22 Multivariate Methods
\(y_1,...,y_p\) are possibly correlated random variables with means \(\mu_1,...,\mu_p\)
\[ \mathbf{y} = \left( \begin{array} {c} y_1 \\ . \\ y_p \\ \end{array} \right) \]
\[ E(\mathbf{y}) = \left( \begin{array} {c} \mu_1 \\ . \\ \mu_p \\ \end{array} \right) \]
Let \(\sigma_{ij} = cov(y_i, y_j)\) for \(i,j = 1,…,p\)
\[ \mathbf{\Sigma} = (\sigma_{ij}) = \left( \begin{array} {cccc} \sigma_{11} & \sigma_{22} & ... & \sigma_{1p} \\ \sigma_{21} & \sigma_{22} & ... & \sigma_{2p} \\ . & . & . & . \\ \sigma_{p1} & \sigma_{p2} & ... & \sigma_{pp} \end{array} \right) \]
where \(\mathbf{\Sigma}\) (symmetric) is the variance-covariance or dispersion matrix
Let \(\mathbf{u}_{p \times 1}\) and \(\mathbf{v}_{q \times 1}\) be random vectors with means \(\mu_u\) and \(\mu_v\) . Then
\[ \mathbf{\Sigma}_{uv} = cov(\mathbf{u,v}) = E[(\mathbf{u} - \mu_u)(\mathbf{v} - \mu_v)'] \]
in which \(\mathbf{\Sigma}_{uv} \neq \mathbf{\Sigma}_{vu}\) and \(\mathbf{\Sigma}_{uv} = \mathbf{\Sigma}_{vu}'\)
Properties of Covariance Matrices
- Symmetric \(\mathbf{\Sigma}' = \mathbf{\Sigma}\)
- Non-negative definite \(\mathbf{a'\Sigma a} \ge 0\) for any \(\mathbf{a} \in R^p\), which is equivalent to eigenvalues of \(\mathbf{\Sigma}\), \(\lambda_1 \ge \lambda_2 \ge ... \ge \lambda_p \ge 0\)
- \(|\mathbf{\Sigma}| = \lambda_1 \lambda_2 ... \lambda_p \ge 0\) (generalized variance) (the bigger this number is, the more variation there is
- \(trace(\mathbf{\Sigma}) = tr(\mathbf{\Sigma}) = \lambda_1 + ... + \lambda_p = \sigma_{11} + ... + \sigma_{pp} =\) sum of variance (total variance)
Note:
- \(\mathbf{\Sigma}\) is typically required to be positive definite, which means all eigenvalues are positive, and \(\mathbf{\Sigma}\) has an inverse \(\mathbf{\Sigma}^{-1}\) such that \(\mathbf{\Sigma}^{-1}\mathbf{\Sigma} = \mathbf{I}_{p \times p} = \mathbf{\Sigma \Sigma}^{-1}\)
Correlation Matrices
\[ \rho_{ij} = \frac{\sigma_{ij}}{\sqrt{\sigma_{ii} \sigma_{jj}}} \]
\[ \mathbf{R} = \left( \begin{array} {cccc} \rho_{11} & \rho_{12} & ... & \rho_{1p} \\ \rho_{21} & \rho_{22} & ... & \rho_{2p} \\ . & . & . &. \\ \rho_{p1} & \rho_{p2} & ... & \rho_{pp} \\ \end{array} \right) \]
where \(\rho_{ij}\) is the correlation, and \(\rho_{ii} = 1\) for all i
Alternatively,
\[ \mathbf{R} = [diag(\mathbf{\Sigma})]^{-1/2}\mathbf{\Sigma}[diag(\mathbf{\Sigma})]^{-1/2} \]
where \(diag(\mathbf{\Sigma})\) is the matrix which has the \(\sigma_{ii}\)’s on the diagonal and 0’s elsewhere
and \(\mathbf{A}^{1/2}\) (the square root of a symmetric matrix) is a symmetric matrix such as \(\mathbf{A} = \mathbf{A}^{1/2}\mathbf{A}^{1/2}\)
Equalities
Let
\(\mathbf{x}\) and \(\mathbf{y}\) be random vectors with means \(\mu_x\) and \(\mu_y\) and variance -variance matrices \(\mathbf{\Sigma}_x\) and \(\mathbf{\Sigma}_y\).
\(\mathbf{A}\) and \(\mathbf{B}\) be matrices of constants and \(\mathbf{c}\) and \(\mathbf{d}\) be vectors of constants
Then
\(E(\mathbf{Ay + c} ) = \mathbf{A} \mu_y + c\)
\(var(\mathbf{Ay + c}) = \mathbf{A} var(\mathbf{y})\mathbf{A}' = \mathbf{A \Sigma_y A}'\)
\(cov(\mathbf{Ay + c, By+ d}) = \mathbf{A\Sigma_y B}'\)
\(E(\mathbf{Ay + Bx + c}) = \mathbf{A \mu_y + B \mu_x + c}\)
\(var(\mathbf{Ay + Bx + c}) = \mathbf{A \Sigma_y A' + B \Sigma_x B' + A \Sigma_{yx}B' + B\Sigma'_{yx}A'}\)
Multivariate Normal Distribution
Let \(\mathbf{y}\) be a multivariate normal (MVN) random variable with mean \(\mu\) and variance \(\mathbf{\Sigma}\). Then the density of \(\mathbf{y}\) is
\[ f(\mathbf{y}) = \frac{1}{(2\pi)^{p/2}|\mathbf{\Sigma}|^{1/2}} \exp(-\frac{1}{2} \mathbf{(y-\mu)'\Sigma^{-1}(y-\mu)} ) \]
\(\mathbf{y} \sim N_p(\mu, \mathbf{\Sigma})\)
22.0.1 Properties of MVN
Let \(\mathbf{A}_{r \times p}\) be a fixed matrix. Then \(\mathbf{Ay} \sim N_r (\mathbf{A \mu, A \Sigma A'})\) . \(r \le p\) and all rows of \(\mathbf{A}\) must be linearly independent to guarantee that \(\mathbf{A \Sigma A}'\) is non-singular.
Let \(\mathbf{G}\) be a matrix such that \(\mathbf{\Sigma}^{-1} = \mathbf{GG}'\). Then \(\mathbf{G'y} \sim N_p(\mathbf{G' \mu, I})\) and \(\mathbf{G'(y-\mu)} \sim N_p (0,\mathbf{I})\)
Any fixed linear combination of \(y_1,...,y_p\) (say \(\mathbf{c'y}\)) follows \(\mathbf{c'y} \sim N_1 (\mathbf{c' \mu, c' \Sigma c})\)
-
Define a partition, \([\mathbf{y}'_1,\mathbf{y}_2']'\) where
\(\mathbf{y}_1\) is \(p_1 \times 1\)
\(\mathbf{y}_2\) is \(p_2 \times 1\),
\(p_1 + p_2 = p\)
\(p_1,p_2 \ge 1\) Then
\[ \left( \begin{array} {c} \mathbf{y}_1 \\ \mathbf{y}_2 \\ \end{array} \right) \sim N \left( \left( \begin{array} {c} \mu_1 \\ \mu_2 \\ \end{array} \right), \left( \begin{array} {cc} \mathbf{\Sigma}_{11} & \mathbf{\Sigma}_{12} \\ \mathbf{\Sigma}_{21} & \mathbf{\Sigma}_{22}\\ \end{array} \right) \right) \]
The marginal distributions of \(\mathbf{y}_1\) and \(\mathbf{y}_2\) are \(\mathbf{y}_1 \sim N_{p1}(\mathbf{\mu_1, \Sigma_{11}})\) and \(\mathbf{y}_2 \sim N_{p2}(\mathbf{\mu_2, \Sigma_{22}})\)
Individual components \(y_1,...,y_p\) are all normally distributed \(y_i \sim N_1(\mu_i, \sigma_{ii})\)
-
The conditional distribution of \(\mathbf{y}_1\) and \(\mathbf{y}_2\) is normal
-
\(\mathbf{y}_1 | \mathbf{y}_2 \sim N_{p1}(\mathbf{\mu_1 + \Sigma_{12} \Sigma_{22}^{-1}(y_2 - \mu_2),\Sigma_{11} - \Sigma_{12} \Sigma_{22}^{-1} \sigma_{21}})\)
- In this formula, we see if we know (have info about) \(\mathbf{y}_2\), we can re-weight \(\mathbf{y}_1\) ’s mean, and the variance is reduced because we know more about \(\mathbf{y}_1\) because we know \(\mathbf{y}_2\)
which is analogous to \(\mathbf{y}_2 | \mathbf{y}_1\). And \(\mathbf{y}_1\) and \(\mathbf{y}_2\) are independently distrusted only if \(\mathbf{\Sigma}_{12} = 0\)
-
If \(\mathbf{y} \sim N(\mathbf{\mu, \Sigma})\) and \(\mathbf{\Sigma}\) is positive definite, then \(\mathbf{(y-\mu)' \Sigma^{-1} (y - \mu)} \sim \chi^2_{(p)}\)
If \(\mathbf{y}_i\) are independent \(N_p (\mathbf{\mu}_i , \mathbf{\Sigma}_i)\) random variables, then for fixed matrices \(\mathbf{A}_{i(m \times p)}\), \(\sum_{i=1}^k \mathbf{A}_i \mathbf{y}_i \sim N_m (\sum_{i=1}^{k} \mathbf{A}_i \mathbf{\mu}_i, \sum_{i=1}^k \mathbf{A}_i \mathbf{\Sigma}_i \mathbf{A}_i)\)
Multiple Regression
\[ \left( \begin{array} {c} Y \\ \mathbf{x} \end{array} \right) \sim N_{p+1} \left( \left[ \begin{array} {c} \mu_y \\ \mathbf{\mu}_x \end{array} \right] , \left[ \begin{array} {cc} \sigma^2_Y & \mathbf{\Sigma}_{yx} \\ \mathbf{\Sigma}_{yx} & \mathbf{\Sigma}_{xx} \end{array} \right] \right) \]
The conditional distribution of Y given x follows a univariate normal distribution with
\[ \begin{aligned} E(Y| \mathbf{x}) &= \mu_y + \mathbf{\Sigma}_{yx} \Sigma_{xx}^{-1} (\mathbf{x}- \mu_x) \\ &= \mu_y - \Sigma_{yx} \Sigma_{xx}^{-1}\mu_x + \Sigma_{yx} \Sigma_{xx}^{-1}\mathbf{x} \\ &= \beta_0 + \mathbf{\beta'x} \end{aligned} \]
where \(\beta = (\beta_1,...,\beta_p)' = \mathbf{\Sigma}_{xx}^{-1} \mathbf{\Sigma}_{yx}'\) (e.g., analogous to \(\mathbf{(x'x)^{-1}x'y}\) but not the same if we consider \(Y_i\) and \(\mathbf{x}_i\), \(i = 1,..,n\) and use the empirical covariance formula: \(var(Y|\mathbf{x}) = \sigma^2_Y - \mathbf{\Sigma_{yx}\Sigma^{-1}_{xx} \Sigma'_{yx}}\))
Samples from Multivariate Normal Populations
A random sample of size n, \(\mathbf{y}_1,.., \mathbf{y}_n\) from \(N_p (\mathbf{\mu}, \mathbf{\Sigma})\). Then
Since \(\mathbf{y}_1,..., \mathbf{y}_n\) are iid, their sample mean, \(\bar{\mathbf{y}} = \sum_{i=1}^n \mathbf{y}_i/n \sim N_p (\mathbf{\mu}, \mathbf{\Sigma}/n)\). that is, \(\bar{\mathbf{y}}\) is an unbiased estimator of \(\mathbf{\mu}\)
-
The \(p \times p\) sample variance-covariance matrix, \(\mathbf{S}\) is \(\mathbf{S} = \frac{1}{n-1}\sum_{i=1}^n (\mathbf{y}_i - \bar{\mathbf{y}})(\mathbf{y}_i - \bar{\mathbf{y}})' = \frac{1}{n-1} (\sum_{i=1}^n \mathbf{y}_i \mathbf{y}_i' - n \bar{\mathbf{y}}\bar{\mathbf{y}}')\)
- where \(\mathbf{S}\) is symmetric, unbiased estimator of \(\mathbf{\Sigma}\) and has \(p(p+1)/2\) random variables.
\((n-1)\mathbf{S} \sim W_p (n-1, \mathbf{\Sigma})\) is a Wishart distribution with n-1 degrees of freedom and expectation \((n-1) \mathbf{\Sigma}\). The Wishart distribution is a multivariate extension of the Chi-squared distribution.
\(\bar{\mathbf{y}}\) and \(\mathbf{S}\) are independent
\(\bar{\mathbf{y}}\) and \(\mathbf{S}\) are sufficient statistics. (All of the info in the data about \(\mathbf{\mu}\) and \(\mathbf{\Sigma}\) is contained in \(\bar{\mathbf{y}}\) and \(\mathbf{S}\) , regardless of sample size).
Large Sample Properties
\(\mathbf{y}_1,..., \mathbf{y}_n\) are a random sample from some population with mean \(\mathbf{\mu}\) and variance-covariance matrix \(\mathbf{\Sigma}\)
\(\bar{\mathbf{y}}\) is a consistent estimator for \(\mu\)
\(\mathbf{S}\) is a consistent estimator for \(\mathbf{\Sigma}\)
Multivariate Central Limit Theorem: Similar to the univariate case, \(\sqrt{n}(\bar{\mathbf{y}} - \mu) \dot{\sim} N_p (\mathbf{0,\Sigma})\) where n is large relative to p (\(n \ge 25p\)), which is equivalent to \(\bar{\mathbf{y}} \dot{\sim} N_p (\mu, \mathbf{\Sigma}/n)\)
Wald’s Theorem: \(n(\bar{\mathbf{y}} - \mu)' \mathbf{S}^{-1} (\bar{\mathbf{y}} - \mu)\) when n is large relative to p.
Maximum Likelihood Estimation for MVN
Suppose iid \(\mathbf{y}_1 ,... \mathbf{y}_n \sim N_p (\mu, \mathbf{\Sigma})\), the likelihood function for the data is
\[ \begin{aligned} L(\mu, \mathbf{\Sigma}) &= \prod_{j=1}^n (\frac{1}{(2\pi)^{p/2}|\mathbf{\Sigma}|^{1/2}} \exp(-\frac{1}{2}(\mathbf{y}_j -\mu)'\mathbf{\Sigma}^{-1})(\mathbf{y}_j -\mu)) \\ &= \frac{1}{(2\pi)^{np/2}|\mathbf{\Sigma}|^{n/2}} \exp(-\frac{1}{2} \sum_{j=1}^n(\mathbf{y}_j -\mu)'\mathbf{\Sigma}^{-1})(\mathbf{y}_j -\mu) \end{aligned} \]
Then, the MLEs are
\[ \hat{\mu} = \bar{\mathbf{y}} \]
\[ \hat{\mathbf{\Sigma}} = \frac{n-1}{n} \mathbf{S} \]
using derivatives of the log of the likelihood function with respect to \(\mu\) and \(\mathbf{\Sigma}\)
Properties of MLEs
Invariance: If \(\hat{\theta}\) is the MLE of \(\theta\), then the MLE of \(h(\theta)\) is \(h(\hat{\theta})\) for any function h(.)
Consistency: MLEs are consistent estimators, but they are usually biased
Efficiency: MLEs are efficient estimators (no other estimator has a smaller variance for large samples)
-
Asymptotic normality: Suppose that \(\hat{\theta}_n\) is the MLE for \(\theta\) based upon n independent observations. Then \(\hat{\theta}_n \dot{\sim} N(\theta, \mathbf{H}^{-1})\)
\(\mathbf{H}\) is the Fisher Information Matrix, which contains the expected values of the second partial derivatives fo the log-likelihood function. the (i,j)th element of \(\mathbf{H}\) is \(-E(\frac{\partial^2 l(\mathbf{\theta})}{\partial \theta_i \partial \theta_j})\)
we can estimate \(\mathbf{H}\) by finding the form determined above, and evaluate it at \(\theta = \hat{\theta}_n\)
-
Likelihood ratio testing: for some null hypothesis, \(H_0\) we can form a likelihood ratio test
The statistic is: \(\Lambda = \frac{\max_{H_0}l(\mathbf{\mu}, \mathbf{\Sigma|Y})}{\max l(\mu, \mathbf{\Sigma | Y})}\)
For large n, \(-2 \log \Lambda \sim \chi^2_{(v)}\) where v is the number of parameters in the unrestricted space minus the number of parameters under \(H_0\)
Test of Multivariate Normality
-
Check univariate normality for each trait (X) separately
Can check \[Normality Assessment\]
The good thing is that if any of the univariate trait is not normal, then the joint distribution is not normal (see again [m]). If a joint multivariate distribution is normal, then the marginal distribution has to be normal.
However, marginal normality of all traits does not imply joint MVN
Easily rule out multivariate normality, but not easy to prove it
-
Mardia’s tests for multivariate normality
Multivariate skewness is\[ \beta_{1,p} = E[(\mathbf{y}- \mathbf{\mu})' \mathbf{\Sigma}^{-1} (\mathbf{x} - \mathbf{\mu})]^3 \]
where \(\mathbf{x}\) and \(\mathbf{y}\) are independent, but have the same distribution (note: \(\beta\) here is not regression coefficient)
Multivariate kurtosis is defined as
\[ \beta_{2,p} - E[(\mathbf{y}- \mathbf{\mu})' \mathbf{\Sigma}^{-1} (\mathbf{x} - \mathbf{\mu})]^2 \]
For the MVN distribution, we have \(\beta_{1,p} = 0\) and \(\beta_{2,p} = p(p+2)\)
-
For a sample of size n, we can estimate
\[ \hat{\beta}_{1,p} = \frac{1}{n^2}\sum_{i=1}^n \sum_{j=1}^n g^2_{ij} \]
\[ \hat{\beta}_{2,p} = \frac{1}{n} \sum_{i=1}^n g^2_{ii} \]
- where \(g_{ij} = (\mathbf{y}_i - \bar{\mathbf{y}})' \mathbf{S}^{-1} (\mathbf{y}_j - \bar{\mathbf{y}})\). Note: \(g_{ii} = d^2_i\) where \(d^2_i\) is the Mahalanobis distance
-
(Mardia 1970) shows for large n
\[ \kappa_1 = \frac{n \hat{\beta}_{1,p}}{6} \dot{\sim} \chi^2_{p(p+1)(p+2)/6} \]
\[ \kappa_2 = \frac{\hat{\beta}_{2,p} - p(p+2)}{\sqrt{8p(p+2)/n}} \sim N(0,1) \]
Hence, we can use \(\kappa_1\) and \(\kappa_2\) to test the null hypothesis of MVN.
When the data are non-normal, normal theory tests on the mean are sensitive to \(\beta_{1,p}\) , while tests on the covariance are sensitive to \(\beta_{2,p}\)
Alternatively, Doornik-Hansen test for multivariate normality (Doornik and Hansen 2008)
-
Chi-square Q-Q plot
Let \(\mathbf{y}_i, i = 1,...,n\) be a random sample sample from \(N_p(\mathbf{\mu}, \mathbf{\Sigma})\)
Then \(\mathbf{z}_i = \mathbf{\Sigma}^{-1/2}(\mathbf{y}_i - \mathbf{\mu}), i = 1,...,n\) are iid \(N_p (\mathbf{0}, \mathbf{I})\). Thus, \(d_i^2 = \mathbf{z}_i' \mathbf{z}_i \sim \chi^2_p , i = 1,...,n\)
plot the ordered \(d_i^2\) values against the qualities of the \(\chi^2_p\) distribution. When normality holds, the plot should approximately resemble a straight lien passing through the origin at a 45 degree
it requires large sample size (i.e., sensitive to sample size). Even if we generate data from a MVN, the tail of the Chi-square Q-Q plot can still be out of line.
-
If the data are not normal, we can
ignore it
use nonparametric methods
use models based upon an approximate distribution (e.g., GLMM)
try performing a transformation
library(heplots)
library(ICSNP)
library(MVN)
library(tidyverse)
trees = read.table("images/trees.dat")
names(trees) <- c("Nitrogen","Phosphorous","Potassium","Ash","Height")
str(trees)
#> 'data.frame': 26 obs. of 5 variables:
#> $ Nitrogen : num 2.2 2.1 1.52 2.88 2.18 1.87 1.52 2.37 2.06 1.84 ...
#> $ Phosphorous: num 0.417 0.354 0.208 0.335 0.314 0.271 0.164 0.302 0.373 0.265 ...
#> $ Potassium : num 1.35 0.9 0.71 0.9 1.26 1.15 0.83 0.89 0.79 0.72 ...
#> $ Ash : num 1.79 1.08 0.47 1.48 1.09 0.99 0.85 0.94 0.8 0.77 ...
#> $ Height : int 351 249 171 373 321 191 225 291 284 213 ...
