 1e4
z <- rnorm(n)
x <- 2 * z + rnorm(n)
y <- x + 2 * rnorm(n)
jtools::export_summs(lm(y ~ x), lm(y ~ x + z))| Model 1 | Model 2 | |
|---|---|---|
| (Intercept) | -0.02 | -0.02 |
| (0.02) | (0.02) | |
| x | 0.99 *** | 1.00 *** |
| (0.01) | (0.02) | |
| z | -0.00 | |
| (0.04) | ||
| N | 10000 | 10000 |
| R2 | 0.55 | 0.55 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
Similar coefficients, but greater SE when controlling for \(Z\)
Another variation is
rm(list = ls())
# DAG
## specify edges
model <- dagitty("dag{x->y; x->z}")
## coordinates for plotting
coordinates(model) <- list(
x = c(x=1, z=1, y=2),
y = c(x=1, z=2, y=1))
## ggplot
ggdag(model) + theme_dag()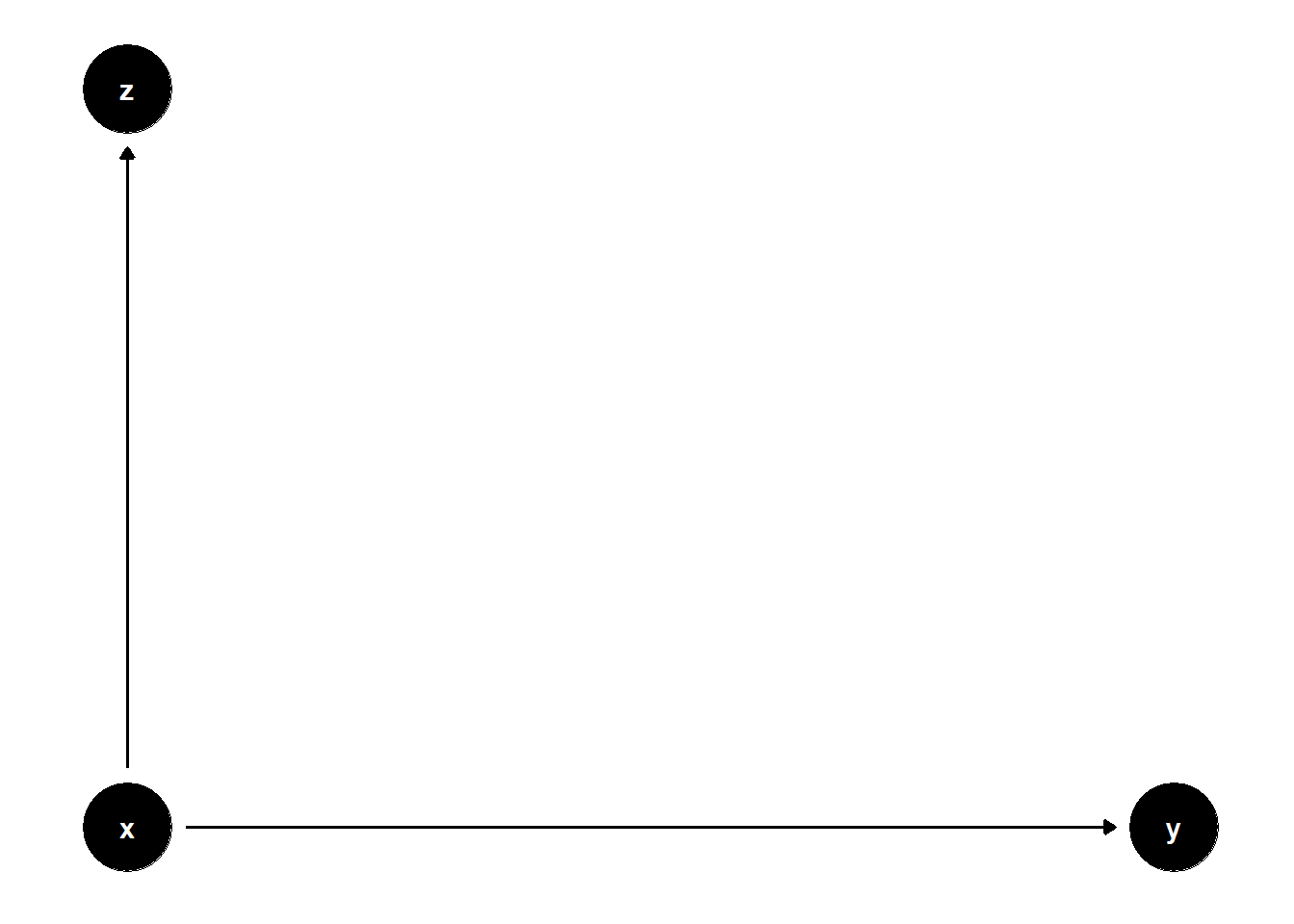
set.seed(1)
n <- 1e4
x <- rnorm(n)
z <- 2 * x + rnorm(n)
y <- x + 2 * rnorm(n)
