7.3 Binomial Regression
Binomial
Here, cancer case = successes, and control case = failures.
data("esoph")
head(esoph, n = 3)
#> agegp alcgp tobgp ncases ncontrols
#> 1 25-34 0-39g/day 0-9g/day 0 40
#> 2 25-34 0-39g/day 10-19 0 10
#> 3 25-34 0-39g/day 20-29 0 6
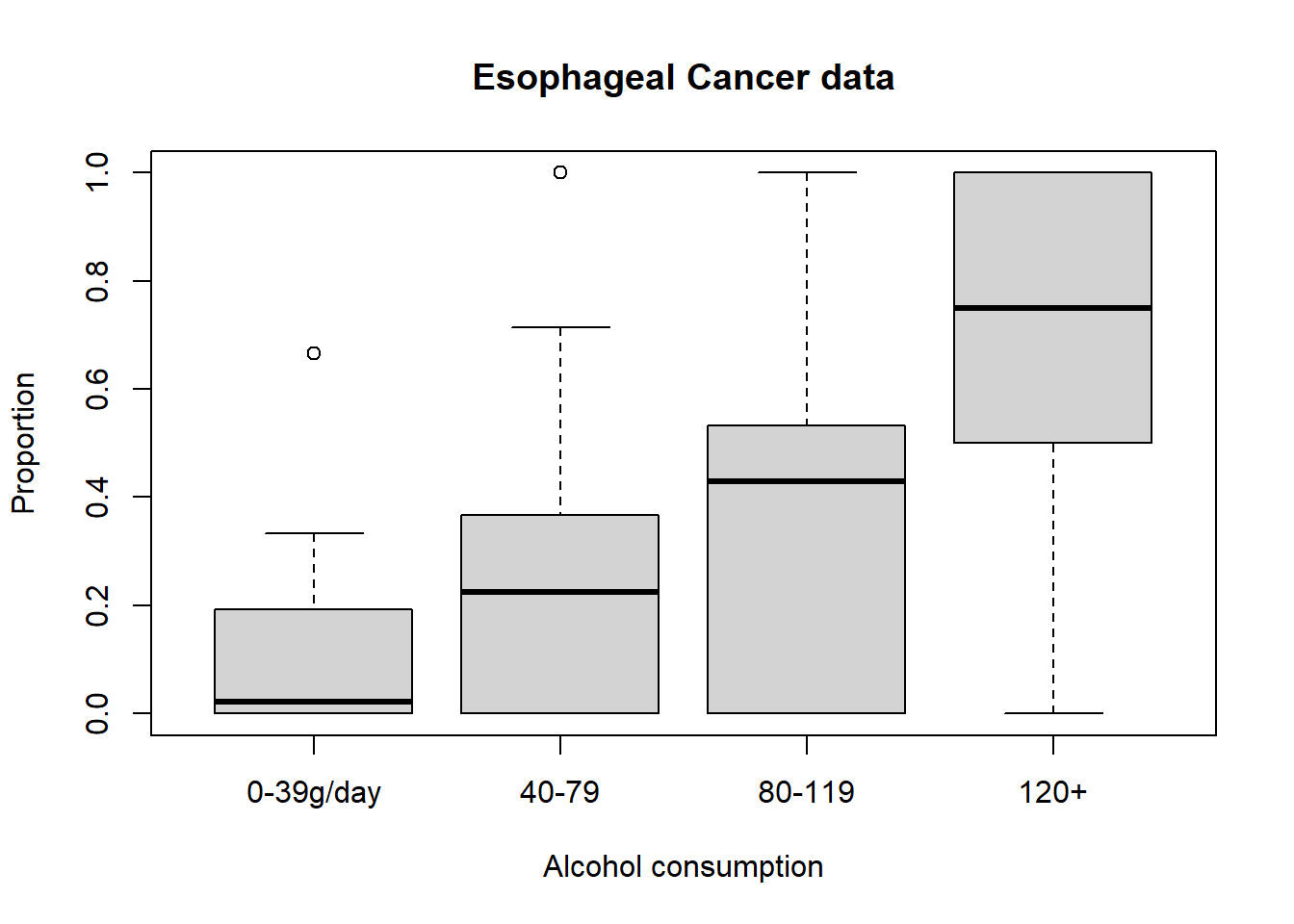
plot(
esoph$ncases / (esoph$ncases + esoph$ncontrols) ~ esoph$alcgp,
ylab = "Proportion",
xlab = 'Alcohol consumption',
main = 'Esophageal Cancer data'
)
# only the alcohol consumption as a predictor
model <- glm(cbind(ncases, ncontrols) ~ alcgp, data = esoph, family = binomial)
summary(model)
#>
#> Call:
#> glm(formula = cbind(ncases, ncontrols) ~ alcgp, family = binomial,
#> data = esoph)
#>
#> Deviance Residuals:
#> Min 1Q Median 3Q Max
#> -4.0759 -1.2037 -0.0183 1.0928 3.7336
#>
#> Coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -2.5885 0.1925 -13.444 < 2e-16 ***
#> alcgp40-79 1.2712 0.2323 5.472 4.46e-08 ***
#> alcgp80-119 2.0545 0.2611 7.868 3.59e-15 ***
#> alcgp120+ 3.3042 0.3237 10.209 < 2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for binomial family taken to be 1)
#>
#> Null deviance: 367.95 on 87 degrees of freedom
#> Residual deviance: 221.46 on 84 degrees of freedom
#> AIC: 344.51
#>
#> Number of Fisher Scoring iterations: 5#Coefficient Odds
coefficients(model) %>% exp
#> (Intercept) alcgp40-79 alcgp80-119 alcgp120+
#> 0.07512953 3.56527094 7.80261593 27.22570533
deviance(model)/df.residual(model)
#> [1] 2.63638
model$aic
#> [1] 344.5109# alcohol consumption and age as predictors
better_model <-
glm(cbind(ncases, ncontrols) ~ agegp + alcgp,
data = esoph,
family = binomial)
summary(better_model)
#>
#> Call:
#> glm(formula = cbind(ncases, ncontrols) ~ agegp + alcgp, family = binomial,
#> data = esoph)
#>
#> Deviance Residuals:
#> Min 1Q Median 3Q Max
#> -2.2395 -0.7186 -0.2324 0.7930 3.3538
#>
#> Coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -6.1472 1.0419 -5.900 3.63e-09 ***
#> agegp35-44 1.6311 1.0800 1.510 0.130973
#> agegp45-54 3.4258 1.0389 3.297 0.000976 ***
#> agegp55-64 3.9435 1.0346 3.811 0.000138 ***