summary(trees)
#> Nitrogen Phosphorous Potassium Ash
#> Min. :1.130 Min. :0.1570 Min. :0.3800 Min. :0.4500
#> 1st Qu.:1.532 1st Qu.:0.1963 1st Qu.:0.6050 1st Qu.:0.6375
#> Median :1.855 Median :0.2250 Median :0.7150 Median :0.9300
#> Mean :1.896 Mean :0.2506 Mean :0.7619 Mean :0.8873
#> 3rd Qu.:2.160 3rd Qu.:0.2975 3rd Qu.:0.8975 3rd Qu.:0.9825
#> Max. :2.880 Max. :0.4170 Max. :1.3500 Max. :1.7900
#> Height
#> Min. : 65.0
#> 1st Qu.:122.5
#> Median :181.0
#> Mean :196.6
#> 3rd Qu.:276.0
#> Max. :373.0
cor(trees, method = "pearson") # correlation matrix
#> Nitrogen Phosphorous Potassium Ash Height
#> Nitrogen 1.0000000 0.6023902 0.5462456 0.6509771 0.8181641
#> Phosphorous 0.6023902 1.0000000 0.7037469 0.6707871 0.7739656
#> Potassium 0.5462456 0.7037469 1.0000000 0.6710548 0.7915683
#> Ash 0.6509771 0.6707871 0.6710548 1.0000000 0.7676771
#> Height 0.8181641 0.7739656 0.7915683 0.7676771 1.0000000
# qq-plot
gg <- trees %>%
pivot_longer(everything(), names_to = "Var", values_to = "Value") %>%
ggplot(aes(sample = Value)) +
geom_qq() +
geom_qq_line() +
facet_wrap("Var", scales = "free")
gg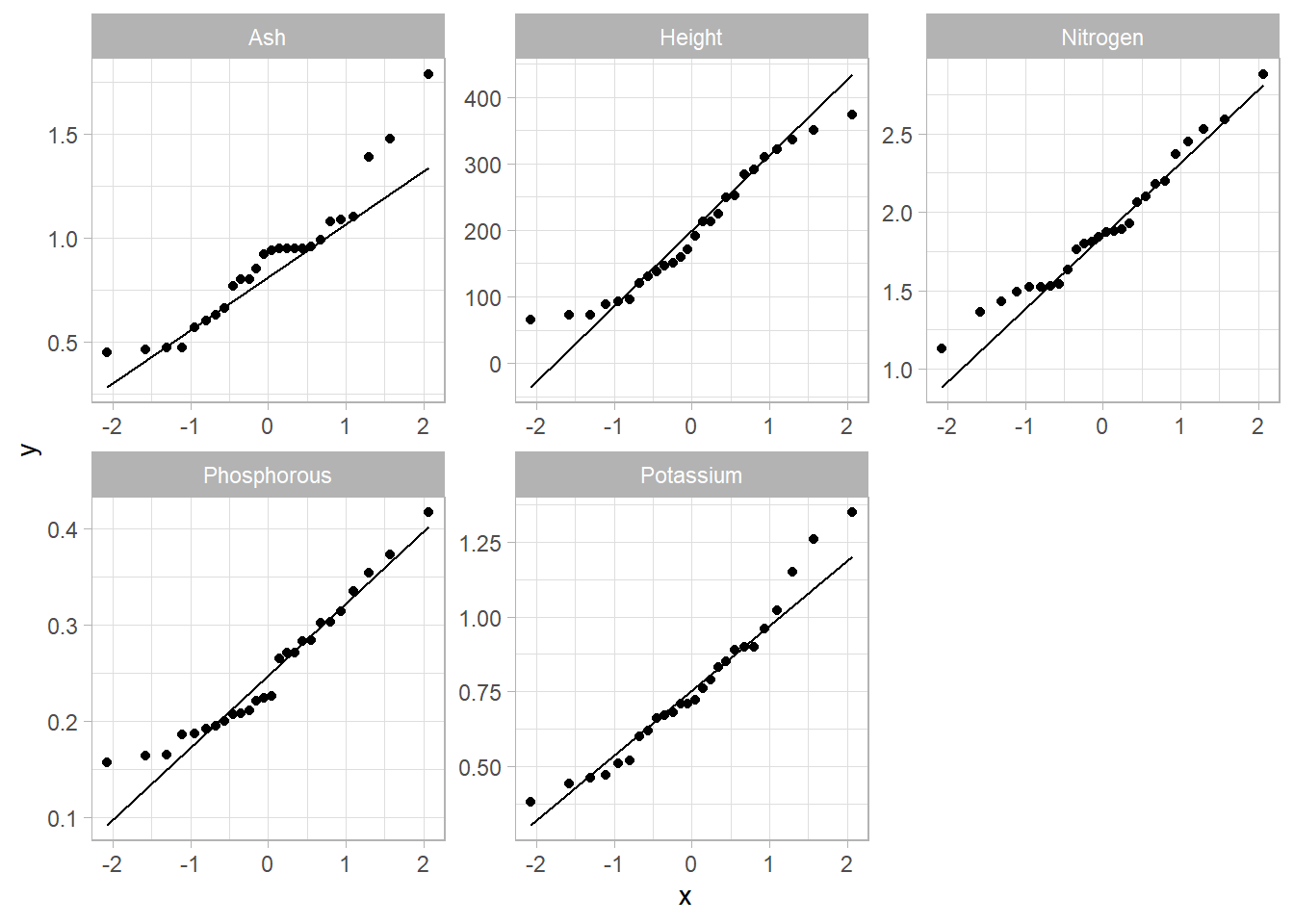
# Univariate normality
sw_tests <- apply(trees, MARGIN = 2, FUN = shapiro.test)
sw_tests
#> $Nitrogen
#>
#> Shapiro-Wilk normality test
#>
#> data: newX[, i]
#> W = 0.96829, p-value = 0.5794
#>
#>
#> $Phosphorous
#>
#> Shapiro-Wilk normality test
#>
#> data: newX[, i]
#> W = 0.93644, p-value = 0.1104
#>
#>
#> $Potassium
#>
#> Shapiro-Wilk normality test
#>
#> data: newX[, i]
#> W = 0.95709, p-value = 0.3375
#>
#>
#> $Ash
#>
#> Shapiro-Wilk normality test
#>
#> data: newX[, i]
#> W = 0.92071, p-value = 0.04671
#>
#>
#> $Height
#>
#> Shapiro-Wilk normality test
#>
#> data: newX[, i]
#> W = 0.94107, p-value = 0.1424
# Kolmogorov-Smirnov test
ks_tests <- map(trees, ~ ks.test(scale(.x),"pnorm"))
ks_tests
#> $Nitrogen
#>
#> Asymptotic one-sample Kolmogorov-Smirnov test
#>
#> data: scale(.x)
#> D = 0.12182, p-value = 0.8351
#> alternative hypothesis: two-sided
#>
#>
#> $Phosphorous
#>
#> Asymptotic one-sample Kolmogorov-Smirnov test
#>
#> data: scale(.x)
#> D = 0.17627, p-value = 0.3944
#> alternative hypothesis: two-sided
#>
#>
#> $Potassium
#>
#> Asymptotic one-sample Kolmogorov-Smirnov test
#>
#> data: scale(.x)
#> D = 0.10542, p-value = 0.9348
#> alternative hypothesis: two-sided
#>
#>
#> $Ash
#>
#> Asymptotic one-sample Kolmogorov-Smirnov test
#>
#> data: scale(.x)
#> D = 0.14503, p-value = 0.6449
#> alternative hypothesis: two-sided
#>
#>
#> $Height
#>
#> Asymptotic one-sample Kolmogorov-Smirnov test
#>
#> data: scale(.x)
#> D = 0.1107, p-value = 0.9076
#> alternative hypothesis: two-sided
# Mardia's test, need large sample size for power
mardia_test <-
mvn(
trees,
mvnTest = "mardia",
covariance = FALSE,
multivariatePlot = "qq"
)
mardia_test$multivariateNormality
#> Test Statistic p value Result
#> 1 Mardia Skewness 29.7248528871795 0.72054426745778 YES
#> 2 Mardia Kurtosis -1.67743173185383 0.0934580886477281 YES
#> 3 MVN <NA> <NA> YES
# Doornik-Hansen's test
dh_test <-
mvn(
trees,
mvnTest = "dh",
covariance = FALSE,
multivariatePlot = "qq"
)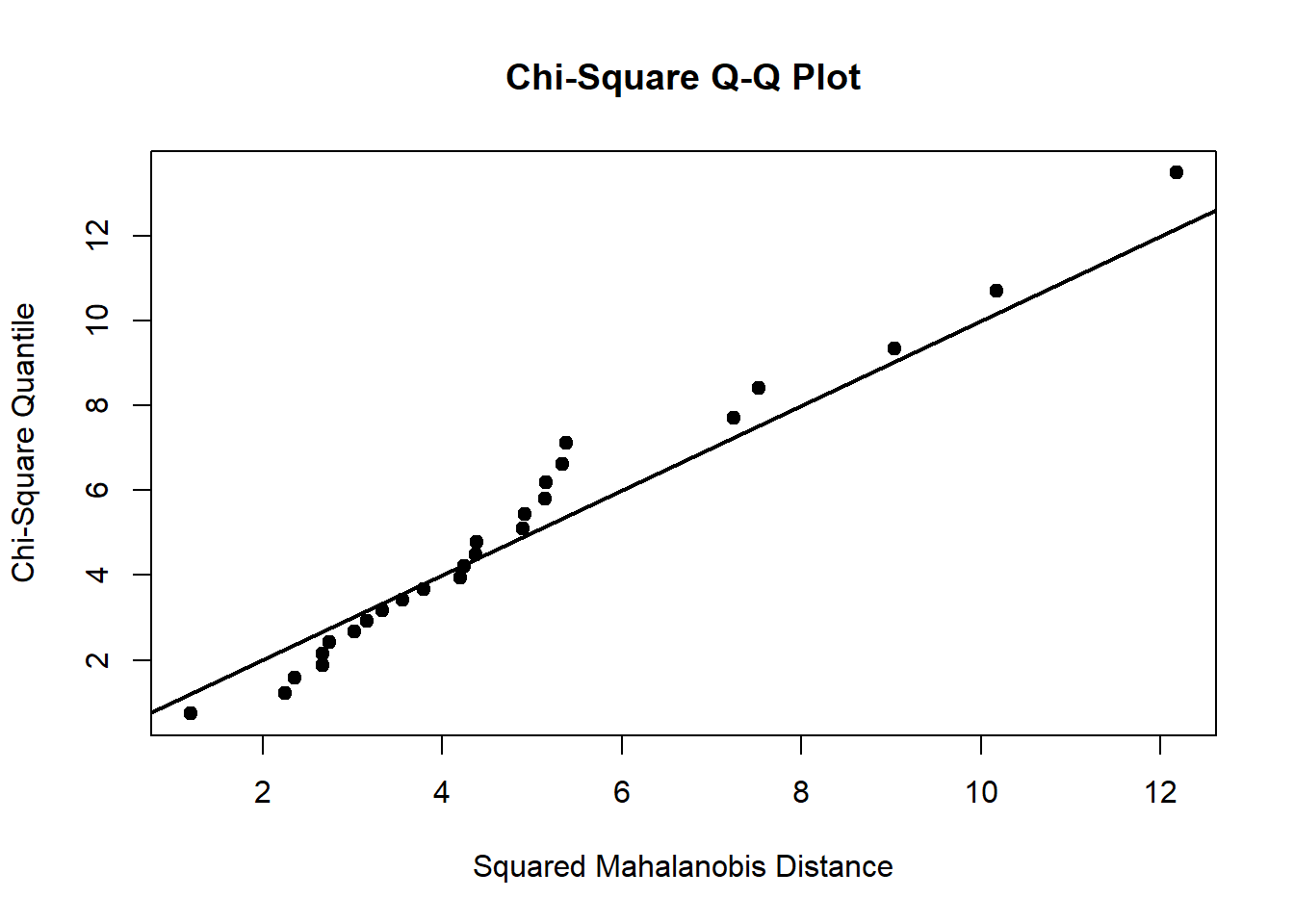
dh_test$multivariateNormality
#> Test E df p value MVN
#> 1 Doornik-Hansen 161.9446 10 1.285352e-29 NO
# Henze-Zirkler's test
hz_test <-
mvn(
trees,
mvnTest = "hz",
covariance = FALSE,
multivariatePlot = "qq"
)
hz_test$multivariateNormality
#> Test HZ p value MVN
#> 1 Henze-Zirkler 0.7591525 0.6398905 YES
# The last column indicates whether dataset follows a multivariate normality or not (i.e, YES or NO) at significance level 0.05.
# Royston's test
# can only apply for 3 < obs < 5000 (because of Shapiro-Wilk's test)
royston_test <-
mvn(
trees,
mvnTest = "royston",
covariance = FALSE,
multivariatePlot = "qq"
)
royston_test$multivariateNormality
#> Test H p value MVN
#> 1 Royston 9.064631 0.08199215 YES
# E-statistic
estat_test <-
mvn(
trees,
mvnTest = "energy",
covariance = FALSE,
multivariatePlot = "qq"
)
estat_test$multivariateNormality
#> Test Statistic p value MVN
#> 1 E-statistic 1.091101 0.532 YES22.0.2 Mean Vector Inference
In the univariate normal distribution, we test \(H_0: \mu =\mu_0\) by using
\[ T = \frac{\bar{y}- \mu_0}{s/\sqrt{n}} \sim t_{n-1} \]
under the null hypothesis. And reject the null if \(|T|\) is large relative to \(t_{(1-\alpha/2,n-1)}\) because it means that seeing a value as large as what we observed is rare if the null is true
Equivalently,
\[ T^2 = \frac{(\bar{y}- \mu_0)^2}{s^2/n} = n(\bar{y}- \mu_0)(s^2)^{-1}(\bar{y}- \mu_0) \sim f_{(1,n-1)} \]
22.0.2.1 Natural Multivariate Generalization
\[ \begin{aligned} &H_0: \mathbf{\mu} = \mathbf{\mu}_0 \\ &H_a: \mathbf{\mu} \neq \mathbf{\mu}_0 \end{aligned} \]
Define Hotelling’s \(T^2\) by
\[ T^2 = n(\bar{\mathbf{y}} - \mathbf{\mu}_0)'\mathbf{S}^{-1}(\bar{\mathbf{y}} - \mathbf{\mu}_0) \]
which can be viewed as a generalized distance between \(\bar{\mathbf{y}}\) and \(\mathbf{\mu}_0\)
Under the assumption of normality,
\[ F = \frac{n-p}{(n-1)p} T^2 \sim f_{(p,n-p)} \]
and reject the null hypothesis when \(F > f_{(1-\alpha, p, n-p)}\)
-
The \(T^2\) test is invariant to changes in measurement units.
- If \(\mathbf{z = Cy + d}\) where \(\mathbf{C}\) and \(\mathbf{d}\) do not depend on \(\mathbf{y}\), then \(T^2(\mathbf{z}) - T^2(\mathbf{y})\)
The \(T^2\) test can be derived as a likelihood ratio test of \(H_0: \mu = \mu_0\)
22.0.2.2 Confidence Intervals
22.0.2.2.1 Confidence Region
An “exact” \(100(1-\alpha)\%\) confidence region for \(\mathbf{\mu}\) is the set of all vectors, \(\mathbf{v}\), which are “close enough” to the observed mean vector, \(\bar{\mathbf{y}}\) to satisfy
\[ n(\bar{\mathbf{y}} - \mathbf{\mu}_0)'\mathbf{S}^{-1}(\bar{\mathbf{y}} - \mathbf{\mu}_0) \le \frac{(n-1)p}{n-p} f_{(1-\alpha, p, n-p)} \]
- \(\mathbf{v}\) are just the mean vectors that are not rejected by the \(T^2\) test when \(\mathbf{\bar{y}}\) is observed.
In case that you have 2 parameters, the confidence region is a “hyper-ellipsoid”.
In this region, it consists of all \(\mathbf{\mu}_0\) vectors for which the \(T^2\) test would not reject \(H_0\) at significance level \(\alpha\)
Even though the confidence region better assesses the joint knowledge concerning plausible values of \(\mathbf{\mu}\) , people typically include confidence statement about the individual component means. We’d like all of the separate confidence statements to hold simultaneously with a specified high probability. Simultaneous confidence intervals: intervals against any statement being incorrect
22.0.2.2.1.1 Simultaneous Confidence Statements
- Intervals based on a rectangular confidence region by projecting the previous region onto the coordinate axes:
\[ \bar{y}_{i} \pm \sqrt{\frac{(n-1)p}{n-p}f_{(1-\alpha, p,n-p)}\frac{s_{ii}}{n}} \]
for all \(i = 1,..,p\)
which implied confidence region is conservative; it has at least \(100(1- \alpha)\%\)
Generally, simultaneous \(100(1-\alpha) \%\) confidence intervals for all linear combinations , \(\mathbf{a}\) of the elements of the mean vector are given by
\[ \mathbf{a'\bar{y}} \pm \sqrt{\frac{(n-1)p}{n-p}f_{(1-\alpha, p,n-p)}\frac{\mathbf{a'Sa}}{n}} \]
works for any arbitrary linear combination \(\mathbf{a'\mu} = a_1 \mu_1 + ... + a_p \mu_p\), which is a projection onto the axis in the direction of \(\mathbf{a}\)
These intervals have the property that the probability that at least one such interval does not contain the appropriate \(\mathbf{a' \mu}\) is no more than \(\alpha\)
These types of intervals can be used for “data snooping” (like \[Scheffe\])
22.0.2.2.1.2 One \(\mu\) at a time
- One at a time confidence intervals:
\[ \bar{y}_i \pm t_{(1 - \alpha/2, n-1} \sqrt{\frac{s_{ii}}{n}} \]
Each of these intervals has a probability of \(1-\alpha\) of covering the appropriate \(\mu_i\)
But they ignore the covariance structure of the \(p\) variables
If we only care about \(k\) simultaneous intervals, we can use “one at a time” method with the \[Bonferroni\] correction.
This method gets more conservative as the number of intervals \(k\) increases.
22.0.3 General Hypothesis Testing
22.0.3.1 One-sample Tests
\[ H_0: \mathbf{C \mu= 0} \]
where
- \(\mathbf{C}\) is a \(c \times p\) matrix of rank c where \(c \le p\)
We can test this hypothesis using the following statistic
\[ F = \frac{n - c}{(n-1)c} T^2 \]
where \(T^2 = n(\mathbf{C\bar{y}})' (\mathbf{CSC'})^{-1} (\mathbf{C\bar{y}})\)
Example:
\[ H_0: \mu_1 = \mu_2 = ... = \mu_p \]
Equivalently,
\[ \begin{aligned} \mu_1 - \mu_2 &= 0 \\ &\vdots \\ \mu_{p-1} - \mu_p &= 0 \end{aligned} \]
a total of \(p-1\) tests. Hence, we have \(\mathbf{C}\) as the \(p - 1 \times p\) matrix
\[ \mathbf{C} = \left( \begin{array} {ccccc} 1 & -1 & 0 & \ldots & 0 \\ 0 & 1 & -1 & \ldots & 0 \\ \vdots & \vdots & \vdots & \ddots & \vdots \\ 0 & 0 & \ldots & 1 & -1 \end{array} \right) \]
number of rows = \(c = p -1\)
Equivalently, we can also compare all of the other means to the first mean. Then, we test \(\mu_1 - \mu_2 = 0, \mu_1 - \mu_3 = 0,..., \mu_1 - \mu_p = 0\), the \((p-1) \times p\) matrix \(\mathbf{C}\) is
\[ \mathbf{C} = \left( \begin{array} {ccccc} -1 & 1 & 0 & \ldots & 0 \\ -1 & 0 & 1 & \ldots & 0 \\ \vdots & \vdots & \vdots & \ddots & \vdots \\ -1 & 0 & \ldots & 0 & 1 \end{array} \right) \]
The value of \(T^2\) is invariant to these equivalent choices of \(\mathbf{C}\)
This is often used for repeated measures designs, where each subject receives each treatment once over successive periods of time (all treatments are administered to each unit).
Example:
Let \(y_{ij}\) be the response from subject i at time j for \(i = 1,..,n, j = 1,...,T\). In this case, \(\mathbf{y}_i = (y_{i1}, ..., y_{iT})', i = 1,...,n\) are a random sample from \(N_T (\mathbf{\mu}, \mathbf{\Sigma})\)
Let \(n=8\) subjects, \(T = 6\). We are interested in \(\mu_1, .., \mu_6\)
\[ H_0: \mu_1 = \mu_2 = ... = \mu_6 \]
Equivalently,
\[ \begin{aligned} \mu_1 - \mu_2 &= 0 \\ \mu_2 - \mu_3 &= 0 \\ &... \\ \mu_5 - \mu_6 &= 0 \end{aligned} \]
We can test orthogonal polynomials for 4 equally spaced time points. To test for example the null hypothesis that quadratic and cubic effects are jointly equal to 0, we would define \(\mathbf{C}\)
\[ \mathbf{C} = \left( \begin{array} {cccc} 1 & -1 & -1 & 1 \\ -1 & 3 & -3 & 1 \end{array} \right) \]
22.0.3.2 Two-Sample Tests
Consider the analogous two sample multivariate tests.
Example: we have data on two independent random samples, one sample from each of two populations
\[ \begin{aligned} \mathbf{y}_{1i} &\sim N_p (\mathbf{\mu_1, \Sigma}) \\ \mathbf{y}_{2j} &\sim N_p (\mathbf{\mu_2, \Sigma}) \end{aligned} \]
We assume
normality
equal variance-covariance matrices
independent random samples
We can summarize our data using the sufficient statistics \(\mathbf{\bar{y}}_1, \mathbf{S}_1, \mathbf{\bar{y}}_2, \mathbf{S}_2\) with respective sample sizes, \(n_1,n_2\)
Since we assume that \(\mathbf{\Sigma}_1 = \mathbf{\Sigma}_2 = \mathbf{\Sigma}\), compute a pooled estimate of the variance-covariance matrix on \(n_1 + n_2 - 2\) df
\[ \mathbf{S} = \frac{(n_1 - 1)\mathbf{S}_1 + (n_2-1) \mathbf{S}_2}{(n_1 -1) + (n_2 - 1)} \]
\[ \begin{aligned} &H_0: \mathbf{\mu}_1 = \mathbf{\mu}_2 \\ &H_a: \mathbf{\mu}_1 \neq \mathbf{\mu}_2 \end{aligned} \]
At least one element of the mean vectors is different
We use
\(\mathbf{\bar{y}}_1 - \mathbf{\bar{y}}_2\) to estimate \(\mu_1 - \mu_2\)
-
\(\mathbf{S}\) to estimate \(\mathbf{\Sigma}\)
Note: because we assume the two populations are independent, there is no covariance
\(cov(\mathbf{\bar{y}}_1 - \mathbf{\bar{y}}_2) = var(\mathbf{\bar{y}}_1) + var(\mathbf{\bar{y}}_2) = \frac{\mathbf{\Sigma_1}}{n_1} + \frac{\mathbf{\Sigma_2}}{n_2} = \mathbf{\Sigma}(\frac{1}{n_1} + \frac{1}{n_2})\)
Reject \(H_0\) if
\[ \begin{aligned} T^2 &= (\mathbf{\bar{y}}_1 - \mathbf{\bar{y}}_2)'\{ \mathbf{S} (\frac{1}{n_1} + \frac{1}{n_2})\}^{-1} (\mathbf{\bar{y}}_1 - \mathbf{\bar{y}}_2)\\ &= \frac{n_1 n_2}{n_1 +n_2} (\mathbf{\bar{y}}_1 - \mathbf{\bar{y}}_2)'\{ \mathbf{S} \}^{-1} (\mathbf{\bar{y}}_1 - \mathbf{\bar{y}}_2)\\ & \ge \frac{(n_1 + n_2 -2)p}{n_1 + n_2 - p - 1} f_{(1- \alpha,n_1 + n_2 - p -1)} \end{aligned} \]
or equivalently, if
\[ F = \frac{n_1 + n_2 - p -1}{(n_1 + n_2 -2)p} T^2 \ge f_{(1- \alpha, p , n_1 + n_2 -p -1)} \]
A \(100(1-\alpha) \%\) confidence region for \(\mu_1 - \mu_2\) consists of all vector \(\delta\) which satisfy
\[ \frac{n_1 n_2}{n_1 + n_2} (\mathbf{\bar{y}}_1 - \mathbf{\bar{y}}_2 - \mathbf{\delta})' \mathbf{S}^{-1}(\mathbf{\bar{y}}_1 - \mathbf{\bar{y}}_2 - \mathbf{\delta}) \le \frac{(n_1 + n_2 - 2)p}{n_1 + n_2 -p - 1}f_{(1-\alpha, p , n_1 + n_2 - p -1)} \]
The simultaneous confidence intervals for all linear combinations of \(\mu_1 - \mu_2\) have the form
\[ \mathbf{a'}(\mathbf{\bar{y}}_1 - \mathbf{\bar{y}}_2) \pm \sqrt{\frac{(n_1 + n_2 -2)p}{n_1 + n_2 - p -1}}f_{(1-\alpha, p, n_1 + n_2 -p -1)} \times \sqrt{\mathbf{a'Sa}(\frac{1}{n_1} + \frac{1}{n_2})} \]
Bonferroni intervals, for k combinations
\[ (\bar{y}_{1i} - \bar{y}_{2i}) \pm t_{(1-\alpha/2k, n_1 + n_2 - 2)}\sqrt{(\frac{1}{n_1} + \frac{1}{n_2})s_{ii}} \]
22.0.3.3 Model Assumptions
If model assumption are not met
-
Unequal Covariance Matrices
If \(n_1 = n_2\) (large samples) there is little effect on the Type I error rate and power fo the two sample test
If \(n_1 > n_2\) and the eigenvalues of \(\mathbf{\Sigma}_1 \mathbf{\Sigma}^{-1}_2\) are less than 1, the Type I error level is inflated
If \(n_1 > n_2\) and some eigenvalues of \(\mathbf{\Sigma}_1 \mathbf{\Sigma}_2^{-1}\) are greater than 1, the Type I error rate is too small, leading to a reduction in power
-
Sample Not Normal
Type I error level of the two sample \(T^2\) test isn’t much affect by moderate departures from normality if the two populations being sampled have similar distributions
One sample \(T^2\) test is much more sensitive to lack of normality, especially when the distribution is skewed.
Intuitively, you can think that in one sample your distribution will be sensitive, but the distribution of the difference between two similar distributions will not be as sensitive.
-
Solutions:
Transform to make the data more normal
-
Large large samples, use the \(\chi^2\) (Wald) test, in which populations don’t need to be normal, or equal sample sizes, or equal variance-covariance matrices
- \(H_0: \mu_1 - \mu_2 =0\) use \((\mathbf{\bar{y}}_1 - \mathbf{\bar{y}}_2)'( \frac{1}{n_1} \mathbf{S}_1 + \frac{1}{n_2}\mathbf{S}_2)^{-1}(\mathbf{\bar{y}}_1 - \mathbf{\bar{y}}_2) \dot{\sim} \chi^2_{(p)}\)
22.0.3.3.1 Equal Covariance Matrices Tests
With independent random samples from k populations of \(p\)-dimensional vectors. We compute the sample covariance matrix for each, \(\mathbf{S}_i\), where \(i = 1,...,k\)
\[ \begin{aligned} &H_0: \mathbf{\Sigma}_1 = \mathbf{\Sigma}_2 = \ldots = \mathbf{\Sigma}_k = \mathbf{\Sigma} \\ &H_a: \text{at least 2 are different} \end{aligned} \]
Assume \(H_0\) is true, we would use a pooled estimate of the common covariance matrix, \(\mathbf{\Sigma}\)
\[ \mathbf{S} = \frac{\sum_{i=1}^k (n_i -1)\mathbf{S}_i}{\sum_{i=1}^k (n_i - 1)} \]
with \(\sum_{i=1}^k (n_i -1)\)
22.0.3.3.1.1 Bartlett’s Test
(a modification of the likelihood ratio test). Define
\[ N = \sum_{i=1}^k n_i \]
and (note: \(| |\) are determinants here, not absolute value)
\[ M = (N - k) \log|\mathbf{S}| - \sum_{i=1}^k (n_i - 1) \log|\mathbf{S}_i| \]
\[ C^{-1} = 1 - \frac{2p^2 + 3p - 1}{6(p+1)(k-1)} \{\sum_{i=1}^k (\frac{1}{n_i - 1}) - \frac{1}{N-k} \} \]
Reject \(H_0\) when \(MC^{-1} > \chi^2_{1- \alpha, (k-1)p(p+1)/2}\)
If not all samples are from normal populations, \(MC^{-1}\) has a distribution which is often shifted to the right of the nominal \(\chi^2\) distribution, which means \(H_0\) is often rejected even when it is true (the Type I error level is inflated). Hence, it is better to test individual normality first, or then multivariate normality before you do Bartlett’s test.
22.0.3.4 Two-Sample Repeated Measurements
Define \(\mathbf{y}_{hi} = (y_{hi1}, ..., y_{hit})'\) to be the observations from the i-th subject in the h-th group for times 1 through T
Assume that \(\mathbf{y}_{11}, ..., \mathbf{y}_{1n_1}\) are iid \(N_t(\mathbf{\mu}_1, \mathbf{\Sigma})\) and that \(\mathbf{y}_{21},...,\mathbf{y}_{2n_2}\) are iid \(N_t(\mathbf{\mu}_2, \mathbf{\Sigma})\)
\(H_0: \mathbf{C}(\mathbf{\mu}_1 - \mathbf{\mu}_2) = \mathbf{0}_c\) where \(\mathbf{C}\) is a \(c \times t\) matrix of rank \(c\) where \(c \le t\)
The test statistic has the form
\[ T^2 = \frac{n_1 n_2}{n_1 + n_2} (\mathbf{\bar{y}}_1 - \mathbf{\bar{y}}_2)' \mathbf{C}'(\mathbf{CSC}')^{-1}\mathbf{C} (\mathbf{\bar{y}}_1 - \mathbf{\bar{y}}_2) \]
where \(\mathbf{S}\) is the pooled covariance estimate. Then,
\[ F = \frac{n_1 + n_2 - c -1}{(n_1 + n_2-2)c} T^2 \sim f_{(c, n_1 + n_2 - c-1)} \]
when \(H_0\) is true
If the null hypothesis \(H_0: \mu_1 = \mu_2\) is rejected. A weaker hypothesis is that the profiles for the two groups are parallel.
\[ \begin{aligned} \mu_{11} - \mu_{21} &= \mu_{12} - \mu_{22} \\ &\vdots \\ \mu_{1t-1} - \mu_{2t-1} &= \mu_{1t} - \mu_{2t} \end{aligned} \]
The null hypothesis matrix term is then
\(H_0: \mathbf{C}(\mu_1 - \mu_2) = \mathbf{0}_c\) , where \(c = t - 1\) and
\[ \mathbf{C} = \left( \begin{array} {ccccc} 1 & -1 & 0 & \ldots & 0 \\ 0 & 1 & -1 & \ldots & 0 \\ \vdots & \vdots & \vdots & \ddots & \vdots \\ 0 & 0 & 0 & \ldots & -1 \end{array} \right)_{(t-1) \times t} \]
# One-sample Hotelling's T^2 test
# Create data frame
plants <- data.frame(
y1 = c(2.11, 2.36, 2.13, 2.78, 2.17),
y2 = c(10.1, 35.0, 2.0, 6.0, 2.0),
y3 = c(3.4, 4.1, 1.9, 3.8, 1.7)
)
# Center the data with
# the hypothesized means and make a matrix
plants_ctr <- plants %>%
transmute(y1_ctr = y1 - 2.85,
y2_ctr = y2 - 15.0,
y3_ctr = y3 - 6.0) %>%
as.matrix()
# Use anova.mlm to calculate Wilks' lambda
onesamp_fit <- anova(lm(plants_ctr ~ 1), test = "Wilks")
onesamp_fit
#> Analysis of Variance Table
#>
#> Df Wilks approx F num Df den Df Pr(>F)
#> (Intercept) 1 0.054219 11.629 3 2 0.08022 .