jtools::export_summs(lm(y ~ x), lm(y ~ x + z))| Model 1 | Model 2 | |
|---|---|---|
| (Intercept) | 0.02 | 0.02 |
| (0.02) | (0.02) | |
| x | 1.00 *** | 0.99 *** |
| (0.02) | (0.05) | |
| z | 0.00 | |
| (0.02) | ||
| N | 10000 | 10000 |
| R2 | 0.20 | 0.20 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
Worse SE when controlling for \(Z\) (\(0.02 < 0.05\))
39.3.4 Bad Selection Bias
# cleans workspace
rm(list = ls())
# DAG
## specify edges
model <- dagitty("dag{x->y; x->z}")
## coordinates for plotting
coordinates(model) <- list(
x = c(x=1, z=2, y=2),
y = c(x=1, z=2, y=1))
## ggplot
ggdag(model) + theme_dag()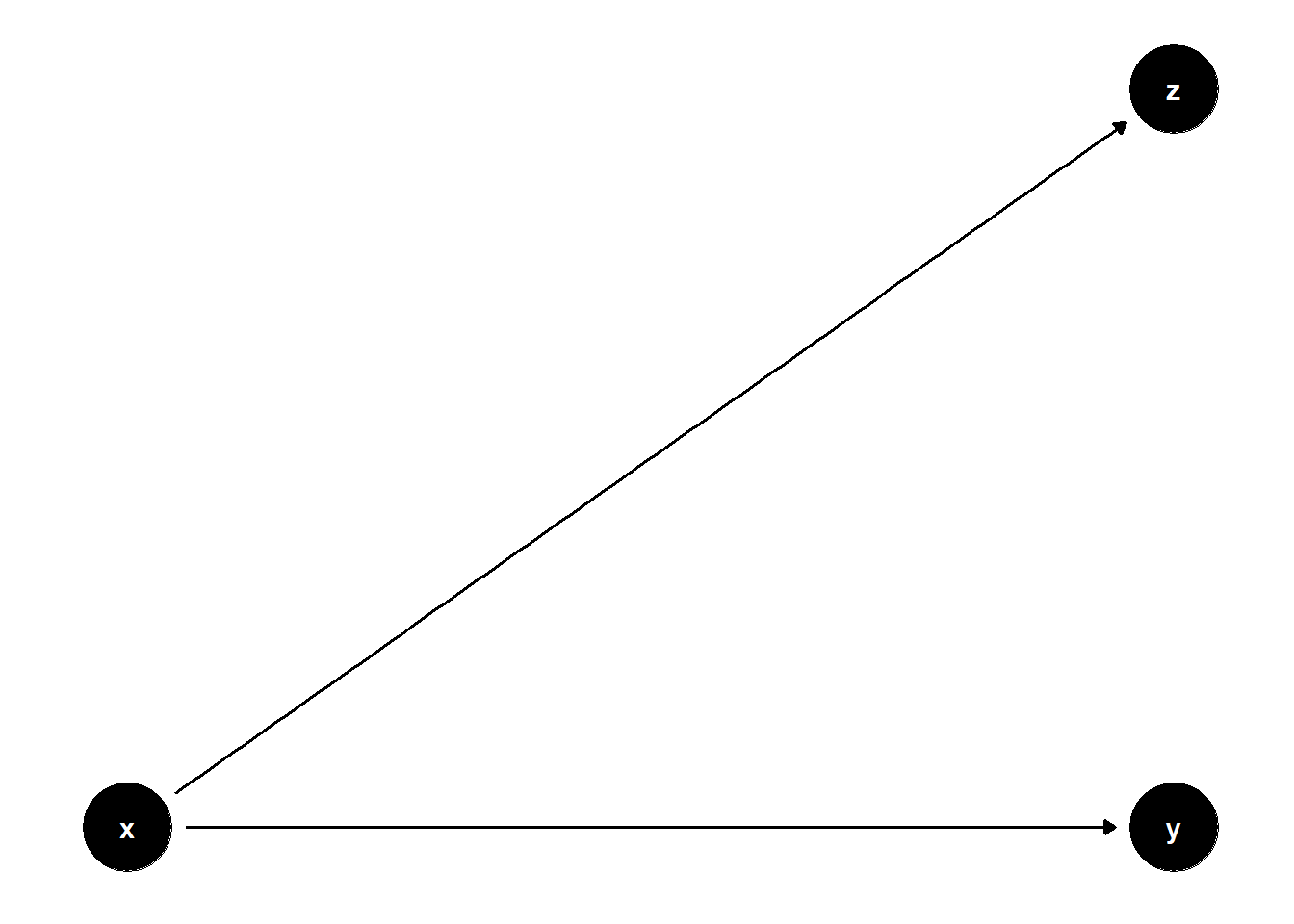
Not all post-treatment variables are bad.
Controlling for \(Z\) is neutral, but it might hurt the precision of the causal effect.