#> agegp65-74 4.3568 1.0413 4.184 2.87e-05 ***
#> agegp75+ 4.4242 1.0914 4.054 5.04e-05 ***
#> alcgp40-79 1.4343 0.2448 5.859 4.64e-09 ***
#> alcgp80-119 2.0071 0.2776 7.230 4.84e-13 ***
#> alcgp120+ 3.6800 0.3763 9.778 < 2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for binomial family taken to be 1)
#>
#> Null deviance: 367.95 on 87 degrees of freedom
#> Residual deviance: 105.88 on 79 degrees of freedom
#> AIC: 238.94
#>
#> Number of Fisher Scoring iterations: 6better_model$aic #smaller AIC is better
#> [1] 238.9361
coefficients(better_model) %>% exp
#> (Intercept) agegp35-44 agegp45-54 agegp55-64 agegp65-74 agegp75+
#> 0.002139482 5.109601844 30.748594216 51.596634690 78.005283850 83.448437749
#> alcgp40-79 alcgp80-119 alcgp120+
#> 4.196747169 7.441782227 39.646885126
pchisq(
q = model$deviance - better_model$deviance,
df = model$df.residual - better_model$df.residual,
lower = FALSE
)
#> [1] 2.713923e-23# specify link function as probit
Prob_better_model <- glm(
cbind(ncases, ncontrols) ~ agegp + alcgp,
data = esoph,
family = binomial(link = probit)
)
summary(Prob_better_model)
#>
#> Call:
#> glm(formula = cbind(ncases, ncontrols) ~ agegp + alcgp, family = binomial(link = probit),
#> data = esoph)
#>
#> Deviance Residuals:
#> Min 1Q Median 3Q Max
#> -2.1325 -0.6877 -0.1661 0.7654 3.3258
#>
#> Coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -3.3741 0.4922 -6.855 7.13e-12 ***
#> agegp35-44 0.8562 0.5081 1.685 0.092003 .
#> agegp45-54 1.7829 0.4904 3.636 0.000277 ***
#> agegp55-64 2.1034 0.4876 4.314 1.61e-05 ***
#> agegp65-74 2.3374 0.4930 4.741 2.13e-06 ***
#> agegp75+ 2.3694 0.5275 4.491 7.08e-06 ***
#> alcgp40-79 0.8080 0.1330 6.076 1.23e-09 ***
#> alcgp80-119 1.1399 0.1558 7.318 2.52e-13 ***
#> alcgp120+ 2.1204 0.2060 10.295 < 2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for binomial family taken to be 1)
#>
#> Null deviance: 367.95 on 87 degrees of freedom
#> Residual deviance: 104.48 on 79 degrees of freedom
#> AIC: 237.53
#>
#> Number of Fisher Scoring iterations: 6