#> Residuals 4
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1can’t reject the null of hypothesized vector of means
# Paired-Sample Hotelling's T^2 test
library(ICSNP)
# Create data frame
waste <- data.frame(
case = 1:11,
com_y1 = c(6, 6, 18, 8, 11, 34, 28, 71, 43, 33, 20),
com_y2 = c(27, 23, 64, 44, 30, 75, 26, 124, 54, 30, 14),
state_y1 = c(25, 28, 36, 35, 15, 44, 42, 54, 34, 29, 39),
state_y2 = c(15, 13, 22, 29, 31, 64, 30, 64, 56, 20, 21)
)
# Calculate the difference between commercial and state labs
waste_diff <- waste %>%
transmute(y1_diff = com_y1 - state_y1,
y2_diff = com_y2 - state_y2)
# Run the test
paired_fit <- HotellingsT2(waste_diff)
# value T.2 in the output corresponds to
# the approximate F-value in the output from anova.mlm
paired_fit
#>
#> Hotelling's one sample T2-test
#>
#> data: waste_diff
#> T.2 = 6.1377, df1 = 2, df2 = 9, p-value = 0.02083
#> alternative hypothesis: true location is not equal to c(0,0)reject the null that the two labs’ measurements are equal
# Independent-Sample Hotelling's T^2 test with Bartlett's test
# Read in data
steel <- read.table("images/steel.dat")
names(steel) <- c("Temp", "Yield", "Strength")
str(steel)
#> 'data.frame': 12 obs. of 3 variables:
#> $ Temp : int 1 1 1 1 1 2 2 2 2 2 ...
#> $ Yield : int 33 36 35 38 40 35 36 38 39 41 ...
#> $ Strength: int 60 61 64 63 65 57 59 59 61 63 ...
# Plot the data
ggplot(steel, aes(x = Yield, y = Strength)) +
geom_text(aes(label = Temp), size = 5) +
geom_segment(aes(
x = 33,
y = 57.5,
xend = 42,
yend = 65
), col = "red")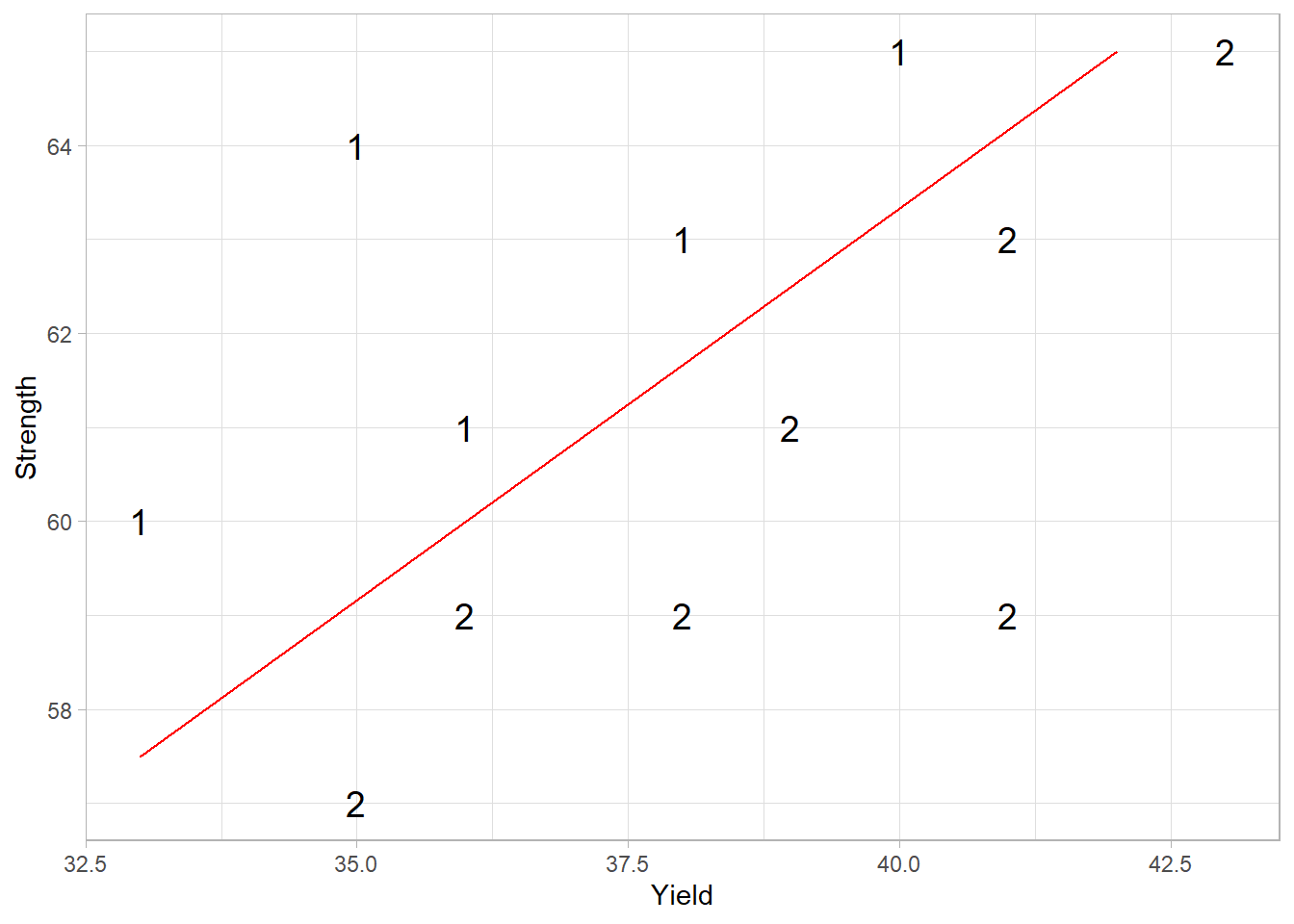
# Bartlett's test for equality of covariance matrices
# same thing as Box's M test in the multivariate setting
bart_test <- boxM(steel[, -1], steel$Temp)
bart_test # fail to reject the null of equal covariances
#>
#> Box's M-test for Homogeneity of Covariance Matrices
#>
#> data: steel[, -1]
#> Chi-Sq (approx.) = 0.38077, df = 3, p-value = 0.9442
# anova.mlm
twosamp_fit <-
anova(lm(cbind(Yield, Strength) ~ factor(Temp),
data = steel),
test = "Wilks")
twosamp_fit
#> Analysis of Variance Table
#>
#> Df Wilks approx F num Df den Df Pr(>F)
#> (Intercept) 1 0.001177 3818.1 2 9 6.589e-14 ***
#> factor(Temp) 1 0.294883 10.8 2 9 0.004106 **
#> Residuals 10
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
# ICSNP package
twosamp_fit2 <-
HotellingsT2(cbind(steel$Yield, steel$Strength) ~
factor(steel$Temp))
twosamp_fit2
#>
#> Hotelling's two sample T2-test
#>
#> data: cbind(steel$Yield, steel$Strength) by factor(steel$Temp)
#> T.2 = 10.76, df1 = 2, df2 = 9, p-value = 0.004106
#> alternative hypothesis: true location difference is not equal to c(0,0)reject null. Hence, there is a difference in the means of the bivariate normal distributions
22.1 MANOVA
Multivariate Analysis of Variance
One-way MANOVA
Compare treatment means for h different populations
Population 1: \(\mathbf{y}_{11}, \mathbf{y}_{12}, \dots, \mathbf{y}_{1n_1} \sim idd N_p (\mathbf{\mu}_1, \mathbf{\Sigma})\)
\(\vdots\)
Population h: \(\mathbf{y}_{h1}, \mathbf{y}_{h2}, \dots, \mathbf{y}_{hn_h} \sim idd N_p (\mathbf{\mu}_h, \mathbf{\Sigma})\)
Assumptions
- Independent random samples from \(h\) different populations
- Common covariance matrices
- Each population is multivariate normal
Calculate the summary statistics \(\mathbf{\bar{y}}_i, \mathbf{S}\) and the pooled estimate of the covariance matrix \(\mathbf{S}\)
Similar to the univariate one-way ANVOA, we can use the effects model formulation \(\mathbf{\mu}_i = \mathbf{\mu} + \mathbf{\tau}_i\), where
\(\mathbf{\mu}_i\) is the population mean for population i
\(\mathbf{\mu}\) is the overall mean effect
\(\mathbf{\tau}_i\) is the treatment effect of the i-th treatment.
For the one-way model: \(\mathbf{y}_{ij} = \mu + \tau_i + \epsilon_{ij}\) for \(i = 1,..,h; j = 1,..., n_i\) and \(\epsilon_{ij} \sim N_p(\mathbf{0, \Sigma})\)
However, the above model is over-parameterized (i.e., infinite number of ways to define \(\mathbf{\mu}\) and the \(\mathbf{\tau}_i\)’s such that they add up to \(\mu_i\). Thus we can constrain by having
\[ \sum_{i=1}^h n_i \tau_i = 0 \]
or
\[ \mathbf{\tau}_h = 0 \]
The observational equivalent of the effects model is
\[ \begin{aligned} \mathbf{y}_{ij} &= \mathbf{\bar{y}} + (\mathbf{\bar{y}}_i - \mathbf{\bar{y}}) + (\mathbf{y}_{ij} - \mathbf{\bar{y}}_i) \\ &= \text{overall sample mean} + \text{treatement effect} + \text{residual} \text{ (under univariate ANOVA)} \end{aligned} \]
After manipulation
\[ \sum_{i = 1}^h \sum_{j = 1}^{n_i} (\mathbf{\bar{y}}_{ij} - \mathbf{\bar{y}})(\mathbf{\bar{y}}_{ij} - \mathbf{\bar{y}})' = \sum_{i = 1}^h n_i (\mathbf{\bar{y}}_i - \mathbf{\bar{y}})(\mathbf{\bar{y}}_i - \mathbf{\bar{y}})' + \sum_{i=1}^h \sum_{j = 1}^{n_i} (\mathbf{\bar{y}}_{ij} - \mathbf{\bar{y}})(\mathbf{\bar{y}}_{ij} - \mathbf{\bar{y}}_i)' \]
LHS = Total corrected sums of squares and cross products (SSCP) matrix
RHS =
1st term = Treatment (or between subjects) sum of squares and cross product matrix (denoted H;B)
2nd term = residual (or within subject) SSCP matrix denoted (E;W)
Note:
\[ \mathbf{E} = (n_1 - 1)\mathbf{S}_1 + ... + (n_h -1) \mathbf{S}_h = (n-h) \mathbf{S} \]
MANOVA table
| Source | SSCP | df |
|---|---|---|
| Treatment | \(\mathbf{H}\) | \(h -1\) |
| Residual (error) | \(\mathbf{E}\) | \(\sum_{i= 1}^h n_i - h\) |
| Total Corrected | \(\mathbf{H + E}\) | \(\sum_{i=1}^h n_i -1\) |
\[ H_0: \tau_1 = \tau_2 = \dots = \tau_h = \mathbf{0} \]
We consider the relative “sizes” of \(\mathbf{E}\) and \(\mathbf{H+E}\)
Wilk’s Lambda
Define Wilk’s Lambda
\[ \Lambda^* = \frac{|\mathbf{E}|}{|\mathbf{H+E}|} \]
Properties:
Wilk’s Lambda is equivalent to the F-statistic in the univariate case
The exact distribution of \(\Lambda^*\) can be determined for especial cases.
For large sample sizes, reject \(H_0\) if
\[ -(\sum_{i=1}^h n_i - 1 - \frac{p+h}{2}) \log(\Lambda^*) > \chi^2_{(1-\alpha, p(h-1))} \]
22.1.1 Testing General Hypotheses
\(h\) different treatments
with the i-th treatment
applied to \(n_i\) subjects that
are observed for \(p\) repeated measures.
Consider this a \(p\) dimensional obs on a random sample from each of \(h\) different treatment populations.
\[ \mathbf{y}_{ij} = \mathbf{\mu} + \mathbf{\tau}_i + \mathbf{\epsilon}_{ij} \]
for \(i = 1,..,h\) and \(j = 1,..,n_i\)
Equivalently,
\[ \mathbf{Y} = \mathbf{XB} + \mathbf{\epsilon} \]
where \(n = \sum_{i = 1}^h n_i\) and with restriction \(\mathbf{\tau}_h = 0\)
\[ \mathbf{Y}_{(n \times p)} = \left[ \begin{array} {c} \mathbf{y}_{11}' \\ \vdots \\ \mathbf{y}_{1n_1}' \\ \vdots \\ \mathbf{y}_{hn_h}' \end{array} \right], \mathbf{B}_{(h \times p)} = \left[ \begin{array} {c} \mathbf{\mu}' \\ \mathbf{\tau}_1' \\ \vdots \\ \mathbf{\tau}_{h-1}' \end{array} \right], \mathbf{\epsilon}_{(n \times p)} = \left[ \begin{array} {c} \epsilon_{11}' \\ \vdots \\ \epsilon_{1n_1}' \\ \vdots \\ \epsilon_{hn_h}' \end{array} \right] \]
\[ \mathbf{X}_{(n \times h)} = \left[ \begin{array} {ccccc} 1 & 1 & 0 & \ldots & 0 \\ \vdots & \vdots & \vdots & & \vdots \\ 1 & 1 & 0 & \ldots & 0 \\ \vdots & \vdots & \vdots & \ldots & \vdots \\ 1 & 0 & 0 & \ldots & 0 \\ \vdots & \vdots & \vdots & & \vdots \\ 1 & 0 & 0 & \ldots & 0 \end{array} \right] \]
Estimation
\[ \mathbf{\hat{B}} = (\mathbf{X'X})^{-1} \mathbf{X'Y} \]
Rows of \(\mathbf{Y}\) are independent (i.e., \(var(\mathbf{Y}) = \mathbf{I}_n \otimes \mathbf{\Sigma}\) , an \(np \times np\) matrix, where \(\otimes\) is the Kronecker product).
\[ \begin{aligned} &H_0: \mathbf{LBM} = 0 \\ &H_a: \mathbf{LBM} \neq 0 \end{aligned} \]
where
\(\mathbf{L}\) is a \(g \times h\) matrix of full row rank (\(g \le h\)) = comparisons across groups
\(\mathbf{M}\) is a \(p \times u\) matrix of full column rank (\(u \le p\)) = comparisons across traits
The general treatment corrected sums of squares and cross product is
\[ \mathbf{H} = \mathbf{M'Y'X(X'X)^{-1}L'[L(X'X)^{-1}L']^{-1}L(X'X)^{-1}X'YM} \]
or for the null hypothesis \(H_0: \mathbf{LBM} = \mathbf{D}\)
\[ \mathbf{H} = (\mathbf{\hat{LBM}} - \mathbf{D})'[\mathbf{X(X'X)^{-1}L}]^{-1}(\mathbf{\hat{LBM}} - \mathbf{D}) \]
The general matrix of residual sums of squares and cross product
\[ \mathbf{E} = \mathbf{M'Y'[I-X(X'X)^{-1}X']YM} = \mathbf{M'[Y'Y - \hat{B}'(X'X)^{-1}\hat{B}]M} \]
We can compute the following statistic eigenvalues of \(\mathbf{HE}^{-1}\)
Wilk’s Criterion: \(\Lambda^* = \frac{|\mathbf{E}|}{|\mathbf{H} + \mathbf{E}|}\) . The df depend on the rank of \(\mathbf{L}, \mathbf{M}, \mathbf{X}\)
Lawley-Hotelling Trace: \(U = tr(\mathbf{HE}^{-1})\)
Pillai Trace: \(V = tr(\mathbf{H}(\mathbf{H}+ \mathbf{E}^{-1})\)
Roy’s Maximum Root: largest eigenvalue of \(\mathbf{HE}^{-1}\)
If \(H_0\) is true and n is large, \(-(n-1- \frac{p+h}{2})\ln \Lambda^* \sim \chi^2_{p(h-1)}\). Some special values of p and h can give exact F-dist under \(H_0\)
# One-way MANOVA
library(car)
library(emmeans)
library(profileR)
library(tidyverse)
## Read in the data
gpagmat <- read.table("images/gpagmat.dat")
## Change the variable names
names(gpagmat) <- c("y1", "y2", "admit")
## Check the structure
str(gpagmat)
#> 'data.frame': 85 obs. of 3 variables:
#> $ y1 : num 2.96 3.14 3.22 3.29 3.69 3.46 3.03 3.19 3.63 3.59 ...
#> $ y2 : int 596 473 482 527 505 693 626 663 447 588 ...
#> $ admit: int 1 1 1 1 1 1 1 1 1 1 ...
## Plot the data
gg <- ggplot(gpagmat, aes(x = y1, y = y2)) +
geom_text(aes(label = admit, col = as.character(admit))) +
scale_color_discrete(name = "Admission",
labels = c("Admit", "Do not admit", "Borderline")) +
scale_x_continuous(name = "GPA") +
scale_y_continuous(name = "GMAT")
## Fit one-way MANOVA
oneway_fit <- manova(cbind(y1, y2) ~ admit, data = gpagmat)
summary(oneway_fit, test = "Wilks")
#> Df Wilks approx F num Df den Df Pr(>F)
#> admit 1 0.6126 25.927 2 82 1.881e-09 ***
#> Residuals 83
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1reject the null of equal multivariate mean vectors between the three admmission groups
# Repeated Measures MANOVA
## Create data frame
stress <- data.frame(
subject = 1:8,
begin = c(3, 2, 5, 6, 1, 5, 1, 5),
middle = c(3, 4, 3, 7, 4, 7, 1, 2),
final = c(6, 7, 4, 7, 6, 7, 3, 5)
)- If independent = time with 3 levels -> univariate ANOVA (require sphericity assumption (i.e., the variances for all differences are equal))
- If each level of independent time as a separate variable -> MANOVA (does not require sphericity assumption)
## MANOVA
stress_mod <- lm(cbind(begin, middle, final) ~ 1, data = stress)
idata <-
data.frame(time = factor(
c("begin", "middle", "final"),
levels = c("begin", "middle", "final")
))
repeat_fit <-
Anova(
stress_mod,
idata = idata,
idesign = ~ time,
icontrasts = "contr.poly"
)
summary(repeat_fit)
#>
#> Type III Repeated Measures MANOVA Tests:
#>
#> ------------------------------------------
#>
#> Term: (Intercept)
#>
#> Response transformation matrix:
#> (Intercept)
#> begin 1
#> middle 1
#> final 1
#>
#> Sum of squares and products for the hypothesis:
#> (Intercept)
#> (Intercept) 1352
#>
#> Multivariate Tests: (Intercept)
#> Df test stat approx F num Df den Df Pr(>F)
#> Pillai 1 0.896552 60.66667 1 7 0.00010808 ***
#> Wilks 1 0.103448 60.66667 1 7 0.00010808 ***
#> Hotelling-Lawley 1 8.666667 60.66667 1 7 0.00010808 ***
#> Roy 1 8.666667 60.66667 1 7 0.00010808 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> ------------------------------------------
#>
#> Term: time
#>
#> Response transformation matrix:
#> time.L time.Q
#> begin -7.071068e-01 0.4082483
#> middle -7.850462e-17 -0.8164966
#> final 7.071068e-01 0.4082483
#>
#> Sum of squares and products for the hypothesis:
#> time.L time.Q
#> time.L 18.062500 6.747781
#> time.Q 6.747781 2.520833
#>
#> Multivariate Tests: time
#> Df test stat approx F num Df den Df Pr(>F)
#> Pillai 1 0.7080717 7.276498 2 6 0.024879 *
#> Wilks 1 0.2919283 7.276498 2 6 0.024879 *
#> Hotelling-Lawley 1 2.4254992 7.276498 2 6 0.024879 *
#> Roy 1 2.4254992 7.276498 2 6 0.024879 *
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Univariate Type III Repeated-Measures ANOVA Assuming Sphericity
#>
#> Sum Sq num Df Error SS den Df F value Pr(>F)
#> (Intercept) 450.67 1 52.00 7 60.6667 0.0001081 ***
#> time 20.58 2 24.75 14 5.8215 0.0144578 *
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#>
#> Mauchly Tests for Sphericity
#>
#> Test statistic p-value
#> time 0.7085 0.35565
#>
#>
#> Greenhouse-Geisser and Huynh-Feldt Corrections
#> for Departure from Sphericity
#>
#> GG eps Pr(>F[GG])
#> time 0.77429 0.02439 *
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> HF eps Pr(>F[HF])
#> time 0.9528433 0.01611634can’t reject the null hypothesis of sphericity, hence univariate ANOVA is also appropriate.We also see linear significant time effect, but no quadratic time effect
## Polynomial contrasts
# What is the reference for the marginal means?
ref_grid(stress_mod, mult.name = "time")
#> 'emmGrid' object with variables:
#> 1 = 1
#> time = multivariate response levels: begin, middle, final
# marginal means for the levels of time
contr_means <- emmeans(stress_mod, ~ time, mult.name = "time")
contrast(contr_means, method = "poly")
#> contrast estimate SE df t.ratio p.value
#> linear 2.12 0.766 7 2.773 0.0276
#> quadratic 1.38 0.944 7 1.457 0.1885
# MANOVA
## Read in Data
heart <- read.table("images/heart.dat")
names(heart) <- c("drug", "y1", "y2", "y3", "y4")
## Create a subject ID nested within drug
heart <- heart %>%
group_by(drug) %>%
mutate(subject = row_number()) %>%
ungroup()
str(heart)
#> tibble [24 × 6] (S3: tbl_df/tbl/data.frame)
#> $ drug : chr [1:24] "ax23" "ax23" "ax23" "ax23" ...
#> $ y1 : int [1:24] 72 78 71 72 66 74 62 69 85 82 ...
#> $ y2 : int [1:24] 86 83 82 83 79 83 73 75 86 86 ...
#> $ y3 : int [1:24] 81 88 81 83 77 84 78 76 83 80 ...
#> $ y4 : int [1:24] 77 82 75 69 66 77 70 70 80 84 ...
#> $ subject: int [1:24] 1 2 3 4 5 6 7 8 1 2 ...
## Create means summary for profile plot,
# pivot longer for plotting with ggplot
heart_means <- heart %>%
group_by(drug) %>%
summarize_at(vars(starts_with("y")), mean) %>%
ungroup() %>%
pivot_longer(-drug, names_to = "time", values_to = "mean") %>%
mutate(time = as.numeric(as.factor(time)))
gg_profile <- ggplot(heart_means, aes(x = time, y = mean)) +
geom_line(aes(col = drug)) +
geom_point(aes(col = drug)) +
ggtitle("Profile Plot") +
scale_y_continuous(name = "Response") +
scale_x_discrete(name = "Time")
gg_profile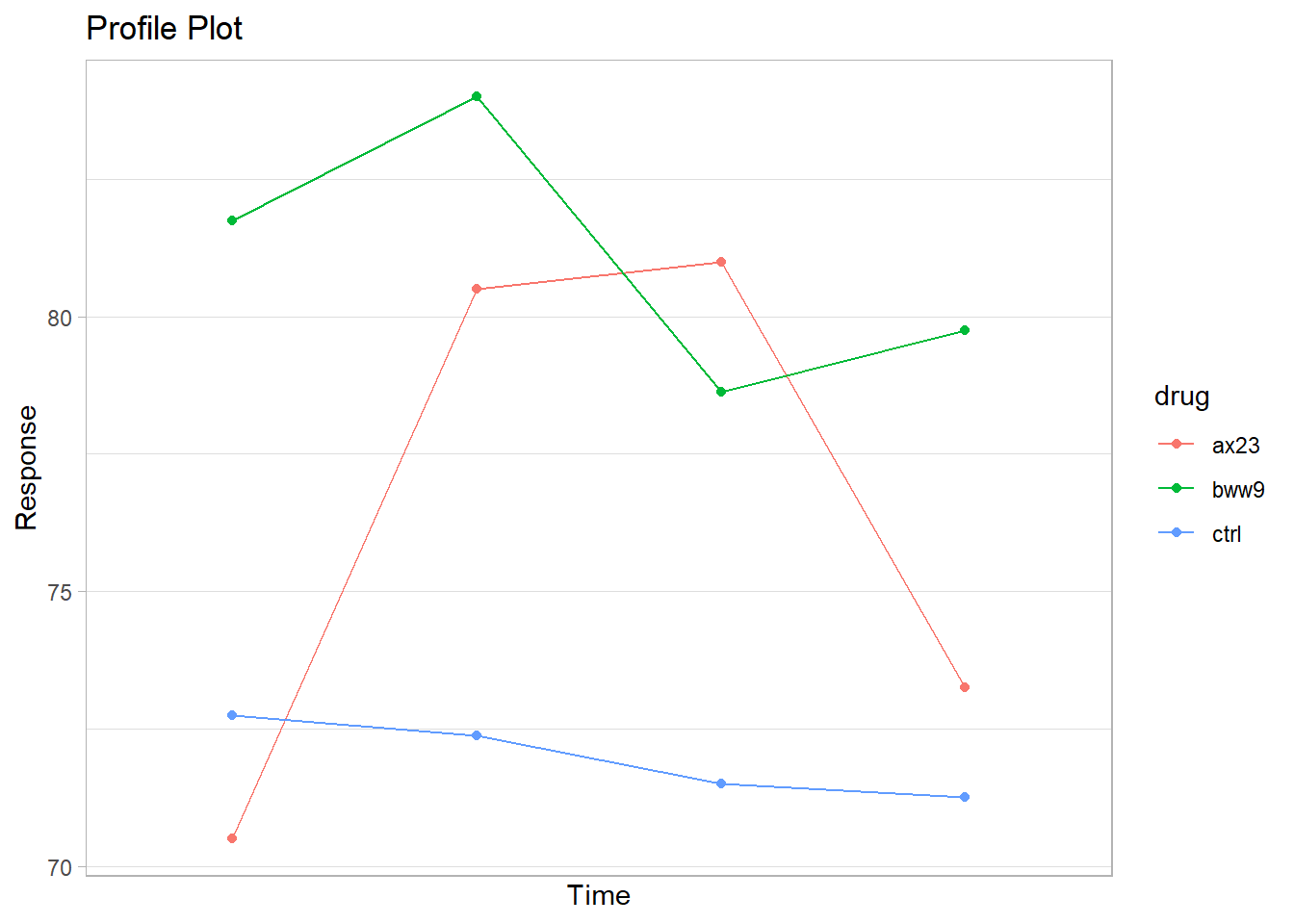
## Fit model
heart_mod <- lm(cbind(y1, y2, y3, y4) ~ drug, data = heart)
man_fit <- car::Anova(heart_mod)
summary(man_fit)
#>
#> Type II MANOVA Tests:
#>
#> Sum of squares and products for error:
#> y1 y2 y3 y4
#> y1 641.00 601.750 535.250 426.00
#> y2 601.75 823.875 615.500 534.25
#> y3 535.25 615.500 655.875 555.25
#> y4 426.00 534.250 555.250 674.50
#>
#> ------------------------------------------
#>
#> Term: drug
#>
#> Sum of squares and products for the hypothesis:
#> y1 y2 y3 y4
#> y1 567.00 335.2500 42.7500 387.0
#> y2 335.25 569.0833 404.5417 367.5
#> y3 42.75 404.5417 391.0833 171.0
#> y4 387.00 367.5000 171.0000 316.0
#>
#> Multivariate Tests: drug
#> Df test stat approx F num Df den Df Pr(>F)
#> Pillai 2 1.283456 8.508082 8 38 1.5010e-06 ***
#> Wilks 2 0.079007 11.509581 8 36 6.3081e-08 ***
#> Hotelling-Lawley 2 7.069384 15.022441 8 34 3.9048e-09 ***
#> Roy 2 6.346509 30.145916 4 19 5.4493e-08 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1reject the null hypothesis of no difference in means between treatments
## Contrasts
heart$drug <- factor(heart$drug)
L <- matrix(c(0, 2,
1, -1,-1, -1), nrow = 3, byrow = T)
colnames(L) <- c("bww9:ctrl", "ax23:rest")
rownames(L) <- unique(heart$drug)
contrasts(heart$drug) <- L
contrasts(heart$drug)
#> bww9:ctrl ax23:rest
#> ax23 0 2
#> bww9 1 -1
#> ctrl -1 -1
# do not set contrast L if you do further analysis (e.g., Anova, lm)
# do M matrix instead
M <- matrix(c(1, -1, 0, 0,
0, 1, -1, 0,
0, 0, 1, -1), nrow = 4)
## update model to test contrasts
heart_mod2 <- update(heart_mod)
coef(heart_mod2)
#> y1 y2 y3 y4
#> (Intercept) 75.00 78.9583333 77.041667 74.75
#> drugbww9:ctrl 4.50 5.8125000 3.562500 4.25
#> drugax23:rest -2.25 0.7708333 1.979167 -0.75
# Hypothesis test for bww9 vs control after transformation M
# same as linearHypothesis(heart_mod, hypothesis.matrix = c(0,1,-1), P = M)
bww9vctrl <-
car::linearHypothesis(heart_mod2,
hypothesis.matrix = c(0, 1, 0),
P = M)
bww9vctrl
#>
#> Response transformation matrix:
#> [,1] [,2] [,3]
#> y1 1 0 0
#> y2 -1 1 0
#> y3 0 -1 1
#> y4 0 0 -1
#>
#> Sum of squares and products for the hypothesis:
#> [,1] [,2] [,3]
#> [1,] 27.5625 -47.25 14.4375
#> [2,] -47.2500 81.00 -24.7500
#> [3,] 14.4375 -24.75 7.5625
#>
#> Sum of squares and products for error:
#> [,1] [,2] [,3]
#> [1,] 261.375 -141.875 28.000
#> [2,] -141.875 248.750 -19.375
#> [3,] 28.000 -19.375 219.875
#>
#> Multivariate Tests:
#> Df test stat approx F num Df den Df Pr(>F)
#> Pillai 1 0.2564306 2.184141 3 19 0.1233
#> Wilks 1 0.7435694 2.184141 3 19 0.1233
#> Hotelling-Lawley 1 0.3448644 2.184141 3 19 0.1233
#> Roy 1 0.3448644 2.184141 3 19 0.1233
bww9vctrl <-
car::linearHypothesis(heart_mod,
hypothesis.matrix = c(0, 1, -1),
P = M)
bww9vctrl
#>
#> Response transformation matrix:
#> [,1] [,2] [,3]
#> y1 1 0 0
#> y2 -1 1 0
#> y3 0 -1 1
#> y4 0 0 -1
#>
#> Sum of squares and products for the hypothesis:
#> [,1] [,2] [,3]
#> [1,] 27.5625 -47.25 14.4375
#> [2,] -47.2500 81.00 -24.7500
#> [3,] 14.4375 -24.75 7.5625
#>
#> Sum of squares and products for error:
#> [,1] [,2] [,3]
#> [1,] 261.375 -141.875 28.000
#> [2,] -141.875 248.750 -19.375
#> [3,] 28.000 -19.375 219.875
#>
#> Multivariate Tests:
#> Df test stat approx F num Df den Df Pr(>F)
#> Pillai 1 0.2564306 2.184141 3 19 0.1233
#> Wilks 1 0.7435694 2.184141 3 19 0.1233
#> Hotelling-Lawley 1 0.3448644 2.184141 3 19 0.1233
#> Roy 1 0.3448644 2.184141 3 19 0.1233there is no significant difference in means between the control and bww9 drug
# Hypothesis test for ax23 vs rest after transformation M
axx23vrest <-
car::linearHypothesis(heart_mod2,
hypothesis.matrix = c(0, 0, 1),
P = M)
axx23vrest
#>
#> Response transformation matrix:
#> [,1] [,2] [,3]
#> y1 1 0 0
#> y2 -1 1 0
#> y3 0 -1 1
#> y4 0 0 -1
#>
#> Sum of squares and products for the hypothesis:
#> [,1] [,2] [,3]
#> [1,] 438.0208 175.20833 -395.7292
#> [2,] 175.2083 70.08333 -158.2917
#> [3,] -395.7292 -158.29167 357.5208
#>
#> Sum of squares and products for error:
#> [,1] [,2] [,3]
#> [1,] 261.375 -141.875 28.000
#> [2,] -141.875 248.750 -19.375
#> [3,] 28.000 -19.375 219.875
#>
#> Multivariate Tests:
#> Df test stat approx F num Df den Df Pr(>F)
#> Pillai 1 0.855364 37.45483 3 19 3.5484e-08 ***
#> Wilks 1 0.144636 37.45483 3 19 3.5484e-08 ***
#> Hotelling-Lawley 1 5.913921 37.45483 3 19 3.5484e-08 ***
#> Roy 1 5.913921 37.45483 3 19 3.5484e-08 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
axx23vrest <-
car::linearHypothesis(heart_mod,
hypothesis.matrix = c(2, -1, 1),
P = M)
axx23vrest
#>
#> Response transformation matrix:
#> [,1] [,2] [,3]
#> y1 1 0 0
#> y2 -1 1 0
#> y3 0 -1 1
#> y4 0 0 -1
#>
#> Sum of squares and products for the hypothesis:
#> [,1] [,2] [,3]
#> [1,] 402.5208 127.41667 -390.9375
#> [2,] 127.4167 40.33333 -123.7500
#> [3,] -390.9375 -123.75000 379.6875
#>
#> Sum of squares and products for error:
#> [,1] [,2] [,3]
#> [1,] 261.375 -141.875 28.000
#> [2,] -141.875 248.750 -19.375
#> [3,] 28.000 -19.375 219.875
#>
#> Multivariate Tests:
#> Df test stat approx F num Df den Df Pr(>F)
#> Pillai 1 0.842450 33.86563 3 19 7.9422e-08 ***
#> Wilks 1 0.157550 33.86563 3 19 7.9422e-08 ***
#> Hotelling-Lawley 1 5.347205 33.86563 3 19 7.9422e-08 ***
#> Roy 1 5.347205 33.86563 3 19 7.9422e-08 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1there is a significant difference in means between ax23 drug treatment and the rest of the treatments
22.1.2 Profile Analysis
Examine similarities between the treatment effects (between subjects), which is useful for longitudinal analysis. Null is that all treatments have the same average effect.
\[ H_0: \mu_1 = \mu_2 = \dots = \mu_h \]
Equivalently,
\[ H_0: \tau_1 = \tau_2 = \dots = \tau_h \]
The exact nature of the similarities and differences between the treatments can be examined under this analysis.
Sequential steps in profile analysis:
- Are the profiles parallel? (i.e., is there no interaction between treatment and time)
- Are the profiles coincidental? (i.e., are the profiles identical?)
- Are the profiles horizontal? (i.e., are there no differences between any time points?)
If we reject the null hypothesis that the profiles are parallel, we can test
Are there differences among groups within some subset of the total time points?
Are there differences among time points in a particular group (or groups)?
Are there differences within some subset of the total time points in a particular group (or groups)?
Example
4 times (p = 4)
3 treatments (h=3)
22.1.2.1 Parallel Profile
Are the profiles for each population identical expect for a mean shift?
\[ \begin{aligned} H_0: \mu_{11} - \mu_{21} - \mu_{12} - \mu_{22} = &\dots = \mu_{1t} - \mu_{2t} \\ \mu_{11} - \mu_{31} - \mu_{12} - \mu_{32} = &\dots = \mu_{1t} - \mu_{3t} \\ &\dots \end{aligned} \]
for \(h-1\) equations
Equivalently,
\[ H_0: \mathbf{LBM = 0} \]
\[ \mathbf{LBM} = \left[ \begin{array} {ccc} 1 & -1 & 0 \\ 1 & 0 & -1 \end{array} \right] \left[ \begin{array} {ccc} \mu_{11} & \dots & \mu_{14} \\ \mu_{21} & \dots & \mu_{24} \\ \mu_{31} & \dots & \mu_{34} \end{array} \right] \left[ \begin{array} {ccc} 1 & 1 & 1 \\ -1 & 0 & 0 \\ 0 & -1 & 0 \\ 0 & 0 & -1 \end{array} \right] = \mathbf{0} \]
where this is the cell means parameterization of \(\mathbf{B}\)
The multiplication of the first 2 matrices \(\mathbf{LB}\) is
\[ \left[ \begin{array} {cccc} \mu_{11} - \mu_{21} & \mu_{12} - \mu_{22} & \mu_{13} - \mu_{23} & \mu_{14} - \mu_{24}\\ \mu_{11} - \mu_{31} & \mu_{12} - \mu_{32} & \mu_{13} - \mu_{33} & \mu_{14} - \mu_{34} \end{array} \right] \]
which is the differences in treatment means at the same time
Multiplying by \(\mathbf{M}\), we get the comparison across time
\[ \left[ \begin{array} {ccc} (\mu_{11} - \mu_{21}) - (\mu_{12} - \mu_{22}) & (\mu_{11} - \mu_{21}) -(\mu_{13} - \mu_{23}) & (\mu_{11} - \mu_{21}) - (\mu_{14} - \mu_{24}) \\ (\mu_{11} - \mu_{31}) - (\mu_{12} - \mu_{32}) & (\mu_{11} - \mu_{31}) - (\mu_{13} - \mu_{33}) & (\mu_{11} - \mu_{31}) -(\mu_{14} - \mu_{34}) \end{array} \right] \]
Alternatively, we can also use the effects parameterization
\[ \mathbf{LBM} = \left[ \begin{array} {cccc} 0 & 1 & -1 & 0 \\ 0 & 1 & 0 & -1 \end{array} \right] \left[ \begin{array} {c} \mu' \\ \tau'_1 \\ \tau_2' \\ \tau_3' \end{array} \right] \left[ \begin{array} {ccc} 1 & 1 & 1 \\ -1 & 0 & 0 \\ 0 & -1 & 0 \\ 0 & 0 & -1 \end{array} \right] = \mathbf{0} \]
In both parameterizations, \(rank(\mathbf{L}) = h-1\) and \(rank(\mathbf{M}) = p-1\)
We could also choose \(\mathbf{L}\) and \(\mathbf{M}\) in other forms
\[ \mathbf{L} = \left[ \begin{array} {cccc} 0 & 1 & 0 & -1 \\ 0 & 0 & 1 & -1 \end{array} \right] \]
and
\[ \mathbf{M} = \left[ \begin{array} {ccc} 1 & 0 & 0 \\ -1 & 1 & 0 \\ 0 & -1 & 1 \\ 0 & 0 & -1 \end{array} \right] \]
and still obtain the same result.
22.1.2.2 Coincidental Profiles
After we have evidence that the profiles are parallel (i.e., fail to reject the parallel profile test), we can ask whether they are identical?
Given profiles are parallel, then if the sums of the components of \(\mu_i\) are identical for all the treatments, then the profiles are identical.
\[ H_0: \mathbf{1'}_p \mu_1 = \mathbf{1'}_p \mu_2 = \dots = \mathbf{1'}_p \mu_h \]
Equivalently,
\[ H_0: \mathbf{LBM} = \mathbf{0} \]
where for the cell means parameterization
\[ \mathbf{L} = \left[ \begin{array} {ccc} 1 & 0 & -1 \\ 0 & 1 & -1 \end{array} \right] \]
and
\[ \mathbf{M} = \left[ \begin{array} {cccc} 1 & 1 & 1 & 1 \end{array} \right]' \]
multiplication yields
\[ \left[ \begin{array} {c} (\mu_{11} + \mu_{12} + \mu_{13} + \mu_{14}) - (\mu_{31} + \mu_{32} + \mu_{33} + \mu_{34}) \\ (\mu_{21} + \mu_{22} + \mu_{23} + \mu_{24}) - (\mu_{31} + \mu_{32} + \mu_{33} + \mu_{34}) \end{array} \right] = \left[ \begin{array} {c} 0 \\ 0 \end{array} \right] \]
Different choices of \(\mathbf{L}\) and \(\mathbf{M}\) can yield the same result
22.1.2.3 Horizontal Profiles
Given that we can’t reject the null hypothesis that all \(h\) profiles are the same, we can ask whether all of the elements of the common profile equal? (i.e., horizontal)
\[ H_0: \mathbf{LBM} = \mathbf{0} \]
\[ \mathbf{L} = \left[ \begin{array} {ccc} 1 & 0 & 0 \end{array} \right] \]
and
\[ \mathbf{M} = \left[ \begin{array} {ccc} 1 & 0 & 0 \\ -1 & 1 & 0 \\ 0 & -1 & 1 \\ 0 & 0 & -1 \end{array} \right] \]
hence,
\[ \left[ \begin{array} {ccc} (\mu_{11} - \mu_{12}) & (\mu_{12} - \mu_{13}) & (\mu_{13} + \mu_{14}) \end{array} \right] = \left[ \begin{array} {ccc} 0 & 0 & 0 \end{array} \right] \]
Note:
- If we fail to reject all 3 hypotheses, then we fail to reject the null hypotheses of both no difference between treatments and no differences between traits.
| Test | Equivalent test for |
|---|---|
| Parallel profile | Interaction |
| Coincidental profile | main effect of between-subjects factor |
| Horizontal profile | main effect of repeated measures factor |
profile_fit <-
pbg(
data = as.matrix(heart[, 2:5]),
group = as.matrix(heart[, 1]),
original.names = TRUE,
profile.plot = FALSE
)
summary(profile_fit)
#> Call:
#> pbg(data = as.matrix(heart[, 2:5]), group = as.matrix(heart[,
#> 1]), original.names = TRUE, profile.plot = FALSE)
#>
#> Hypothesis Tests:
#> $`Ho: Profiles are parallel`
#> Multivariate.Test Statistic Approx.F num.df den.df p.value
#> 1 Wilks 0.1102861 12.737599 6 38 7.891497e-08
#> 2 Pillai 1.0891707 7.972007 6 40 1.092397e-05
#> 3 Hotelling-Lawley 6.2587852 18.776356 6 36 9.258571e-10
#> 4 Roy 5.9550887 39.700592 3 20 1.302458e-08
#>
#> $`Ho: Profiles have equal levels`
#> Df Sum Sq Mean Sq F value Pr(>F)
#> group 2 328.7 164.35 5.918 0.00915 **
#> Residuals 21 583.2 27.77
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> $`Ho: Profiles are flat`
#> F df1 df2 p-value
#> 1 14.30928 3 19 4.096803e-05
# reject null hypothesis of parallel profiles
# reject the null hypothesis of coincidental profiles
# reject the null hypothesis that the profiles are flat22.2 Principal Components
- Unsupervised learning
- find important features
- reduce the dimensions of the data set
- “decorrelate” multivariate vectors that have dependence.
- uses eigenvector/eigvenvalue decomposition of covariance (correlation) matrices.
According to the “spectral decomposition theorem”, if \(\mathbf{\Sigma}_{p \times p}\) i s a positive semi-definite, symmetric, real matrix, then there exists an orthogonal matrix \(\mathbf{A}\) such that \(\mathbf{A'\Sigma A} = \Lambda\) where \(\Lambda\) is a diagonal matrix containing the eigenvalues \(\mathbf{\Sigma}\)
\[ \mathbf{\Lambda} = \left( \begin{array} {cccc} \lambda_1 & 0 & \ldots & 0 \\ 0 & \lambda_2 & \ldots & 0 \\ \vdots & \vdots & \ddots & \vdots \\ 0 & 0 & \ldots & \lambda_p \end{array} \right) \]
\[ \mathbf{A} = \left( \begin{array} {cccc} \mathbf{a}_1 & \mathbf{a}_2 & \ldots & \mathbf{a}_p \end{array} \right) \]
the i-th column of \(\mathbf{A}\) , \(\mathbf{a}_i\), is the i-th \(p \times 1\) eigenvector of \(\mathbf{\Sigma}\) that corresponds to the eigenvalue, \(\lambda_i\) , where \(\lambda_1 \ge \lambda_2 \ge \ldots \ge \lambda_p\) . Alternatively, express in matrix decomposition:
\[ \mathbf{\Sigma} = \mathbf{A \Lambda A}' \]
\[ \mathbf{\Sigma} = \mathbf{A} \left( \begin{array} {cccc} \lambda_1 & 0 & \ldots & 0 \\ 0 & \lambda_2 & \ldots & 0 \\ \vdots & \vdots& \ddots & \vdots \\ 0 & 0 & \ldots & \lambda_p \end{array} \right) \mathbf{A}' = \sum_{i=1}^p \lambda_i \mathbf{a}_i \mathbf{a}_i' \]
where the outer product \(\mathbf{a}_i \mathbf{a}_i'\) is a \(p \times p\) matrix of rank 1.
For example,
\(\mathbf{x} \sim N_2(\mathbf{\mu}, \mathbf{\Sigma})\)
\[ \mathbf{\mu} = \left( \begin{array} {c} 5 \\ 12 \end{array} \right); \mathbf{\Sigma} = \left( \begin{array} {cc} 4 & 1 \\ 1 & 2 \end{array} \right) \]
library(MASS)
mu = as.matrix(c(5, 12))
Sigma = matrix(c(4, 1, 1, 2), nrow = 2, byrow = T)
sim <- mvrnorm(n = 1000, mu = mu, Sigma = Sigma)
plot(sim[, 1], sim[, 2])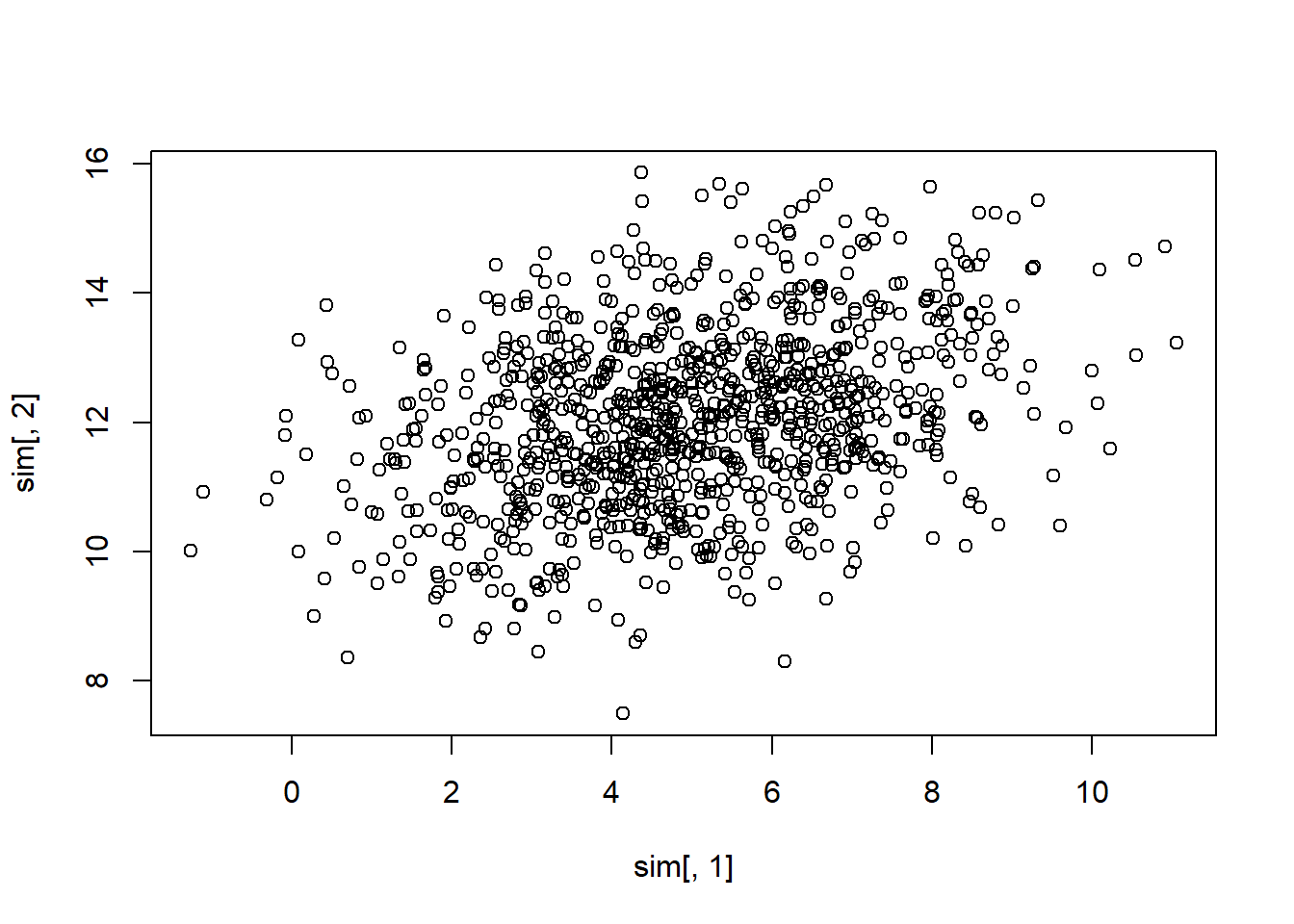
Here,
\[ \mathbf{A} = \left( \begin{array} {cc} 0.9239 & -0.3827 \\ 0.3827 & 0.9239 \\ \end{array} \right) \]
Columns of \(\mathbf{A}\) are the eigenvectors for the decomposition
Under matrix multiplication (\(\mathbf{A'\Sigma A}\) or \(\mathbf{A'A}\) ), the off-diagonal elements equal to 0
Multiplying data by this matrix (i.e., projecting the data onto the orthogonal axes); the distribution of the resulting data (i.e., “scores”) is
\[ N_2 (\mathbf{A'\mu,A'\Sigma A}) = N_2 (\mathbf{A'\mu, \Lambda}) \]
Equivalently,
\[ \mathbf{y} = \mathbf{A'x} \sim N \left[ \left( \begin{array} {c} 9.2119 \\ 9.1733 \end{array} \right), \left( \begin{array} {cc} 4.4144 & 0 \\ 0 & 1.5859 \end{array} \right) \right] \]
A_matrix = matrix(c(0.9239, -0.3827, 0.3827, 0.9239),
nrow = 2,
byrow = T)
t(A_matrix) %*% A_matrix
#> [,1] [,2]
#> [1,] 1.000051 0.000000
#> [2,] 0.000000 1.000051
sim1 <-
mvrnorm(
n = 1000,
mu = t(A_matrix) %*% mu,
Sigma = t(A_matrix) %*% Sigma %*% A_matrix
)
plot(sim1[, 1], sim1[, 2])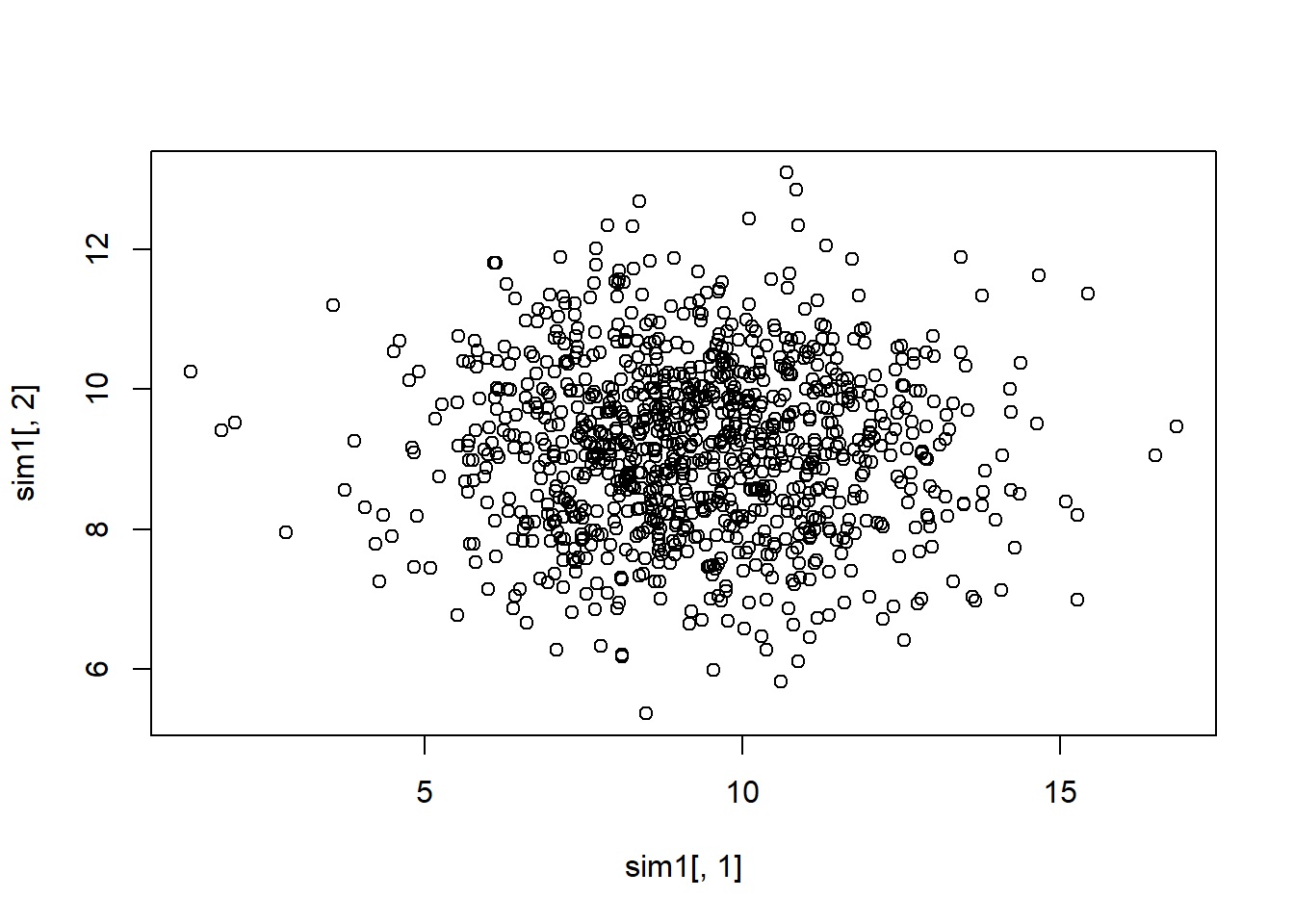
No more dependence in the data structure, plot
Notes:
The i-th eigenvalue is the variance of a linear combination of the elements of \(\mathbf{x}\) ; \(var(y_i) = var(\mathbf{a'_i x}) = \lambda_i\)
The values on the transformed set of axes (i.e., the \(y_i\)’s) are called the scores. These are the orthogonal projections of the data onto the “new principal component axes
Variances of \(y_1\) are greater than those for any other possible projection
Covariance matrix decomposition and projection onto orthogonal axes = PCA
22.2.1 Population Principal Components
\(p \times 1\) vectors \(\mathbf{x}_1, \dots , \mathbf{x}_n\) which are iid with \(var(\mathbf{x}_i) = \mathbf{\Sigma}\)
The first PC is the linear combination \(y_1 = \mathbf{a}_1' \mathbf{x} = a_{11}x_1 + \dots + a_{1p}x_p\) with \(\mathbf{a}_1' \mathbf{a}_1 = 1\) such that \(var(y_1)\) is the maximum of all linear combinations of \(\mathbf{x}\) which have unit length
The second PC is the linear combination \(y_1 = \mathbf{a}_2' \mathbf{x} = a_{21}x_1 + \dots + a_{2p}x_p\) with \(\mathbf{a}_2' \mathbf{a}_2 = 1\) such that \(var(y_1)\) is the maximum of all linear combinations of \(\mathbf{x}\) which have unit length and uncorrelated with \(y_1\) (i.e., \(cov(\mathbf{a}_1' \mathbf{x}, \mathbf{a}'_2 \mathbf{x}) =0\)
continues for all \(y_i\) to \(y_p\)
\(\mathbf{a}_i\)’s are those that make up the matrix \(\mathbf{A}\) in the symmetric decomposition \(\mathbf{A'\Sigma A} = \mathbf{\Lambda}\) , where \(var(y_1) = \lambda_1, \dots , var(y_p) = \lambda_p\) And the total variance of \(\mathbf{x}\) is
\[ \begin{aligned} var(x_1) + \dots + var(x_p) &= tr(\Sigma) = \lambda_1 + \dots + \lambda_p \\ &= var(y_1) + \dots + var(y_p) \end{aligned} \]
Data Reduction
To reduce the dimension of data from p (original) to k dimensions without much “loss of information”, we can use properties of the population principal components
Suppose \(\mathbf{\Sigma} \approx \sum_{i=1}^k \lambda_i \mathbf{a}_i \mathbf{a}_i'\) . Even thought the true variance-covariance matrix has rank \(p\) , it can be be well approximate by a matrix of rank k (k <p)
New “traits” are linear combinations of the measured traits. We can attempt to make meaningful interpretation fo the combinations (with orthogonality constraints).
The proportion of the total variance accounted for by the j-th principal component is
\[ \frac{var(y_j)}{\sum_{i=1}^p var(y_i)} = \frac{\lambda_j}{\sum_{i=1}^p \lambda_i} \]
The proportion of the total variation accounted for by the first k principal components is \(\frac{\sum_{i=1}^k \lambda_i}{\sum_{i=1}^p \lambda_i}\)
Above example , we have \(4.4144/(4+2) = .735\) of the total variability can be explained by the first principal component
22.2.2 Sample Principal Components
Since \(\mathbf{\Sigma}\) is unknown, we use
\[ \mathbf{S} = \frac{1}{n-1}\sum_{i=1}^n (\mathbf{x}_i - \bar{\mathbf{x}})(\mathbf{x}_i - \bar{\mathbf{x}})' \]
Let \(\hat{\lambda}_1 \ge \hat{\lambda}_2 \ge \dots \ge \hat{\lambda}_p \ge 0\) be the eigenvalues of \(\mathbf{S}\) and \(\hat{\mathbf{a}}_1, \hat{\mathbf{a}}_2, \dots, \hat{\mathbf{a}}_p\) denote the eigenvectors of \(\mathbf{S}\)
Then, the i-th sample principal component score (or principal component or score) is
\[ \hat{y}_{ij} = \sum_{k=1}^p \hat{a}_{ik}x_{kj} = \hat{\mathbf{a}}_i'\mathbf{x}_j \]
Properties of Sample Principal Components
The estimated variance of \(y_i = \hat{\mathbf{a}}_i'\mathbf{x}_j\) is \(\hat{\lambda}_i\)
The sample covariance between \(\hat{y}_i\) and \(\hat{y}_{i'}\) is 0 when \(i \neq i'\)
The proportion of the total sample variance accounted for by the i-th sample principal component is \(\frac{\hat{\lambda}_i}{\sum_{k=1}^p \hat{\lambda}_k}\)
The estimated correlation between the \(i\)-th principal component score and the \(l\)-th attribute of \(\mathbf{x}\) is
\[ r_{x_l , \hat{y}_i} = \frac{\hat{a}_{il}\sqrt{\lambda_i}}{\sqrt{s_{ll}}} \]
The correlation coefficient is typically used to interpret the components (i.e., if this correlation is high then it suggests that the l-th original trait is important in the i-th principle component). According to R. A. Johnson, Wichern, et al. (2002), pp.433-434, \(r_{x_l, \hat{y}_i}\) only measures the univariate contribution of an individual X to a component Y without taking into account the presence of the other X’s. Hence, some prefer \(\hat{a}_{il}\) coefficient to interpret the principal component.
\(r_{x_l, \hat{y}_i} ; \hat{a}_{il}\) are referred to as “loadings”
To use k principal components, we must calculate the scores for each data vector in the sample
\[ \mathbf{y}_j = \left( \begin{array} {c} y_{1j} \\ y_{2j} \\ \vdots \\ y_{kj} \end{array} \right) = \left( \begin{array} {c} \hat{\mathbf{a}}_1' \mathbf{x}_j \\ \hat{\mathbf{a}}_2' \mathbf{x}_j \\ \vdots \\ \hat{\mathbf{a}}_k' \mathbf{x}_j \end{array} \right) = \left( \begin{array} {c} \hat{\mathbf{a}}_1' \\ \hat{\mathbf{a}}_2' \\ \vdots \\ \hat{\mathbf{a}}_k' \end{array} \right) \mathbf{x}_j \]
Issues:
Large sample theory exists for eigenvalues and eigenvectors of sample covariance matrices if inference is necessary. But we do not do inference with PCA, we only use it as exploratory or descriptive analysis.
-
PC is not invariant to changes in scale (Exception: if all trait are rescaled by multiplying by the same constant, such as feet to inches).
PCA based on the correlation matrix \(\mathbf{R}\) is different than that based on the covariance matrix \(\mathbf{\Sigma}\)
PCA for the correlation matrix is just rescaling each trait to have unit variance
Transform \(\mathbf{x}\) to \(\mathbf{z}\) where \(z_{ij} = (x_{ij} - \bar{x}_i)/\sqrt{s_{ii}}\) where the denominator affects the PCA
After transformation, \(cov(\mathbf{z}) = \mathbf{R}\)
PCA on \(\mathbf{R}\) is calculated in the same way as that on \(\mathbf{S}\) (where \(\hat{\lambda}{}_1 + \dots + \hat{\lambda}{}_p = p\) )
-
The use of \(\mathbf{R}, \mathbf{S}\) depends on the purpose of PCA.
- If the scale of the observations if different, covariance matrix is more preferable. but if they are dramatically different, analysis can still be dominated by the large variance traits.
-
How many PCs to use can be guided by
Scree Graphs: plot the eigenvalues against their indices. Look for the “elbow” where the steep decline in the graph suddenly flattens out; or big gaps.
minimum Percent of total variation (e.g., choose enough components to have 50% or 90%). can be used for interpretations.
Kaiser’s rule: use only those PC with eigenvalues larger than 1 (applied to PCA on the correlation matrix) - ad hoc
Compare to the eigenvalue scree plot of data to the scree plot when the data are randomized.
22.2.3 Application
PCA on the covariance matrix is usually not preferred due to the fact that PCA is not invariant to changes in scale. Hence, PCA on the correlation matrix is more preferred
This also addresses the problem of multicollinearity
The eigvenvectors may differ by a multiplication of -1 for different implementation, but same interpretation.
library(tidyverse)
## Read in and check data
stock <- read.table("images/stock.dat")
names(stock) <- c("allied", "dupont", "carbide", "exxon", "texaco")
str(stock)
#> 'data.frame': 100 obs. of 5 variables:
#> $ allied : num 0 0.027 0.1228 0.057 0.0637 ...
#> $ dupont : num 0 -0.04485 0.06077 0.02995 -0.00379 ...
#> $ carbide: num 0 -0.00303 0.08815 0.06681 -0.03979 ...
#> $ exxon : num 0.0395 -0.0145 0.0862 0.0135 -0.0186 ...
#> $ texaco : num 0 0.0435 0.0781 0.0195 -0.0242 ...
## Covariance matrix of data
cov(stock)
#> allied dupont carbide exxon texaco
#> allied 0.0016299269 0.0008166676 0.0008100713 0.0004422405 0.0005139715
#> dupont 0.0008166676 0.0012293759 0.0008276330 0.0003868550 0.0003109431
#> carbide 0.0008100713 0.0008276330 0.0015560763 0.0004872816 0.0004624767
#> exxon 0.0004422405 0.0003868550 0.0004872816 0.0008023323 0.0004084734
#> texaco 0.0005139715 0.0003109431 0.0004624767 0.0004084734 0.0007587370
## Correlation matrix of data
cor(stock)
#> allied dupont carbide exxon texaco
#> allied 1.0000000 0.5769244 0.5086555 0.3867206 0.4621781
#> dupont 0.5769244 1.0000000 0.5983841 0.3895191 0.3219534
#> carbide 0.5086555 0.5983841 1.0000000 0.4361014 0.4256266
#> exxon 0.3867206 0.3895191 0.4361014 1.0000000 0.5235293
#> texaco 0.4621781 0.3219534 0.4256266 0.5235293 1.0000000
# cov(scale(stock)) # give the same result
## PCA with covariance
cov_pca <- prcomp(stock)
# uses singular value decomposition for calculation and an N -1 divisor
# alternatively, princomp can do PCA via spectral decomposition,
# but it has worse numerical accuracy
# eigen values
cov_results <- data.frame(eigen_values = cov_pca$sdev ^ 2)
cov_results %>%
mutate(proportion = eigen_values / sum(eigen_values),
cumulative = cumsum(proportion))
#> eigen_values proportion cumulative
#> 1 0.0035953867 0.60159252 0.6015925
#> 2 0.0007921798 0.13255027 0.7341428
#> 3 0.0007364426 0.12322412 0.8573669
#> 4 0.0005086686 0.08511218 0.9424791
#> 5 0.0003437707 0.05752091 1.0000000
# first 2 PCs account for 73% variance in the data
# eigen vectors
cov_pca$rotation # prcomp calls rotation
#> PC1 PC2 PC3 PC4 PC5
#> allied 0.5605914 0.73884565 -0.1260222 0.28373183 -0.20846832
#> dupont 0.4698673 -0.09286987 -0.4675066 -0.68793190 0.28069055
#> carbide 0.5473322 -0.65401929 -0.1140581 0.50045312 -0.09603973
#> exxon 0.2908932 -0.11267353 0.6099196 -0.43808002 -0.58203935
#> texaco 0.2842017 0.07103332 0.6168831 0.06227778 0.72784638
# princomp calls loadings.
# first PC = overall average
# second PC compares Allied to Carbide
## PCA with correlation
#same as scale(stock) %>% prcomp
cor_pca <- prcomp(stock, scale = T)
# eigen values
cor_results <- data.frame(eigen_values = cor_pca$sdev ^ 2)
cor_results %>%
mutate(proportion = eigen_values / sum(eigen_values),
cumulative = cumsum(proportion))
#> eigen_values proportion cumulative
#> 1 2.8564869 0.57129738 0.5712974
#> 2 0.8091185 0.16182370 0.7331211
#> 3 0.5400440 0.10800880 0.8411299
#> 4 0.4513468 0.09026936 0.9313992
#> 5 0.3430038 0.06860076 1.0000000
# first egiven values corresponds to less variance
# than PCA based on the covariance matrix
# eigen vectors
cor_pca$rotation
#> PC1 PC2 PC3 PC4 PC5
#> allied 0.4635405 -0.2408499 0.6133570 -0.3813727 0.4532876
#> dupont 0.4570764 -0.5090997 -0.1778996 -0.2113068 -0.6749814
#> carbide 0.4699804 -0.2605774 -0.3370355 0.6640985 0.3957247
#> exxon 0.4216770 0.5252647 -0.5390181 -0.4728036 0.1794482
#> texaco 0.4213291 0.5822416 0.4336029 0.3812273 -0.3874672
# interpretation of PC2 is different from above:
# it is a comparison of Allied, Dupont and Carbid to Exxon and Texaco Covid Example
To reduce collinearity problem in this dataset, we can use principal components as regressors.
load('images/MOcovid.RData')
covidpca <- prcomp(ndat[,-1],scale = T,center = T)
covidpca$rotation[,1:2]
#> PC1 PC2
#> X..Population.in.Rural.Areas 0.32865838 0.05090955
#> Area..sq..miles. 0.12014444 -0.28579183
#> Population.density..sq..miles. -0.29670124 0.28312922
#> Literacy.rate -0.12517700 -0.08999542
#> Families -0.25856941 0.16485752
#> Area.of.farm.land..sq..miles. 0.02101106 -0.31070363
#> Number.of.farms -0.03814582 -0.44809679
#> Average.value.of.all.property.per.farm..dollars. -0.05410709 0.14404306
#> Estimation.of.rurality.. -0.19040210 0.12089501
#> Male.. 0.02182394 -0.09568768
#> Number.of.Physcians.per.100.000 -0.31451606 0.13598026
#> average.age 0.29414708 0.35593459
#> X0.4.age.proportion -0.11431336 -0.23574057
#> X20.44.age.proportion -0.32802128 -0.22718550
#> X65.and.over.age.proportion 0.30585033 0.32201626
#> prop..White..nonHisp 0.35627561 -0.14142646
#> prop..Hispanic -0.16655381 -0.15105342
#> prop..Black -0.33333359 0.24405802
# Variability of each principal component: pr.var
pr.var <- covidpca$sdev ^ 2
# Variance explained by each principal component: pve
pve <- pr.var / sum(pr.var)
plot(
pve,
xlab = "Principal Component",
ylab = "Proportion of Variance Explained",
ylim = c(0, 0.5),
type = "b"
)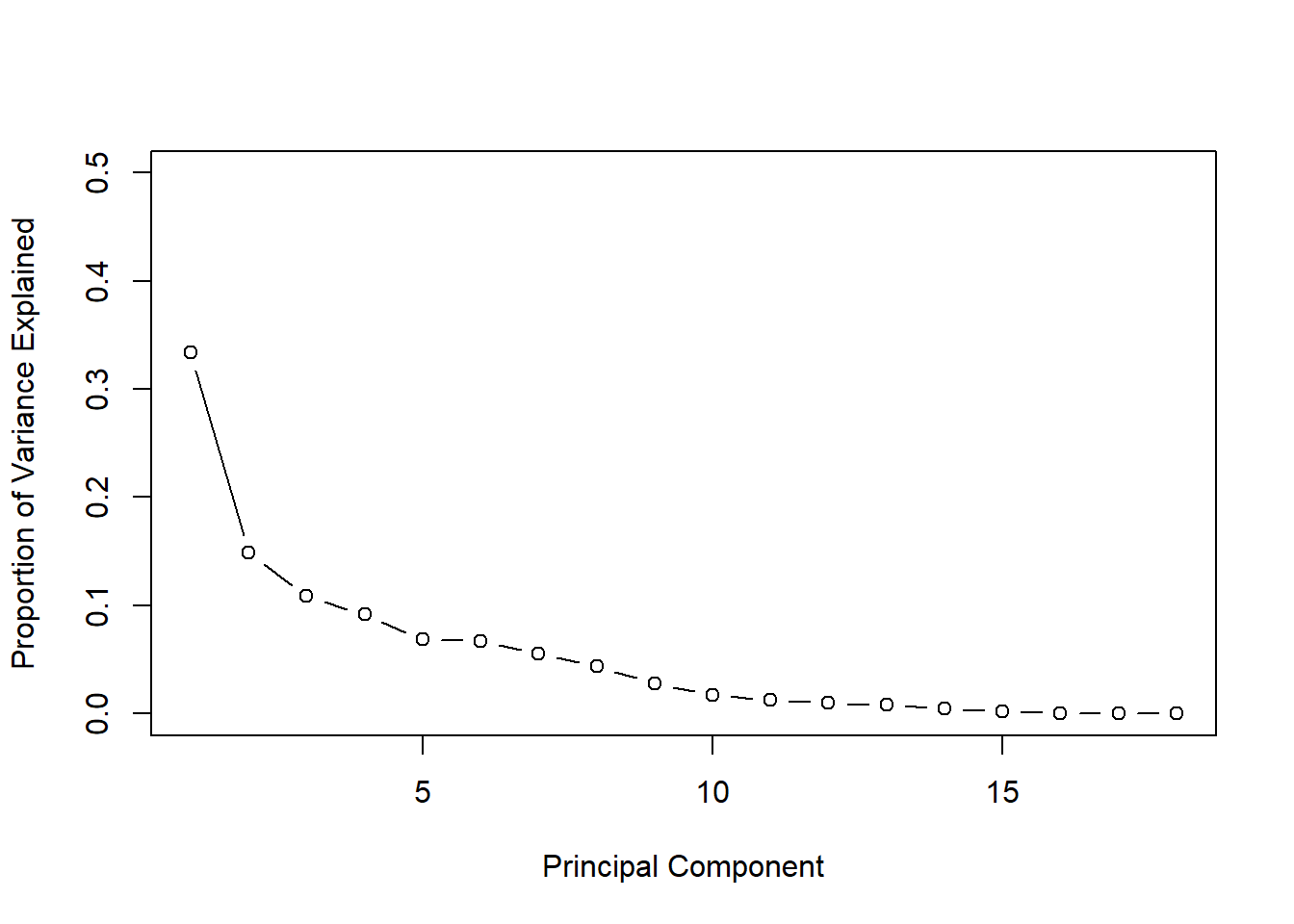
plot(
cumsum(pve),
xlab = "Principal Component",
ylab = "Cumulative Proportion of Variance Explained",
ylim = c(0, 1),
type = "b"
)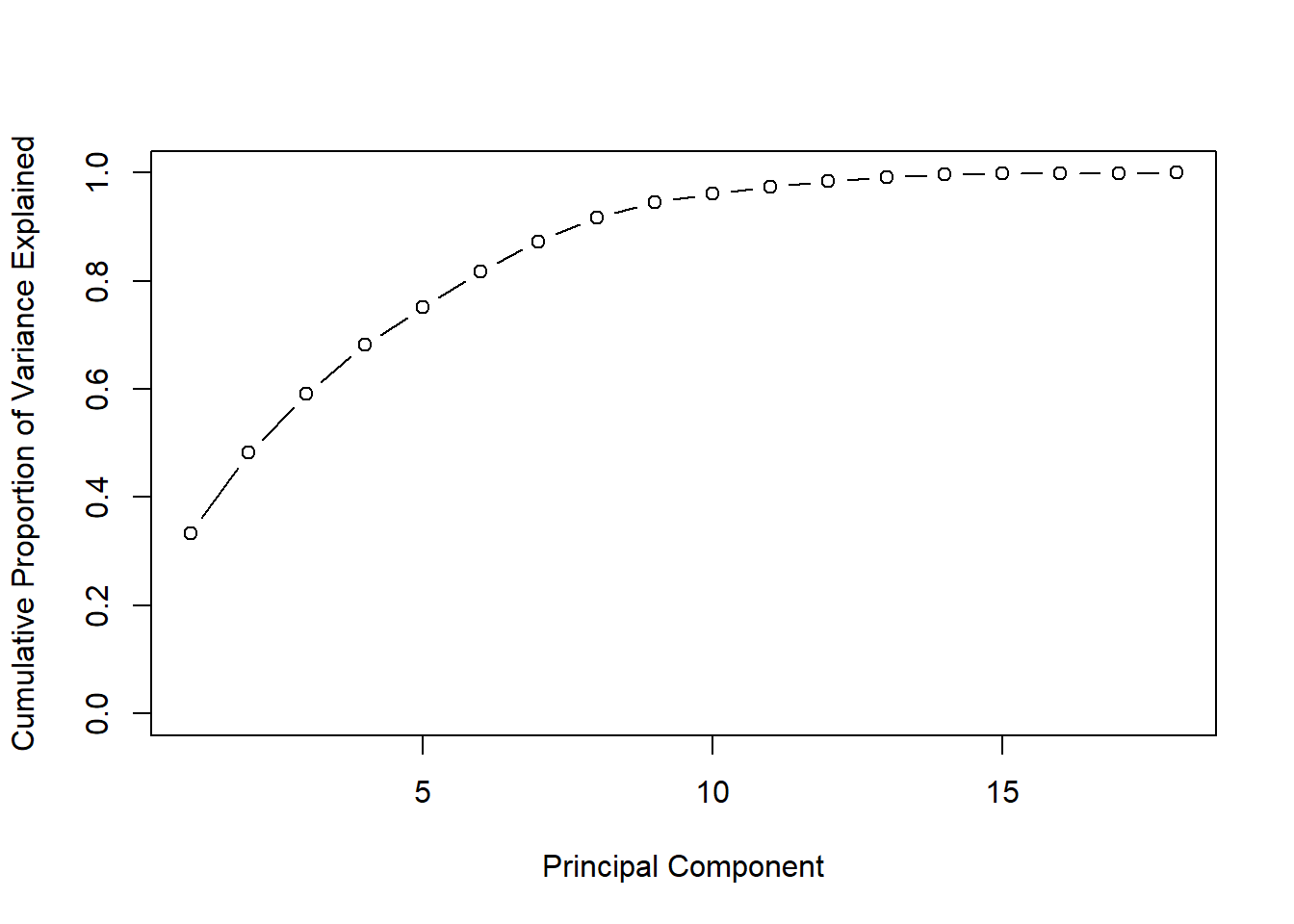
# the first six principe account for around 80% of the variance.
#using base lm function for PC regression
pcadat <- data.frame(covidpca$x[, 1:6])
pcadat$y <- ndat$Y
pcr.man <- lm(log(y) ~ ., pcadat)
mean(pcr.man$residuals ^ 2)
#> [1] 0.03453371
#comparison to lm w/o prin comps
lm.fit <- lm(log(Y) ~ ., data = ndat)
mean(lm.fit$residuals ^ 2)
#> [1] 0.02335128MSE for the PC-based model is larger than regular regression, because models with a large degree of collinearity can still perform well.
pcr function in pls can be used for fitting PC regression (it will select the optimal number of components in the model).
22.3 Factor Analysis
Purpose
Using a few linear combinations of underlying unobservable (latent) traits, we try to describe the covariance relationship among a large number of measured traits
Similar to PCA, but factor analysis is model based
More details can be found on PSU stat or UMN stat
Let \(\mathbf{y}\) be the set of \(p\) measured variables
\(E(\mathbf{y}) = \mathbf{\mu}\)
\(var(\mathbf{y}) = \mathbf{\Sigma}\)
We have
\[ \begin{aligned} \mathbf{y} - \mathbf{\mu} &= \mathbf{Lf} + \epsilon \\ &= \left( \begin{array} {c} l_{11}f_1 + l_{12}f_2 + \dots + l_{tm}f_m \\ \vdots \\ l_{p1}f_1 + l_{p2}f_2 + \dots + l_{pm} f_m \end{array} \right) + \left( \begin{array} {c} \epsilon_1 \\ \vdots \\ \epsilon_p \end{array} \right) \end{aligned} \]
where
\(\mathbf{y} - \mathbf{\mu}\) = the p centered measurements
\(\mathbf{L}\) = \(p \times m\) matrix of factor loadings
\(\mathbf{f}\) = unobserved common factors for the population
\(\mathbf{\epsilon}\) = random errors (i.e., variation that is not accounted for by the common factors).
We want \(m\) (the number of factors) to be much smaller than \(p\) (the number of measured attributes)
Restrictions on the model
\(E(\epsilon) = \mathbf{0}\)
\(var(\epsilon) = \Psi_{p \times p} = diag( \psi_1, \dots, \psi_p)\)
\(\mathbf{\epsilon}, \mathbf{f}\) are independent
Additional assumption could be \(E(\mathbf{f}) = \mathbf{0}, var(\mathbf{f}) = \mathbf{I}_{m \times m}\) (known as the orthogonal factor model) , which imposes the following covariance structure on \(\mathbf{y}\)
\[ \begin{aligned} var(\mathbf{y}) = \mathbf{\Sigma} &= var(\mathbf{Lf} + \mathbf{\epsilon}) \\ &= var(\mathbf{Lf}) + var(\epsilon) \\ &= \mathbf{L} var(\mathbf{f}) \mathbf{L}' + \mathbf{\Psi} \\ &= \mathbf{LIL}' + \mathbf{\Psi} \\ &= \mathbf{LL}' + \mathbf{\Psi} \end{aligned} \]
Since \(\mathbf{\Psi}\) is diagonal, the off-diagonal elements of \(\mathbf{LL}'\) are \(\sigma_{ij}\), the co variances in \(\mathbf{\Sigma}\), which means \(cov(y_i, y_j) = \sum_{k=1}^m l_{ik}l_{jk}\) and the covariance of \(\mathbf{y}\) is completely determined by the m factors ( \(m <<p\))
\(var(y_i) = \sum_{k=1}^m l_{ik}^2 + \psi_i\) where \(\psi_i\) is the specific variance and the summation term is the i-th communality (i.e., portion of the variance of the i-th variable contributed by the \(m\) common factors (\(h_i^2 = \sum_{k=1}^m l_{ik}^2\))
The factor model is only uniquely determined up to an orthogonal transformation of the factors.
Let \(\mathbf{T}_{m \times m}\) be an orthogonal matrix \(\mathbf{TT}' = \mathbf{T'T} = \mathbf{I}\) then
\[ \begin{aligned} \mathbf{y} - \mathbf{\mu} &= \mathbf{Lf} + \epsilon \\ &= \mathbf{LTT'f} + \epsilon \\ &= \mathbf{L}^*(\mathbf{T'f}) + \epsilon & \text{where } \mathbf{L}^* = \mathbf{LT} \end{aligned} \]
and
\[ \begin{aligned} \mathbf{\Sigma} &= \mathbf{LL}' + \mathbf{\Psi} \\ &= \mathbf{LTT'L} + \mathbf{\Psi} \\ &= (\mathbf{L}^*)(\mathbf{L}^*)' + \mathbf{\Psi} \end{aligned} \]
Hence, any orthogonal transformation of the factors is an equally good description of the correlations among the observed traits.
Let \(\mathbf{y} = \mathbf{Cx}\) , where \(\mathbf{C}\) is any diagonal matrix, then \(\mathbf{L}_y = \mathbf{CL}_x\) and \(\mathbf{\Psi}_y = \mathbf{C\Psi}_x\mathbf{C}\)
Hence, we can see that factor analysis is also invariant to changes in scale
22.3.1 Methods of Estimation
To estimate \(\mathbf{L}\)
22.3.1.1 Principal Component Method
Spectral decomposition
\[ \begin{aligned} \mathbf{\Sigma} &= \lambda_1 \mathbf{a}_1 \mathbf{a}_1' + \dots + \lambda_p \mathbf{a}_p \mathbf{a}_p' \\ &= \mathbf{A\Lambda A}' \\ &= \sum_{k=1}^m \lambda+k \mathbf{a}_k \mathbf{a}_k' + \sum_{k= m+1}^p \lambda_k \mathbf{a}_k \mathbf{a}_k' \\ &= \sum_{k=1}^m l_k l_k' + \sum_{k=m+1}^p \lambda_k \mathbf{a}_k \mathbf{a}_k' \end{aligned} \]
where \(l_k = \mathbf{a}_k \sqrt{\lambda_k}\) and the second term is not diagonal in general.
Assume
\[ \psi_i = \sigma_{ii} - \sum_{k=1}^m l_{ik}^2 = \sigma_{ii} - \sum_{k=1}^m \lambda_i a_{ik}^2 \]
then
\[ \mathbf{\Sigma} \approx \mathbf{LL}' + \mathbf{\Psi} \]
To estimate \(\mathbf{L}\) and \(\Psi\) , we use the expected eigenvalues and eigenvectors from \(\mathbf{S}\) or \(\mathbf{R}\)
The estimated factor loadings don’t change as the number of actors increases
The diagonal elements of \(\hat{\mathbf{L}}\hat{\mathbf{L}}' + \hat{\mathbf{\Psi}}\) are equal to the diagonal elements of \(\mathbf{S}\) and \(\mathbf{R}\), but the covariances may not be exactly reproduced
We select \(m\) so that the off-diagonal elements close to the values in \(\mathbf{S}\) (or to make the off-diagonal elements of \(\mathbf{S} - \hat{\mathbf{L}} \hat{\mathbf{L}}' + \hat{\mathbf{\Psi}}\) small)
22.3.1.2 Principal Factor Method
Consider modeling the correlation matrix, \(\mathbf{R} = \mathbf{L} \mathbf{L}' + \mathbf{\Psi}\) . Then
\[ \mathbf{L} \mathbf{L}' = \mathbf{R} - \mathbf{\Psi} = \left( \begin{array} {cccc} h_1^2 & r_{12} & \dots & r_{1p} \\ r_{21} & h_2^2 & \dots & r_{2p} \\ \vdots & \vdots & \ddots & \vdots \\ r_{p1} & r_{p2} & \dots & h_p^2 \end{array} \right) \]
where \(h_i^2 = 1- \psi_i\) (the communality)
Suppose that initial estimates are available for the communalities, \((h_1^*)^2,(h_2^*)^2, \dots , (h_p^*)^2\), then we can regress each trait on all the others, and then use the \(r^2\) as \(h^2\)
The estimate of \(\mathbf{R} - \mathbf{\Psi}\) at step k is
\[ (\mathbf{R} - \mathbf{\Psi})_k = \left( \begin{array} {cccc} (h_1^*)^2 & r_{12} & \dots & r_{1p} \\ r_{21} & (h_2^*)^2 & \dots & r_{2p} \\ \vdots & \vdots & \ddots & \vdots \\ r_{p1} & r_{p2} & \dots & (h_p^*)^2 \end{array} \right) = \mathbf{L}_k^*(\mathbf{L}_k^*)' \]
where
\[ \mathbf{L}_k^* = (\sqrt{\hat{\lambda}_1^*\hat{\mathbf{a}}_1^* , \dots \hat{\lambda}_m^*\hat{\mathbf{a}}_m^*}) \]
and
\[ \hat{\psi}_{i,k}^* = 1 - \sum_{j=1}^m \hat{\lambda}_i^* (\hat{a}_{ij}^*)^2 \]
we used the spectral decomposition on the estimated matrix \((\mathbf{R}- \mathbf{\Psi})\) to calculate the \(\hat{\lambda}_i^* s\) and the \(\mathbf{\hat{a}}_i^* s\)
After updating the values of \((\hat{h}_i^*)^2 = 1 - \hat{\psi}_{i,k}^*\) we will use them to form a new \(\mathbf{L}_{k+1}^*\) via another spectral decomposition. Repeat the process
Notes:
The matrix \((\mathbf{R} - \mathbf{\Psi})_k\) is not necessarily positive definite
The principal component method is similar to principal factor if one considers the initial communalities are \(h^2 = 1\)
-
if \(m\) is too large, some communalities may become larger than 1, causing the iterations to terminate. To combat, we can
fix any communality that is greater than 1 at 1 and then continues.
continue iterations regardless of the size of the communalities. However, results can be outside fo the parameter space.
22.3.1.3 Maximum Likelihood Method
Since we need the likelihood function, we make the additional (critical) assumption that
\(\mathbf{y}_j \sim N(\mathbf{\mu},\mathbf{\Sigma})\) for \(j = 1,..,n\)
\(\mathbf{f} \sim N(\mathbf{0}, \mathbf{I})\)
\(\epsilon_j \sim N(\mathbf{0}, \mathbf{\Psi})\)
and restriction
- \(\mathbf{L}' \mathbf{\Psi}^{-1}\mathbf{L} = \mathbf{\Delta}\) where \(\mathbf{\Delta}\) is a diagonal matrix. (since the factor loading matrix is not unique, we need this restriction).
Notes:
Finding MLE can be computationally expensive
we typically use other methods for exploratory data analysis
Likelihood ratio tests could be used for testing hypotheses in this framework (i.e., Confirmatory Factor Analysis)
22.3.2 Factor Rotation
\(\mathbf{T}_{m \times m}\) is an orthogonal matrix that has the property that
\[ \hat{\mathbf{L}} \hat{\mathbf{L}}' + \hat{\mathbf{\Psi}} = \hat{\mathbf{L}}^*(\hat{\mathbf{L}}^*)' + \hat{\mathbf{\Psi}} \]
where \(\mathbf{L}^* = \mathbf{LT}\)
This means that estimated specific variances and communalities are not altered by the orthogonal transformation.
Since there are an infinite number of choices for \(\mathbf{T}\), some selection criterion is necessary
For example, we can find the orthogonal transformation that maximizes the objective function
\[ \sum_{j = 1}^m [\frac{1}{p}\sum_{i=1}^p (\frac{l_{ij}^{*2}}{h_i})^2 - \{\frac{\gamma}{p} \sum_{i=1}^p (\frac{l_{ij}^{*2}}{h_i})^2 \}^2] \]
where \(\frac{l_{ij}^{*2}}{h_i}\) are “scaled loadings”, which gives variables with small communalities more influence.
Different choices of \(\gamma\) in the objective function correspond to different orthogonal rotation found in the literature;
Varimax \(\gamma = 1\) (rotate the factors so that each of the \(p\) variables should have a high loading on only one factor, but this is not always possible).
Quartimax \(\gamma = 0\)
Equimax \(\gamma = m/2\)
Parsimax \(\gamma = \frac{p(m-1)}{p+m-2}\)
Promax: non-orthogonal or olique transformations
Harris-Kaiser (HK): non-orthogonal or oblique transformations
22.3.3 Estimation of Factor Scores
Recall
\[ (\mathbf{y}_j - \mathbf{\mu}) = \mathbf{L}_{p \times m}\mathbf{f}_j + \epsilon_j \]
If the factor model is correct then
\[ var(\epsilon_j) = \mathbf{\Psi} = diag (\psi_1, \dots , \psi_p) \]
Thus we could consider using weighted least squares to estimate \(\mathbf{f}_j\) , the vector of factor scores for the j-th sampled unit by
\[ \begin{aligned} \hat{\mathbf{f}} &= (\mathbf{L}'\mathbf{\Psi}^{-1} \mathbf{L})^{-1} \mathbf{L}' \mathbf{\Psi}^{-1}(\mathbf{y}_j - \mathbf{\mu}) \\ & \approx (\mathbf{L}'\mathbf{\Psi}^{-1} \mathbf{L})^{-1} \mathbf{L}' \mathbf{\Psi}^{-1}(\mathbf{y}_j - \mathbf{\bar{y}}) \end{aligned} \]
22.3.3.1 The Regression Method
Alternatively, we can use the regression method to estimate the factor scores
Consider the joint distribution of \((\mathbf{y}_j - \mathbf{\mu})\) and \(\mathbf{f}_j\) assuming multivariate normality, as in the maximum likelihood approach. then,
\[ \left( \begin{array} {c} \mathbf{y}_j - \mathbf{\mu} \\ \mathbf{f}_j \end{array} \right) \sim N_{p + m} \left( \left[ \begin{array} {cc} \mathbf{LL}' + \mathbf{\Psi} & \mathbf{L} \\ \mathbf{L}' & \mathbf{I}_{m\times m} \end{array} \right] \right) \]
when the \(m\) factor model is correct
Hence,
\[ E(\mathbf{f}_j | \mathbf{y}_j - \mathbf{\mu}) = \mathbf{L}' (\mathbf{LL}' + \mathbf{\Psi})^{-1}(\mathbf{y}_j - \mathbf{\mu}) \]
notice that \(\mathbf{L}' (\mathbf{LL}' + \mathbf{\Psi})^{-1}\) is an \(m \times p\) matrix of regression coefficients
Then, we use the estimated conditional mean vector to estimate the factor scores
\[ \mathbf{\hat{f}}_j = \mathbf{\hat{L}}'(\mathbf{\hat{L}}\mathbf{\hat{L}}' + \mathbf{\hat{\Psi}})^{-1}(\mathbf{y}_j - \mathbf{\bar{y}}) \]
Alternatively, we could reduce the effect of possible incorrect determination fo the number of factors \(m\) by using \(\mathbf{S}\) as a substitute for \(\mathbf{\hat{L}}\mathbf{\hat{L}}' + \mathbf{\hat{\Psi}}\) then
\[ \mathbf{\hat{f}}_j = \mathbf{\hat{L}}'\mathbf{S}^{-1}(\mathbf{y}_j - \mathbf{\bar{y}}) \]
where \(j = 1,\dots,n\)
22.3.4 Model Diagnostic
Plots
Check for outliers (recall that \(\mathbf{f}_j \sim iid N(\mathbf{0}, \mathbf{I}_{m \times m})\))
Check for multivariate normality assumption
Use univariate tests for normality to check the factor scores
Confirmatory Factor Analysis: formal testing of hypotheses about loadings, use MLE and full/reduced model testing paradigm and measures of model fit
22.3.5 Application
In the psych package,
h2 = the communalities
u2 = the uniqueness
com = the complexity
library(psych)
library(tidyverse)
## Load the data from the psych package
data(Harman.5)
Harman.5
#> population schooling employment professional housevalue
#> Tract1 5700 12.8 2500 270 25000
#> Tract2 1000 10.9 600 10 10000
#> Tract3 3400 8.8 1000 10 9000
#> Tract4 3800 13.6 1700 140 25000
#> Tract5 4000 12.8 1600 140 25000
#> Tract6 8200 8.3 2600 60 12000
#> Tract7 1200 11.4 400 10 16000
#> Tract8 9100 11.5 3300 60 14000
#> Tract9 9900 12.5 3400 180 18000
#> Tract10 9600 13.7 3600 390 25000
#> Tract11 9600 9.6 3300 80 12000
#> Tract12 9400 11.4 4000 100 13000
# Correlation matrix
cor_mat <- cor(Harman.5)
cor_mat
#> population schooling employment professional housevalue
#> population 1.00000000 0.00975059 0.9724483 0.4388708 0.02241157
#> schooling 0.00975059 1.00000000 0.1542838 0.6914082 0.86307009
#> employment 0.97244826 0.15428378 1.0000000 0.5147184 0.12192599
#> professional 0.43887083 0.69140824 0.5147184 1.0000000 0.77765425
#> housevalue 0.02241157 0.86307009 0.1219260 0.7776543 1.00000000
## Principal Component Method with Correlation
cor_pca <- prcomp(Harman.5, scale = T)
# eigen values
cor_results <- data.frame(eigen_values = cor_pca$sdev ^ 2)
cor_results <- cor_results %>%
mutate(
proportion = eigen_values / sum(eigen_values),
cumulative = cumsum(proportion),
number = row_number()
)
cor_results
#> eigen_values proportion cumulative number
#> 1 2.87331359 0.574662719 0.5746627 1
#> 2 1.79666009 0.359332019 0.9339947 2
#> 3 0.21483689 0.042967377 0.9769621 3
#> 4 0.09993405 0.019986811 0.9969489 4
#> 5 0.01525537 0.003051075 1.0000000 5
# Scree plot of Eigenvalues
scree_gg <- ggplot(cor_results, aes(x = number, y = eigen_values)) +
geom_line(alpha = 0.5) +
geom_text(aes(label = number)) +
scale_x_continuous(name = "Number") +
scale_y_continuous(name = "Eigenvalue") +
theme_bw()
scree_gg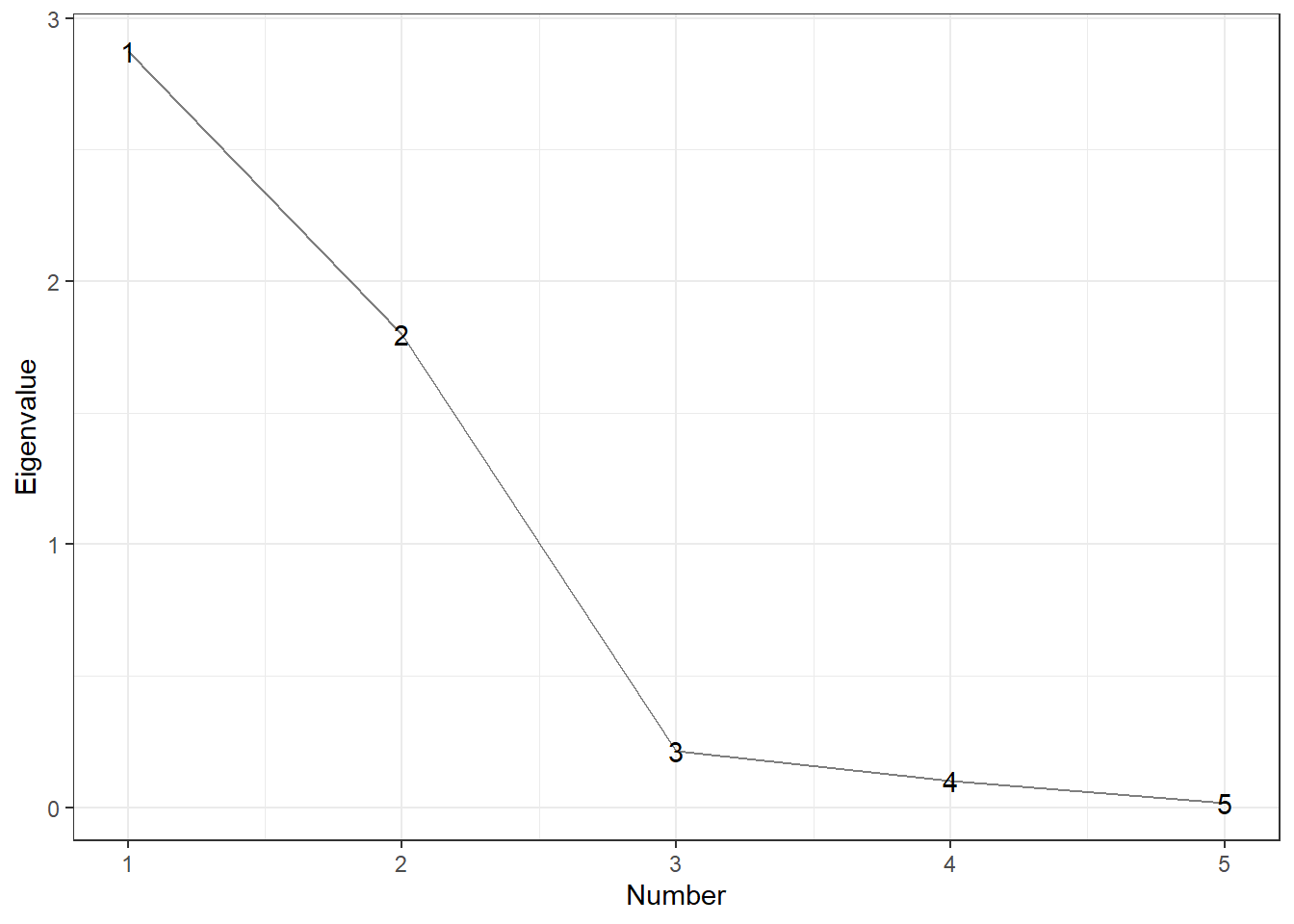
screeplot(cor_pca, type = 'lines')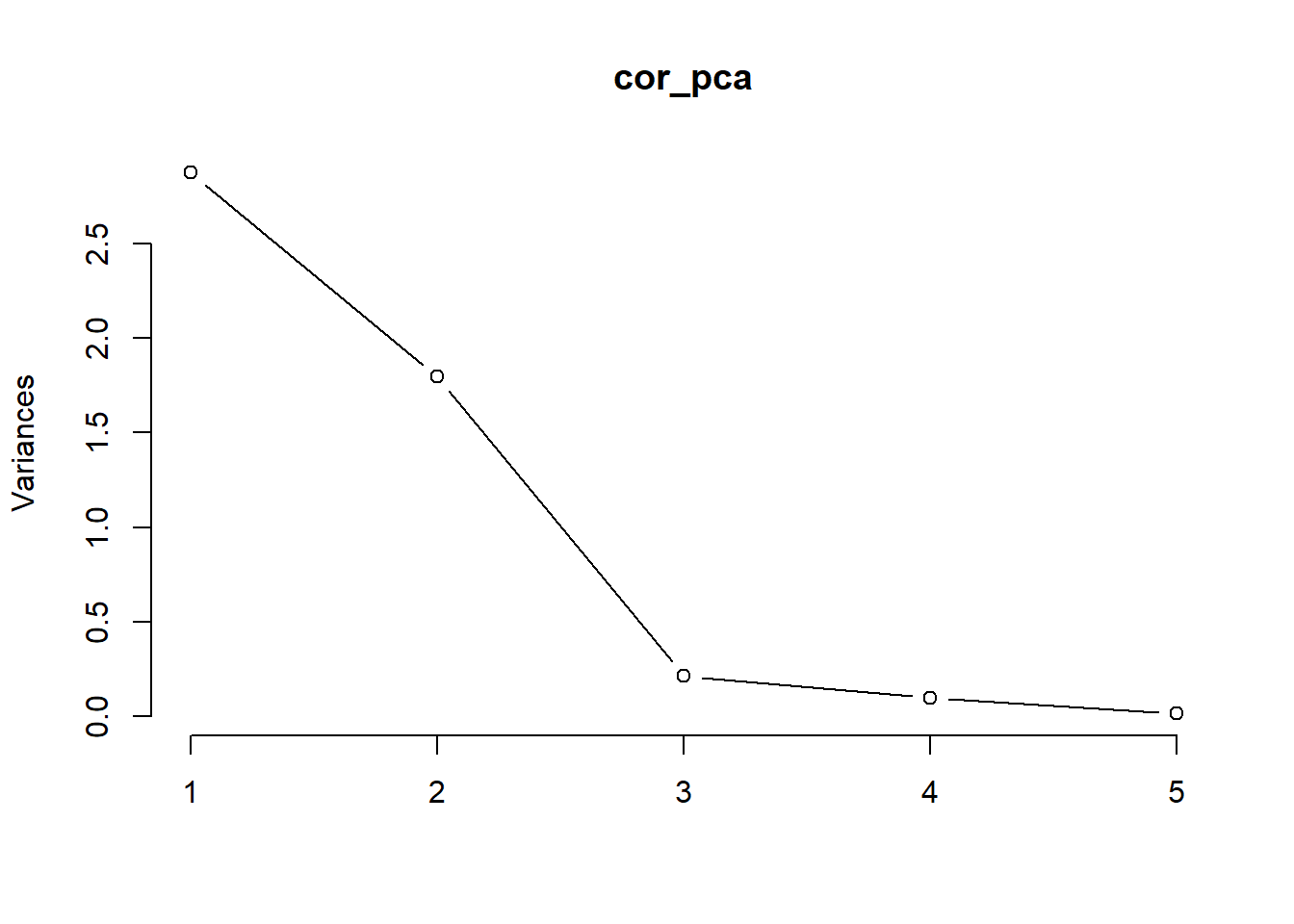
## Keep 2 factors based on scree plot and eigenvalues
factor_pca <- principal(Harman.5, nfactors = 2, rotate = "none")
factor_pca
#> Principal Components Analysis
#> Call: principal(r = Harman.5, nfactors = 2, rotate = "none")
#> Standardized loadings (pattern matrix) based upon correlation matrix
#> PC1 PC2 h2 u2 com
#> population 0.58 0.81 0.99 0.012 1.8
#> schooling 0.77 -0.54 0.89 0.115 1.8
#> employment 0.67 0.73 0.98 0.021 2.0
#> professional 0.93 -0.10 0.88 0.120 1.0
#> housevalue 0.79 -0.56 0.94 0.062 1.8
#>
#> PC1 PC2
#> SS loadings 2.87 1.80
#> Proportion Var 0.57 0.36
#> Cumulative Var 0.57 0.93
#> Proportion Explained 0.62 0.38
#> Cumulative Proportion 0.62 1.00
#>
#> Mean item complexity = 1.7
#> Test of the hypothesis that 2 components are sufficient.
#>
#> The root mean square of the residuals (RMSR) is 0.03
#> with the empirical chi square 0.29 with prob < 0.59
#>
#> Fit based upon off diagonal values = 1
# factor 1 = overall socioeconomic health
# factor 2 = contrast of the population and employment against school and house value
## Ssquared multiple correlation (SMC) prior, no rotation
factor_pca_smc <- fa(
Harman.5,
nfactors = 2,
fm = "pa",
rotate = "none",
SMC = TRUE
)
factor_pca_smc
#> Factor Analysis using method = pa
#> Call: fa(r = Harman.5, nfactors = 2, rotate = "none", SMC = TRUE, fm = "pa")
#> Standardized loadings (pattern matrix) based upon correlation matrix
#> PA1 PA2 h2 u2 com
#> population 0.62 0.78 1.00 -0.0027 1.9
#> schooling 0.70 -0.53 0.77 0.2277 1.9
#> employment 0.70 0.68 0.96 0.0413 2.0
#> professional 0.88 -0.15 0.80 0.2017 1.1
#> housevalue 0.78 -0.60 0.96 0.0361 1.9
#>
#> PA1 PA2
#> SS loadings 2.76 1.74
#> Proportion Var 0.55 0.35
#> Cumulative Var 0.55 0.90
#> Proportion Explained 0.61 0.39
#> Cumulative Proportion 0.61 1.00
#>
#> Mean item complexity = 1.7
#> Test of the hypothesis that 2 factors are sufficient.
#>
#> df null model = 10 with the objective function = 6.38 with Chi Square = 54.25
#> df of the model are 1 and the objective function was 0.34
#>
#> The root mean square of the residuals (RMSR) is 0.01
#> The df corrected root mean square of the residuals is 0.03
#>
#> The harmonic n.obs is 12 with the empirical chi square 0.02 with prob < 0.88
#> The total n.obs was 12 with Likelihood Chi Square = 2.44 with prob < 0.12
#>
#> Tucker Lewis Index of factoring reliability = 0.596
#> RMSEA index = 0.336 and the 90 % confidence intervals are 0 0.967
#> BIC = -0.04
#> Fit based upon off diagonal values = 1
## SMC prior, Promax rotation
factor_pca_smc_pro <- fa(
Harman.5,
nfactors = 2,
fm = "pa",
rotate = "Promax",
SMC = TRUE
)
factor_pca_smc_pro
#> Factor Analysis using method = pa
#> Call: fa(r = Harman.5, nfactors = 2, rotate = "Promax", SMC = TRUE,
#> fm = "pa")
#> Standardized loadings (pattern matrix) based upon correlation matrix
#> PA1 PA2 h2 u2 com
#> population -0.11 1.02 1.00 -0.0027 1.0
#> schooling 0.90 -0.11 0.77 0.2277 1.0
#> employment 0.02 0.97 0.96 0.0413 1.0
#> professional 0.75 0.33 0.80 0.2017 1.4
#> housevalue 1.01 -0.14 0.96 0.0361 1.0
#>
#> PA1 PA2
#> SS loadings 2.38 2.11
#> Proportion Var 0.48 0.42
#> Cumulative Var 0.48 0.90
#> Proportion Explained 0.53 0.47
#> Cumulative Proportion 0.53 1.00
#>
#> With factor correlations of
#> PA1 PA2
#> PA1 1.00 0.25
#> PA2 0.25 1.00
#>
#> Mean item complexity = 1.1
#> Test of the hypothesis that 2 factors are sufficient.
#>
#> df null model = 10 with the objective function = 6.38 with Chi Square = 54.25
#> df of the model are 1 and the objective function was 0.34
#>
#> The root mean square of the residuals (RMSR) is 0.01
#> The df corrected root mean square of the residuals is 0.03
#>
#> The harmonic n.obs is 12 with the empirical chi square 0.02 with prob < 0.88
#> The total n.obs was 12 with Likelihood Chi Square = 2.44 with prob < 0.12
#>
#> Tucker Lewis Index of factoring reliability = 0.596
#> RMSEA index = 0.336 and the 90 % confidence intervals are 0 0.967
#> BIC = -0.04
#> Fit based upon off diagonal values = 1
## SMC prior, varimax rotation
factor_pca_smc_var <- fa(
Harman.5,
nfactors = 2,
fm = "pa",
rotate = "varimax",
SMC = TRUE
)
## Make a data frame of the loadings for ggplot2
factors_df <-
bind_rows(
data.frame(
y = rownames(factor_pca_smc$loadings),
unclass(factor_pca_smc$loadings)
),
data.frame(
y = rownames(factor_pca_smc_pro$loadings),
unclass(factor_pca_smc_pro$loadings)
),
data.frame(
y = rownames(factor_pca_smc_var$loadings),
unclass(factor_pca_smc_var$loadings)
),
.id = "Rotation"
)
flag_gg <- ggplot(factors_df) +
geom_vline(aes(xintercept = 0)) +
geom_hline(aes(yintercept = 0)) +
geom_point(aes(
x = PA2,
y = PA1,
col = y,
shape = y
), size = 2) +
scale_x_continuous(name = "Factor 2", limits = c(-1.1, 1.1)) +
scale_y_continuous(name = "Factor1", limits = c(-1.1, 1.1)) +
facet_wrap("Rotation", labeller = labeller(Rotation = c(
"1" = "Original", "2" = "Promax", "3" = "Varimax"
))) +
coord_fixed(ratio = 1) # make aspect ratio of each facet 1
flag_gg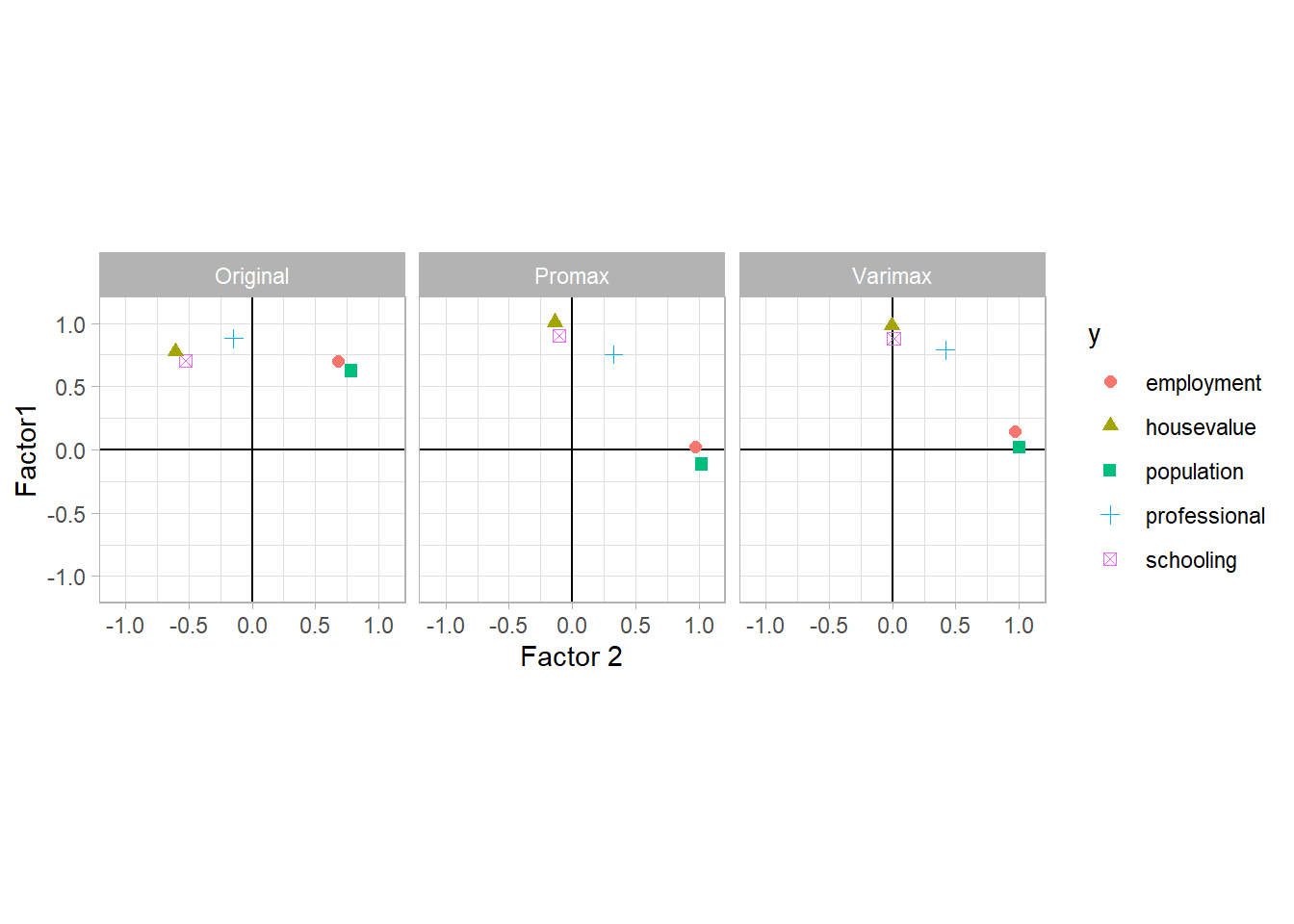
# promax and varimax did a good job to assign trait to a particular factor
factor_mle_1 <- fa(
Harman.5,
nfactors = 1,
fm = "mle",
rotate = "none",
SMC = TRUE
)
factor_mle_1
#> Factor Analysis using method = ml
#> Call: fa(r = Harman.5, nfactors = 1, rotate = "none", SMC = TRUE, fm = "mle")
#> Standardized loadings (pattern matrix) based upon correlation matrix
#> ML1 h2 u2 com
#> population 0.97 0.950 0.0503 1
#> schooling 0.14 0.021 0.9791 1
#> employment 1.00 0.995 0.0049 1
#> professional 0.51 0.261 0.7388 1
#> housevalue 0.12 0.014 0.9864 1
#>
#> ML1
#> SS loadings 2.24
#> Proportion Var 0.45
#>
#> Mean item complexity = 1
#> Test of the hypothesis that 1 factor is sufficient.
#>
#> df null model = 10 with the objective function = 6.38 with Chi Square = 54.25
#> df of the model are 5 and the objective function was 3.14
#>
#> The root mean square of the residuals (RMSR) is 0.41
#> The df corrected root mean square of the residuals is 0.57
#>
#> The harmonic n.obs is 12 with the empirical chi square 39.41 with prob < 2e-07
#> The total n.obs was 12 with Likelihood Chi Square = 24.56 with prob < 0.00017
#>
#> Tucker Lewis Index of factoring reliability = 0.022
#> RMSEA index = 0.564 and the 90 % confidence intervals are 0.374 0.841
#> BIC = 12.14
#> Fit based upon off diagonal values = 0.5
#> Measures of factor score adequacy
#> ML1
#> Correlation of (regression) scores with factors 1.00
#> Multiple R square of scores with factors 1.00
#> Minimum correlation of possible factor scores 0.99
factor_mle_2 <- fa(
Harman.5,
nfactors = 2,
fm = "mle",
rotate = "none",
SMC = TRUE
)
factor_mle_2
#> Factor Analysis using method = ml
#> Call: fa(r = Harman.5, nfactors = 2, rotate = "none", SMC = TRUE, fm = "mle")
#> Standardized loadings (pattern matrix) based upon correlation matrix
#> ML2 ML1 h2 u2 com
#> population -0.03 1.00 1.00 0.005 1.0
#> schooling 0.90 0.04 0.81 0.193 1.0
#> employment 0.09 0.98 0.96 0.036 1.0
#> professional 0.78 0.46 0.81 0.185 1.6
#> housevalue 0.96 0.05 0.93 0.074 1.0
#>
#> ML2 ML1
#> SS loadings 2.34 2.16
#> Proportion Var 0.47 0.43
#> Cumulative Var 0.47 0.90
#> Proportion Explained 0.52 0.48
#> Cumulative Proportion 0.52 1.00
#>
#> Mean item complexity = 1.1
#> Test of the hypothesis that 2 factors are sufficient.
#>
#> df null model = 10 with the objective function = 6.38 with Chi Square = 54.25
#> df of the model are 1 and the objective function was 0.31
#>
#> The root mean square of the residuals (RMSR) is 0.01
#> The df corrected root mean square of the residuals is 0.05
#>
#> The harmonic n.obs is 12 with the empirical chi square 0.05 with prob < 0.82
#> The total n.obs was 12 with Likelihood Chi Square = 2.22 with prob < 0.14
#>
#> Tucker Lewis Index of factoring reliability = 0.658
#> RMSEA index = 0.307 and the 90 % confidence intervals are 0 0.945
#> BIC = -0.26
#> Fit based upon off diagonal values = 1
#> Measures of factor score adequacy
#> ML2 ML1
#> Correlation of (regression) scores with factors 0.98 1.00
#> Multiple R square of scores with factors 0.95 1.00
#> Minimum correlation of possible factor scores 0.91 0.99
factor_mle_3 <- fa(
Harman.5,
nfactors = 3,
fm = "mle",
rotate = "none",
SMC = TRUE
)
factor_mle_3
#> Factor Analysis using method = ml
#> Call: fa(r = Harman.5, nfactors = 3, rotate = "none", SMC = TRUE, fm = "mle")
#> Standardized loadings (pattern matrix) based upon correlation matrix
#> ML2 ML1 ML3 h2 u2 com
#> population -0.12 0.98 -0.11 0.98 0.0162 1.1
#> schooling 0.89 0.15 0.29 0.90 0.0991 1.3
#> employment 0.00 1.00 0.04 0.99 0.0052 1.0
#> professional 0.72 0.52 -0.10 0.80 0.1971 1.9
#> housevalue 0.97 0.13 -0.09 0.97 0.0285 1.1
#>
#> ML2 ML1 ML3
#> SS loadings 2.28 2.26 0.11
#> Proportion Var 0.46 0.45 0.02
#> Cumulative Var 0.46 0.91 0.93
#> Proportion Explained 0.49 0.49 0.02
#> Cumulative Proportion 0.49 0.98 1.00
#>
#> Mean item complexity = 1.2
#> Test of the hypothesis that 3 factors are sufficient.
#>
#> df null model = 10 with the objective function = 6.38 with Chi Square = 54.25
#> df of the model are -2 and the objective function was 0
#>
#> The root mean square of the residuals (RMSR) is 0
#> The df corrected root mean square of the residuals is NA
#>
#> The harmonic n.obs is 12 with the empirical chi square 0 with prob < NA
#> The total n.obs was 12 with Likelihood Chi Square = 0 with prob < NA
#>
#> Tucker Lewis Index of factoring reliability = 1.318
#> Fit based upon off diagonal values = 1
#> Measures of factor score adequacy
#> ML2 ML1 ML3
#> Correlation of (regression) scores with factors 0.99 1.00 0.82
#> Multiple R square of scores with factors 0.98 1.00 0.68
#> Minimum correlation of possible factor scores 0.96 0.99 0.36The output info for the null hypothesis of no common factors is in the statement “The degrees of freedom for the null model ..”
The output info for the null hypothesis that number of factors is sufficient is in the statement “The total number of observations was …”
One factor is not enough, two is sufficient, and not enough data for 3 factors (df of -2 and NA for p-value). Hence, we should use 2-factor model.
22.4 Discriminant Analysis
Suppose we have two or more different populations from which observations could come from. Discriminant analysis seeks to determine which of the possible population an observation comes from while making as few mistakes as possible
-
This is an alternative to logistic approaches with the following advantages:
when there is clear separation between classes, the parameter estimates for the logic regression model can be surprisingly unstable, while discriminant approaches do not suffer
If X is normal in each of the classes and the sample size is small, then discriminant approaches can be more accurate
Notation
Similar to MANOVA, let \(\mathbf{y}_{j1},\mathbf{y}_{j2},\dots, \mathbf{y}_{in_j} \sim iid f_j (\mathbf{y})\) for \(j = 1,\dots, h\)
Let \(f_j(\mathbf{y})\) be the density function for population j . Note that each vector \(\mathbf{y}\) contain measurements on all \(p\) traits
- Assume that each observation is from one of \(h\) possible populations.
- We want to form a discriminant rule that will allocate an observation \(\mathbf{y}\) to population j when \(\mathbf{y}\) is in fact from this population
22.4.1 Known Populations
The maximum likelihood discriminant rule for assigning an observation \(\mathbf{y}\) to one of the \(h\) populations allocates \(\mathbf{y}\) to the population that gives the largest likelihood to \(\mathbf{y}\)
Consider the likelihood for a single observation \(\mathbf{y}\), which has the form \(f_j (\mathbf{y})\) where j is the true population.
Since \(j\) is unknown, to make the likelihood as large as possible, we should choose the value j which causes \(f_j (\mathbf{y})\) to be as large as possible
Consider a simple univariate example. Suppose we have data from one of two binomial populations.
The first population has \(n= 10\) trials with success probability \(p = .5\)
The second population has \(n= 10\) trials with success probability \(p = .7\)
to which population would we assign an observation of \(y = 7\)
-
Note:
\(f(y = 7|n = 10, p = .5) = .117\)
\(f(y = 7|n = 10, p = .7) = .267\) where \(f(.)\) is the binomial likelihood.
Hence, we choose the second population
Another example
We have 2 populations, where
First population: \(N(\mu_1, \sigma^2_1)\)
Second population: \(N(\mu_2, \sigma^2_2)\)
The likelihood for a single observation is
\[ f_j (y) = (2\pi \sigma^2_j)^{-1/2} \exp\{ -\frac{1}{2}(\frac{y - \mu_j}{\sigma_j})^2\} \]
Consider a likelihood ratio rule
\[ \begin{aligned} \Lambda &= \frac{\text{likelihood of y from pop 1}}{\text{likelihood of y from pop 2}} \\ &= \frac{f_1(y)}{f_2(y)} \\ &= \frac{\sigma_2}{\sigma_1} \exp\{-\frac{1}{2}[(\frac{y - \mu_1}{\sigma_1})^2- (\frac{y - \mu_2}{\sigma_2})^2] \} \end{aligned} \]
Hence, we classify into
pop 1 if \(\Lambda >1\)
pop 2 if \(\Lambda <1\)
for ties, flip a coin
Another way to think:
we classify into population 1 if the “standardized distance” of y from \(\mu_1\) is less than the “standardized distance” of y from \(\mu_2\) which is referred to as a quadratic discriminant rule.
(Significant simplification occurs in th special case where \(\sigma_1 = \sigma_2 = \sigma^2\))
Thus, we classify into population 1 if
\[ (y - \mu_2)^2 > (y - \mu_1)^2 \]
or
\[ |y- \mu_2| > |y - \mu_1| \]
and
\[ -2 \log (\Lambda) = -2y \frac{(\mu_1 - \mu_2)}{\sigma^2} + \frac{(\mu_1^2 - \mu_2^2)}{\sigma^2} = \beta y + \alpha \]
Thus, we classify into population 1 if this is less than 0.
Discriminant classification rule is linear in y in this case.
22.4.1.1 Multivariate Expansion
Suppose that there are 2 populations
\(N_p(\mathbf{\mu}_1, \mathbf{\Sigma}_1)\)
\(N_p(\mathbf{\mu}_2, \mathbf{\Sigma}_2)\)
\[ \begin{aligned} -2 \log(\frac{f_1 (\mathbf{x})}{f_2 (\mathbf{x})}) &= \log|\mathbf{\Sigma}_1| + (\mathbf{x} - \mathbf{\mu}_1)' \mathbf{\Sigma}^{-1}_1 (\mathbf{x} - \mathbf{\mu}_1) \\ &- [\log|\mathbf{\Sigma}_2|+ (\mathbf{x} - \mathbf{\mu}_2)' \mathbf{\Sigma}^{-1}_2 (\mathbf{x} - \mathbf{\mu}_2) ] \end{aligned} \]
Again, we classify into population 1 if this is less than 0, otherwise, population 2. And like the univariate case with non-equal variances, this is a quadratic discriminant rule.
And if the covariance matrices are equal: \(\mathbf{\Sigma}_1 = \mathbf{\Sigma}_2 = \mathbf{\Sigma}_1\) classify into population 1 if
\[ (\mathbf{\mu}_1 - \mathbf{\mu}_2)' \mathbf{\Sigma}^{-1}\mathbf{x} - \frac{1}{2} (\mathbf{\mu}_1 - \mathbf{\mu}_2)' \mathbf{\Sigma}^{-1} (\mathbf{\mu}_1 - \mathbf{\mu}_2) \ge 0 \]
This linear discriminant rule is also referred to as Fisher’s linear discriminant function
By assuming the covariance matrices are equal, we assume that the shape and orientation fo the two populations must be the same (which can be a strong restriction)
In other words, for each variable, it can have different mean but the same variance.
- Note: LDA Bayes decision boundary is linear. Hence, quadratic decision boundary might lead to better classification. Moreover, the assumption of same variance/covariance matrix across all classes for Gaussian densities imposes the linear rule, if we allow the predictors in each class to follow MVN distribution with class-specific mean vectors and variance/covariance matrices, then it is Quadratic Discriminant Analysis. But then, you will have more parameters to estimate (which gives more flexibility than LDA) at the cost of more variance (bias -variance tradeoff).
When \(\mathbf{\mu}_1, \mathbf{\mu}_2, \mathbf{\Sigma}\) are known, the probability of misclassification can be determined:
\[ \begin{aligned} P(2|1) &= P(\text{calssify into pop 2| x is from pop 1}) \\ &= P((\mathbf{\mu}_1 - \mathbf{\mu}_2)' \mathbf{\Sigma}^{-1} \mathbf{x} \le \frac{1}{2} (\mathbf{\mu}_1 - \mathbf{\mu}_2)' \mathbf{\Sigma}^{-1} (\mathbf{\mu}_1 - \mathbf{\mu}_2)|\mathbf{x} \sim N(\mu_1, \mathbf{\Sigma}) \\ &= \Phi(-\frac{1}{2} \delta) \end{aligned} \]
where
\(\delta^2 = (\mathbf{\mu}_1 - \mathbf{\mu}_2)' \mathbf{\Sigma}^{-1} (\mathbf{\mu}_1 - \mathbf{\mu}_2)\)
\(\Phi\) is the standard normal CDF
Suppose there are \(h\) possible populations, which are distributed as \(N_p (\mathbf{\mu}_p, \mathbf{\Sigma})\). Then, the maximum likelihood (linear) discriminant rule allocates \(\mathbf{y}\) to population j where j minimizes the squared Mahalanobis distance
\[ (\mathbf{y} - \mathbf{\mu}_j)' \mathbf{\Sigma}^{-1} (\mathbf{y} - \mathbf{\mu}_j) \]
22.4.1.2 Bayes Discriminant Rules
If we know that population j has prior probabilities \(\pi_j\) (assume \(\pi_j >0\)) we can form the Bayes discriminant rule.
This rule allocates an observation \(\mathbf{y}\) to the population for which \(\pi_j f_j (\mathbf{y})\) is maximized.
Note:
- Maximum likelihood discriminant rule is a special case of the Bayes discriminant rule, where it sets all the \(\pi_j = 1/h\)
Optimal Properties of Bayes Discriminant Rules
let \(p_{ii}\) be the probability of correctly assigning an observation from population i
then one rule (with probabilities \(p_{ii}\) ) is as good as another rule (with probabilities \(p_{ii}'\) ) if \(p_{ii} \ge p_{ii}'\) for all \(i = 1,\dots, h\)
The first rule is better than the alternative if \(p_{ii} > p_{ii}'\) for at least one i.
A rule for which there is no better alternative is called admissible
Bayes Discriminant Rules are admissible
If we utilized prior probabilities, then we can form the posterior probability of a correct allocation, \(\sum_{i=1}^h \pi_i p_{ii}\)
Bayes Discriminant Rules have the largest possible posterior probability of correct allocation with respect to the prior
These properties show that Bayes Discriminant rule is our best approach.
Unequal Cost
-
We want to consider the cost misallocation
- Define \(c_{ij}\) to be the cost associated with allocation a member of population j to population i.
-
Assume that
\(c_{ij} >0\) for all \(i \neq j\)
\(c_{ij} = 0\) if \(i = j\)
We could determine the expected amount of loss for an observation allocated to population i as \(\sum_j c_{ij} p_{ij}\) where the \(p_{ij}s\) are the probabilities of allocating an observation from population j into population i
We want to minimize the amount of loss expected for our rule. Using a Bayes Discrimination, allocate \(\mathbf{y}\) to the population j which minimizes \(\sum_{k \neq j} c_{ij} \pi_k f_k(\mathbf{y})\)
We could assign equal probabilities to each group and get a maximum likelihood type rule. here, we would allocate \(\mathbf{y}\) to population j which minimizes \(\sum_{k \neq j}c_{jk} f_k(\mathbf{y})\)
Example:
Two binomial populations, each of size 10, with probabilities \(p_1 = .5\) and \(p_2 = .7\)
And the probability of being in the first population is .9
However, suppose the cost of inappropriately allocating into the first population is 1 and the cost of incorrectly allocating into the second population is 5.
In this case, we pick population 1 over population 2
In general, we consider two regions, \(R_1\) and \(R_2\) associated with population 1 and 2:
\[ R_1: \frac{f_1 (\mathbf{x})}{f_2 (\mathbf{x})} \ge \frac{c_{12} \pi_2}{c_{21} \pi_1} \]
\[ R_2: \frac{f_1 (\mathbf{x})}{f_2 (\mathbf{x})} < \frac{c_{12} \pi_2}{c_{21} \pi_1} \]
where \(c_{12}\) is the cost of assigning a member of population 2 to population 1.
22.4.1.3 Discrimination Under Estimation
Suppose we know the form of the distributions for populations of interests, but we still have to estimate the parameters.
Example:
we know the distributions are multivariate normal, but we have to estimate the means and variances
The maximum likelihood discriminant rule allocates an observation \(\mathbf{y}\) to population j when j maximizes the function
\[ f_j (\mathbf{y} |\hat{\theta}) \]
where \(\hat{\theta}\) are the maximum likelihood estimates of the unknown parameters
For instance, we have 2 multivariate normal populations with distinct means, but common variance covariance matrix
MLEs for \(\mathbf{\mu}_1\) and \(\mathbf{\mu}_2\) are \(\mathbf{\bar{y}}_1\) and \(\mathbf{\bar{y}}_2\)and common \(\mathbf{\Sigma}\) is \(\mathbf{S}\).
Thus, an estimated discriminant rule could be formed by substituting these sample values for the population values
22.4.1.4 Native Bayes
The challenge with classification using Bayes’ is that we don’t know the (true) densities, \(f_k, k = 1, \dots, K\), while LDA and QDA make strong multivariate normality assumptions to deal with this.
Naive Bayes makes only one assumption: within the k-th class, the p predictors are independent (i.e,, for \(k = 1,\dots, K\)
\[ f_k(x) = f_{k1}(x_1) \times f_{k2}(x_2) \times \dots \times f_{kp}(x_p) \]
where \(f_{kj}\) is the density function of the j-th predictor among observation in the k-th class.
This assumption allows the use of joint distribution without the need to account for dependence between observations. However, this (native) assumption can be unrealistic, but still works well in cases where the number of sample (n) is not large relative to the number of features (p).
With this assumption, we have
\[ P(Y=k|X=x) = \frac{\pi_k \times f_{k1}(x_1) \times \dots \times f_{kp}(x_p)}{\sum_{l=1}^K \pi_l \times f_{l1}(x_1)\times \dots f_{lp}(x_p)} \]
we only need to estimate the one-dimensional density function \(f_{kj}\) with either of these approaches:
When \(X_j\) is quantitative, assume it has a univariate normal distribution (with independence): \(X_j | Y = k \sim N(\mu_{jk}, \sigma^2_{jk})\) which is more restrictive than QDA because it assumes predictors are independent (e.g., a diagonal covariance matrix)
When \(X_j\) is quantitative, use a kernel density estimator Kernel Methods ; which is a smoothed histogram
When \(X_j\) is qualitative, we count the promotion of training observations for the j-th predictor corresponding to each class.
22.4.1.5 Comparison of Classification Methods
Assuming we have K classes and K is the baseline from (James , Witten, Hastie, and Tibshirani book)
Comparing the log odds relative to the K class
22.4.1.5.1 Logistic Regression
\[ \log(\frac{P(Y=k|X = x)}{P(Y = K| X = x)}) = \beta_{k0} + \sum_{j=1}^p \beta_{kj}x_j \]
22.4.1.5.2 LDA
\[ \log(\frac{P(Y = k | X = x)}{P(Y = K | X = x)} = a_k + \sum_{j=1}^p b_{kj} x_j \]
where \(a_k\) and \(b_{kj}\) are functions of \(\pi_k, \pi_K, \mu_k , \mu_K, \mathbf{\Sigma}\)
Similar to logistic regression, LDA assumes the log odds is linear in \(x\)
Even though they look like having the same form, the parameters in logistic regression are estimated by MLE, where as LDA linear parameters are specified by the prior and normal distributions
We expect LDA to outperform logistic regression when the normality assumption (approximately) holds, and logistic regression to perform better when it does not
22.4.1.5.3 QDA
\[ \log(\frac{P(Y=k|X=x}{P(Y=K | X = x}) = a_k + \sum_{j=1}^{p}b_{kj}x_{j} + \sum_{j=1}^p \sum_{l=1}^p c_{kjl}x_j x_l \]
where \(a_k, b_{kj}, c_{kjl}\) are functions \(\pi_k , \pi_K, \mu_k, \mu_K ,\mathbf{\Sigma}_k, \mathbf{\Sigma}_K\)
22.4.1.5.4 Naive Bayes
\[ \log (\frac{P(Y = k | X = x)}{P(Y = K | X = x}) = a_k + \sum_{j=1}^p g_{kj} (x_j) \]
where \(a_k = \log (\pi_k / \pi_K)\) and \(g_{kj}(x_j) = \log(\frac{f_{kj}(x_j)}{f_{Kj}(x_j)})\) which is the form of generalized additive model
22.4.1.5.5 Summary
LDA is a special case of QDA
LDA is robust when it comes to high dimensions
Any classifier with a linear decision boundary is a special case of naive Bayes with \(g_{kj}(x_j) = b_{kj} x_j\), which means LDA is a special case of naive Bayes. LDA assumes that the features are normally distributed with a common within-class covariance matrix, and naive Bayes assumes independence of the features.
Naive bayes is also a special case of LDA with \(\mathbf{\Sigma}\) restricted to a diagonal matrix with diagonals, \(\sigma^2\) (another notation \(diag (\mathbf{\Sigma})\) ) assuming \(f_{kj}(x_j) = N(\mu_{kj}, \sigma^2_j)\)
QDA and naive Bayes are not special case of each other. In principal,e naive Bayes can produce a more flexible fit by the choice of \(g_{kj}(x_j)\) , but it’s restricted to only purely additive fit, but QDA includes multiplicative terms of the form \(c_{kjl}x_j x_l\)
None of these methods uniformly dominates the others: the choice of method depends on the true distribution of the predictors in each of the K classes, n and p (i.e., related to the bias-variance tradeoff).
Compare to the non-parametric method (KNN)
KNN would outperform both LDA and logistic regression when the decision boundary is highly nonlinear, but can’t say which predictors are most important, and requires many observations
KNN is also limited in high-dimensions due to the curse of dimensionality
Since QDA is a special type of nonlinear decision boundary (quadratic), it can be considered as a compromise between the linear methods and KNN classification. QDA can have fewer training observations than KNN but not as flexible.
From simulation:
| True decision boundary | Best performance |
|---|---|
| Linear | LDA + Logistic regression |
| Moderately nonlinear | QDA + Naive Bayes |
| Highly nonlinear (many training, p is not large) | KNN |
- like linear regression, we can also introduce flexibility by including transformed features \(\sqrt{X}, X^2, X^3\)
22.4.2 Probabilities of Misclassification
When the distribution are exactly known, we can determine the misclassification probabilities exactly. however, when we need to estimate the population parameters, we have to estimate the probability of misclassification
-
Naive method
Plugging the parameters estimates into the form for the misclassification probabilities results to derive at the estimates of the misclassification probability.
But this will tend to be optimistic when the number of samples in one or more populations is small.
-
Resubstitution method
Use the proportion of the samples from population i that would be allocated to another population as an estimate of the misclassification probability
But also optimistic when the number of samples is small
-
Jack-knife estimates:
The above two methods use observation to estimate both parameters and also misclassification probabilities based upon the discriminant rule
Alternatively, we determine the discriminant rule based upon all of the data except the k-th observation from the j-th population
then, determine if the k-th observation would be misclassified under this rule
perform this process for all \(n_j\) observation in population j . An estimate fo the misclassification probability would be the fraction of \(n_j\) observations which were misclassified
repeat the process for other \(i \neq j\) populations
This method is more reliable than the others, but also computationally intensive
Cross-Validation
Summary
Consider the group-specific densities \(f_j (\mathbf{x})\) for multivariate vector \(\mathbf{x}\).
Assume equal misclassifications costs, the Bayes classification probability of \(\mathbf{x}\) belonging to the j-th population is
\[ p(j |\mathbf{x}) = \frac{\pi_j f_j (\mathbf{x})}{\sum_{k=1}^h \pi_k f_k (\mathbf{x})} \]
\(j = 1,\dots, h\)
where there are \(h\) possible groups.
We then classify into the group for which this probability of membership is largest
Alternatively, we can write this in terms of a generalized squared distance formation
\[ D_j^2 (\mathbf{x}) = d_j^2 (\mathbf{x})+ g_1(j) + g_2 (j) \]
where
-
\(d_j^2(\mathbf{x}) = (\mathbf{x} - \mathbf{\mu}_j)' \mathbf{V}_j^{-1} (\mathbf{x} - \mathbf{\mu}_j)\) is the squared Mahalanobis distance from \(\mathbf{x}\) to the centroid of group j, and
\(\mathbf{V}_j = \mathbf{S}_j\) if the within group covariance matrices are not equal
\(\mathbf{V}_j = \mathbf{S}_p\) if a pooled covariance estimate is appropriate
and
\[ g_1(j) = \begin{cases} \ln |\mathbf{S}_j| & \text{within group covariances are not equal} \\ 0 & \text{pooled covariance} \end{cases} \]
\[ g_2(j) = \begin{cases} -2 \ln \pi_j & \text{prior probabilities are not equal} \\ 0 & \text{prior probabilities are equal} \end{cases} \]
then, the posterior probability of belonging to group j is
\[ p(j| \mathbf{x}) = \frac{\exp(-.5 D_j^2(\mathbf{x}))}{\sum_{k=1}^h \exp(-.5 D^2_k (\mathbf{x}))} \]
where \(j = 1,\dots , h\)
and \(\mathbf{x}\) is classified into group j if \(p(j | \mathbf{x})\) is largest for \(j = 1,\dots,h\) (or, \(D_j^2(\mathbf{x})\) is smallest).
22.4.2.1 Assessing Classification Performance
For binary classification, confusion matrix
| Predicted class | ||||
|---|---|---|---|---|
| - or Null | + or Null | Total | ||
| True Class | - or Null | True Neg (TN) | False Pos (FP) | N |
| + or Null | False Neg (FN) | True Pos (TP) | P | |
| Total | N* | P* |
and table 4.6 from (James et al. 2013)
| Name | Definition | Synonyms |
|---|---|---|
| False Pos rate | FP/N | Type I error, 1 0 Specificity |
| True Pos. rate | TP/P | 1 - Type II error, power, sensitivity, recall |
| Pos Pred. value | TP/P* | Precision, 1 - false discovery promotion |
| Neg. Pred. value | TN/N* |
ROC curve (receiver Operating Characteristics) is a graphical comparison between sensitivity (true positive) and specificity ( = 1 - false positive)
y-axis = true positive rate
x-axis = false positive rate
as we change the threshold rate for classifying an observation as from 0 to 1
AUC (area under the ROC) ideally would equal to 1, a bad classifier would have AUC = 0.5 (pure chance)
22.4.3 Unknown Populations/ Nonparametric Discrimination
When your multivariate data are not Gaussian, or known distributional form at all, we can use the following methods
22.4.3.1 Kernel Methods
We approximate \(f_j (\mathbf{x})\) by a kernel density estimate
\[ \hat{f}_j(\mathbf{x}) = \frac{1}{n_j} \sum_{i = 1}^{n_j} K_j (\mathbf{x} - \mathbf{x}_i) \]
where
\(K_j (.)\) is a kernel function satisfying \(\int K_j(\mathbf{z})d\mathbf{z} =1\)
\(\mathbf{x}_i\) , \(i = 1,\dots , n_j\) is a random sample from the j-th population.
Thus, after finding \(\hat{f}_j (\mathbf{x})\) for each of the \(h\) populations, the posterior probability of group membership is
\[ p(j |\mathbf{x}) = \frac{\pi_j \hat{f}_j (\mathbf{x})}{\sum_{k-1}^h \pi_k \hat{f}_k (\mathbf{x})} \]
where \(j = 1,\dots, h\)
There are different choices for the kernel function:
Uniform
Normal
Epanechnikov
Biweight
Triweight
We these kernels, we have to pick the “radius” (or variance, width, window width, bandwidth) of the kernel, which is a smoothing parameter (the larger the radius, the more smooth the kernel estimate of the density).
To select the smoothness parameter, we can use the following method
If we believe the populations were close to multivariate normal, then
\[ R = (\frac{4/(2p+1)}{n_j})^{1/(p+1} \]
But since we do not know for sure, we might choose several different values and select one that vies the best out of sample or cross-validation discrimination.
Moreover, you also have to decide whether to use different kernel smoothness for different populations, which is similar to the individual and pooled covariances in the classical methodology.
22.4.3.2 Nearest Neighbor Methods
The nearest neighbor (also known as k-nearest neighbor) method performs the classification of a new observation vector based on the group membership of its nearest neighbors. In practice, we find
\[ d_{ij}^2 (\mathbf{x}, \mathbf{x}_i) = (\mathbf{x}, \mathbf{x}_i) V_j^{-1}(\mathbf{x}, \mathbf{x}_i) \]
which is the distance between the vector \(\mathbf{x}\) and the \(i\)-th observation in group \(j\)
We consider different choices for \(\mathbf{V}_j\)
For example,
\[ \begin{aligned} \mathbf{V}_j &= \mathbf{S}_p \\ \mathbf{V}_j &= \mathbf{S}_j \\ \mathbf{V}_j &= \mathbf{I} \\ \mathbf{V}_j &= diag (\mathbf{S}_p) \end{aligned} \]
We find the \(k\) observations that are closest to \(\mathbf{x}\) (where users pick \(k\)). Then we classify into the most common population, weighted by the prior.
22.4.3.3 Modern Discriminant Methods
Note:
Logistic regression (with or without random effects) is a flexible model-based procedure for classification between two populations.
The extension of logistic regression to the multi-group setting is polychotomous logistic regression (or, mulinomial regression).
The machine learning and pattern recognition are growing with strong focus on nonlinear discriminant analysis methods such as:
radial basis function networks
support vector machines
multiplayer perceptrons (neural networks)
The general framework
\[ g_j (\mathbf{x}) = \sum_{l = 1}^m w_{jl}\phi_l (\mathbf{x}; \mathbf{\theta}_l) + w_{j0} \]
where
\(j = 1,\dots, h\)
\(m\) nonlinear basis functions \(\phi_l\), each of which has \(n_m\) parameters given by \(\theta_l = \{ \theta_{lk}: k = 1, \dots , n_m \}\)
We assign \(\mathbf{x}\) to the \(j\)-th population if \(g_j(\mathbf{x})\) is the maximum for all \(j = 1,\dots, h\)
Development usually focuses on the choice and estimation of the basis functions, \(\phi_l\) and the estimation of the weights \(w_{jl}\)
More details can be found (Webb, Copsey, and Cawley 2011)
22.4.4 Application
library(class)
library(klaR)
library(MASS)
library(tidyverse)
## Read in the data
crops <- read.table("images/crops.txt")
names(crops) <- c("crop", "y1", "y2", "y3", "y4")
str(crops)
#> 'data.frame': 36 obs. of 5 variables:
#> $ crop: chr "Corn" "Corn" "Corn" "Corn" ...
#> $ y1 : int 16 15 16 18 15 15 12 20 24 21 ...
#> $ y2 : int 27 23 27 20 15 32 15 23 24 25 ...
#> $ y3 : int 31 30 27 25 31 32 16 23 25 23 ...
#> $ y4 : int 33 30 26 23 32 15 73 25 32 24 ...
## Read in test data
crops_test <- read.table("images/crops_test.txt")
names(crops_test) <- c("crop", "y1", "y2", "y3", "y4")
str(crops_test)
#> 'data.frame': 5 obs. of 5 variables:
#> $ crop: chr "Corn" "Soybeans" "Cotton" "Sugarbeets" ...
#> $ y1 : int 16 21 29 54 32
#> $ y2 : int 27 25 24 23 32
#> $ y3 : int 31 23 26 21 62
#> $ y4 : int 33 24 28 54 1622.4.4.1 LDA
Default prior is proportional to sample size and lda and qda do not fit a constant or intercept term
## Linear discriminant analysis
lda_mod <- lda(crop ~ y1 + y2 + y3 + y4,
data = crops)
lda_mod
#> Call:
#> lda(crop ~ y1 + y2 + y3 + y4, data = crops)
#>
#> Prior probabilities of groups:
#> Clover Corn Cotton Soybeans Sugarbeets
#> 0.3055556 0.1944444 0.1666667 0.1666667 0.1666667
#>
#> Group means:
#> y1 y2 y3 y4
#> Clover 46.36364 32.63636 34.18182 36.63636
#> Corn 15.28571 22.71429 27.42857 33.14286
#> Cotton 34.50000 32.66667 35.00000 39.16667
#> Soybeans 21.00000 27.00000 23.50000 29.66667
#> Sugarbeets 31.00000 32.16667 20.00000 40.50000
#>
#> Coefficients of linear discriminants:
#> LD1 LD2 LD3 LD4
#> y1 -6.147360e-02 0.009215431 -0.02987075 -0.014680566
#> y2 -2.548964e-02 0.042838972 0.04631489 0.054842132
#> y3 1.642126e-02 -0.079471595 0.01971222 0.008938745
#> y4 5.143616e-05 -0.013917423 0.05381787 -0.025717667
#>
#> Proportion of trace:
#> LD1 LD2 LD3 LD4
#> 0.7364 0.1985 0.0576 0.0075
## Look at accuracy on the training data
lda_fitted <- predict(lda_mod,newdata = crops)
# Contingency table
lda_table <- table(truth = crops$crop, fitted = lda_fitted$class)
lda_table
#> fitted
#> truth Clover Corn Cotton Soybeans Sugarbeets
#> Clover 6 0 3 0 2
#> Corn 0 6 0 1 0
#> Cotton 3 0 1 2 0
#> Soybeans 0 1 1 3 1
#> Sugarbeets 1 1 0 2 2
# accuracy of 0.5 is just random (not good)
## Posterior probabilities of membership
crops_post <- cbind.data.frame(crops,
crop_pred = lda_fitted$class,
lda_fitted$posterior)
crops_post <- crops_post %>%
mutate(missed = crop != crop_pred)
head(crops_post)
#> crop y1 y2 y3 y4 crop_pred Clover Corn Cotton Soybeans
#> 1 Corn 16 27 31 33 Corn 0.08935164 0.4054296 0.1763189 0.2391845
#> 2 Corn 15 23 30 30 Corn 0.07690181 0.4558027 0.1420920 0.2530101
#> 3 Corn 16 27 27 26 Corn 0.09817815 0.3422454 0.1365315 0.3073105
#> 4 Corn 18 20 25 23 Corn 0.10521511 0.3633673 0.1078076 0.3281477
#> 5 Corn 15 15 31 32 Corn 0.05879921 0.5753907 0.1173332 0.2086696
#> 6 Corn 15 32 32 15 Soybeans 0.09723648 0.3278382 0.1318370 0.3419924
#> Sugarbeets missed
#> 1 0.08971545 FALSE
#> 2 0.07219340 FALSE
#> 3 0.11573442 FALSE
#> 4 0.09546233 FALSE
#> 5 0.03980738 FALSE
#> 6 0.10109590 TRUE
# posterior shows that posterior of corn membership is much higher than the prior
## LOOCV
# leave-one-out cross validation for linear discriminant analysis
# cannot run the predict function using the object with CV = TRUE
# because it returns the within sample predictions
lda_cv <- lda(crop ~ y1 + y2 + y3 + y4,
data = crops, CV = TRUE)
# Contingency table
lda_table_cv <- table(truth = crops$crop, fitted = lda_cv$class)
lda_table_cv
#> fitted
#> truth Clover Corn Cotton Soybeans Sugarbeets
#> Clover 4 3 1 0 3
#> Corn 0 4 1 2 0
#> Cotton 3 0 0 2 1
#> Soybeans 0 1 1 3 1
#> Sugarbeets 2 1 0 2 1
## Predict the test data
lda_pred <- predict(lda_mod, newdata = crops_test)
## Make a contingency table with truth and most likely class
table(truth=crops_test$crop, predict=lda_pred$class)
#> predict
#> truth Clover Corn Cotton Soybeans Sugarbeets
#> Clover 0 0 1 0 0
#> Corn 0 1 0 0 0
#> Cotton 0 0 0 1 0
#> Soybeans 0 0 0 1 0
#> Sugarbeets 1 0 0 0 0LDA didn’t do well on both within sample and out-of-sample data.
22.4.4.2 QDA
## Quadratic discriminant analysis
qda_mod <- qda(crop ~ y1 + y2 + y3 + y4,
data = crops)
## Look at accuracy on the training data
qda_fitted <- predict(qda_mod, newdata = crops)
# Contingency table
qda_table <- table(truth = crops$crop, fitted = qda_fitted$class)
qda_table
#> fitted
#> truth Clover Corn Cotton Soybeans Sugarbeets
#> Clover 9 0 0 0 2
#> Corn 0 7 0 0 0
#> Cotton 0 0 6 0 0
#> Soybeans 0 0 0 6 0
#> Sugarbeets 0 0 1 1 4
## LOOCV
qda_cv <- qda(crop ~ y1 + y2 + y3 + y4,
data = crops, CV = TRUE)
# Contingency table
qda_table_cv <- table(truth = crops$crop, fitted = qda_cv$class)
qda_table_cv
#> fitted
#> truth Clover Corn Cotton Soybeans Sugarbeets
#> Clover 9 0 0 0 2
#> Corn 3 2 0 0 2
#> Cotton 3 0 2 0 1
#> Soybeans 3 0 0 2 1
#> Sugarbeets 3 0 1 1 1
## Predict the test data
qda_pred <- predict(qda_mod, newdata = crops_test)
## Make a contingency table with truth and most likely class
table(truth = crops_test$crop, predict = qda_pred$class)
#> predict
#> truth Clover Corn Cotton Soybeans Sugarbeets
#> Clover 1 0 0 0 0
#> Corn 0 1 0 0 0
#> Cotton 0 0 1 0 0
#> Soybeans 0 0 0 1 0
#> Sugarbeets 0 0 0 0 122.4.4.3 KNN
knn uses design matrices of the features.
## Design matrices
X_train <- crops %>%
dplyr::select(-crop)
X_test <- crops_test %>%
dplyr::select(-crop)
Y_train <- crops$crop
Y_test <- crops_test$crop
## Nearest neighbors with 2 neighbors
knn_2 <- knn(X_train, X_train, Y_train, k = 2)
table(truth = Y_train, fitted = knn_2)
#> fitted
#> truth Clover Corn Cotton Soybeans Sugarbeets
#> Clover 7 0 2 1 1
#> Corn 0 7 0 0 0
#> Cotton 0 0 4 0 2
#> Soybeans 0 0 0 4 2
#> Sugarbeets 1 0 2 0 3
## Accuracy
mean(Y_train==knn_2)
#> [1] 0.6944444
## Performance on test data
knn_2_test <- knn(X_train, X_test, Y_train, k = 2)
table(truth = Y_test, predict = knn_2_test)
#> predict
#> truth Clover Corn Cotton Soybeans Sugarbeets
#> Clover 1 0 0 0 0
#> Corn 0 1 0 0 0
#> Cotton 0 0 0 0 1
#> Soybeans 0 0 0 1 0
#> Sugarbeets 0 0 0 0 1
## Accuracy
mean(Y_test==knn_2_test)
#> [1] 0.8
## Nearest neighbors with 3 neighbors
knn_3 <- knn(X_train, X_train, Y_train, k = 3)
table(truth = Y_train, fitted = knn_3)
#> fitted
#> truth Clover Corn Cotton Soybeans Sugarbeets
#> Clover 8 0 1 1 1
#> Corn 0 4 1 2 0
#> Cotton 1 1 3 0 1
#> Soybeans 0 1 1 4 0
#> Sugarbeets 0 0 0 2 4
## Accuracy
mean(Y_train==knn_3)
#> [1] 0.6388889
## Performance on test data
knn_3_test <- knn(X_train, X_test, Y_train, k = 3)
table(truth = Y_test, predict = knn_3_test)
#> predict
#> truth Clover Corn Cotton Soybeans Sugarbeets
#> Clover 1 0 0 0 0
#> Corn 0 1 0 0 0
#> Cotton 0 0 1 0 0
#> Soybeans 0 0 0 1 0
#> Sugarbeets 0 0 0 0 1
## Accuracy
mean(Y_test==knn_3_test)
#> [1] 122.4.4.4 Stepwise
Stepwise discriminant analysis using the stepclass in function in the klaR package.
step <- stepclass(
crop ~ y1 + y2 + y3 + y4,
data = crops,
method = "qda",
improvement = 0.15
)
#> correctness rate: 0.45; in: "y1"; variables (1): y1
#>
#> hr.elapsed min.elapsed sec.elapsed
#> 0.00 0.00 0.16
step$process
#> step var varname result.pm
#> 0 start 0 -- 0.00
#> 1 in 1 y1 0.45
step$performance.measure
#> [1] "correctness rate"Iris Data
library(dplyr)
data('iris')
set.seed(1)
samp <-
sample.int(nrow(iris), size = floor(0.70 * nrow(iris)), replace = F)
train.iris <- iris[samp,] %>% mutate_if(is.numeric,scale)
test.iris <- iris[-samp,] %>% mutate_if(is.numeric,scale)
library(ggplot2)
iris.model <- lda(Species ~ ., data = train.iris)
#pred
pred.lda <- predict(iris.model, test.iris)
table(truth = test.iris$Species, prediction = pred.lda$class)
#> prediction
#> truth setosa versicolor virginica
#> setosa 15 0 0
#> versicolor 0 17 0
#> virginica 0 0 13
plot(iris.model)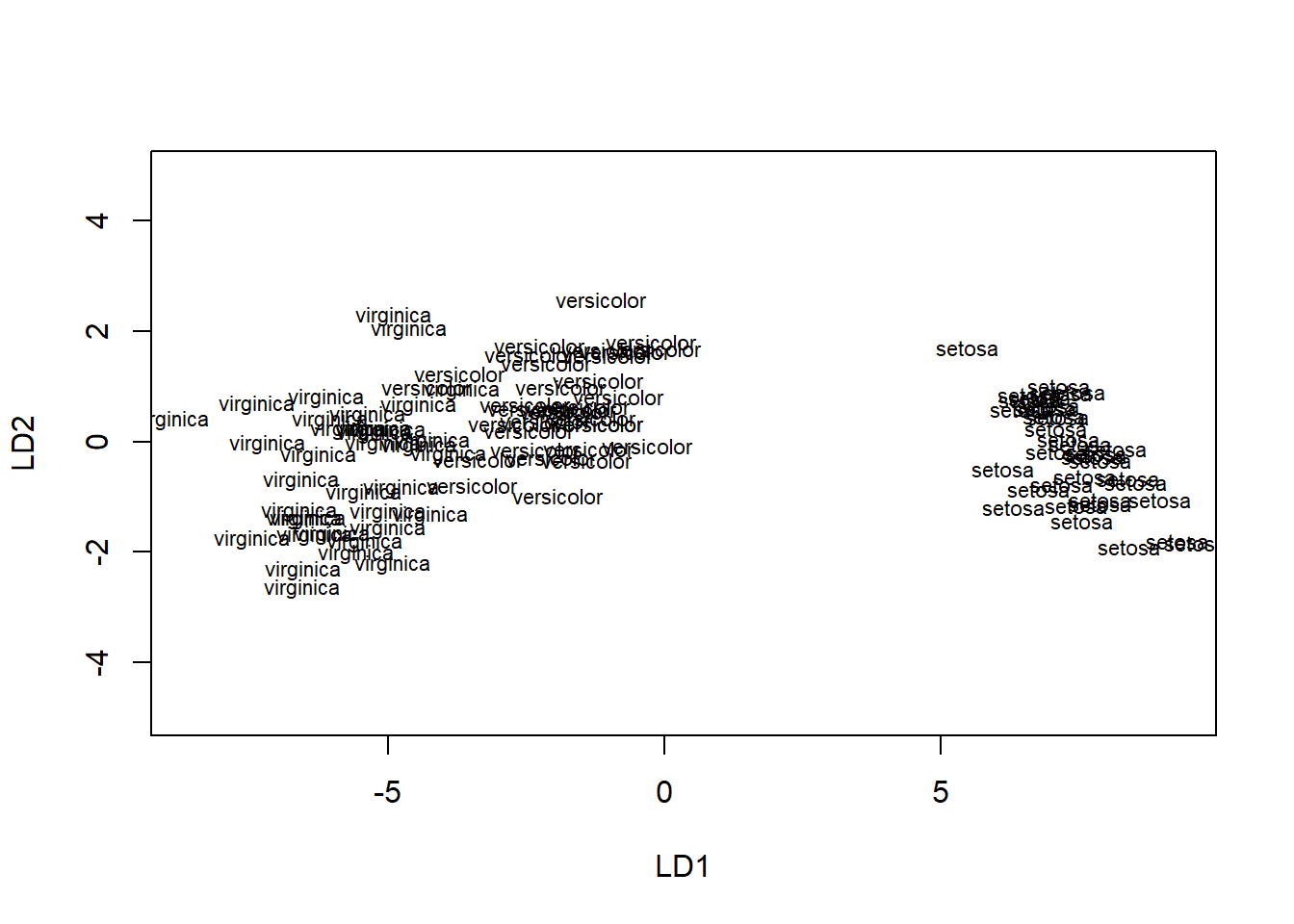
22.4.4.5 PCA with Discriminant Analysis
we can use both PCA for dimension reduction in discriminant analysis
zeros <- as.matrix(read.table("images/mnist0_train_b.txt"))
nines <- as.matrix(read.table("images/mnist9_train_b.txt"))
train <- rbind(zeros[1:1000, ], nines[1:1000, ])
train <- train / 255 #divide by 255 per notes (so ranges from 0 to 1)
train <- t(train) #each column is an observation
image(matrix(train[, 1], nrow = 28), main = 'Example image, unrotated')
test <- rbind(zeros[2501:3000, ], nines[2501:3000, ])
test <- test / 255
test <- t(test)
y.train <- c(rep(0, 1000), rep(9, 1000))
y.test <- c(rep(0, 500), rep(9, 500))
library(MASS)
pc <- prcomp(t(train))
train.large <- data.frame(cbind(y.train, pc$x[, 1:10]))
large <- lda(y.train ~ ., data = train.large)
#the test data set needs to be constucted w/ the same 10 princomps
test.large <- data.frame(cbind(y.test, predict(pc, t(test))[, 1:10]))
pred.lda <- predict(large, test.large)
table(truth = test.large$y.test, prediction = pred.lda$class)
#> prediction
#> truth 0 9
#> 0 491 9
#> 9 5 495
large.qda <- qda(y.train~.,data=train.large)
#prediction
pred.qda <- predict(large.qda,test.large)
table(truth=test.large$y.test,prediction=pred.qda$class)
#> prediction
#> truth 0 9
#> 0 493 7
#> 9 3 497