39.2 Good Controls
39.2.1 Omitted Variable Bias Correction
This is when \(Z\) can block all back-door paths.
rm(list = ls())
model <- dagitty("dag{x->y; z->x; z->y}")
## coordinates for plotting
coordinates(model) <- list(
x = c(x=1, y=3, z=2),
y = c(x=1, y=1, z=2))
## ggplot
ggdag(model) + theme_dag()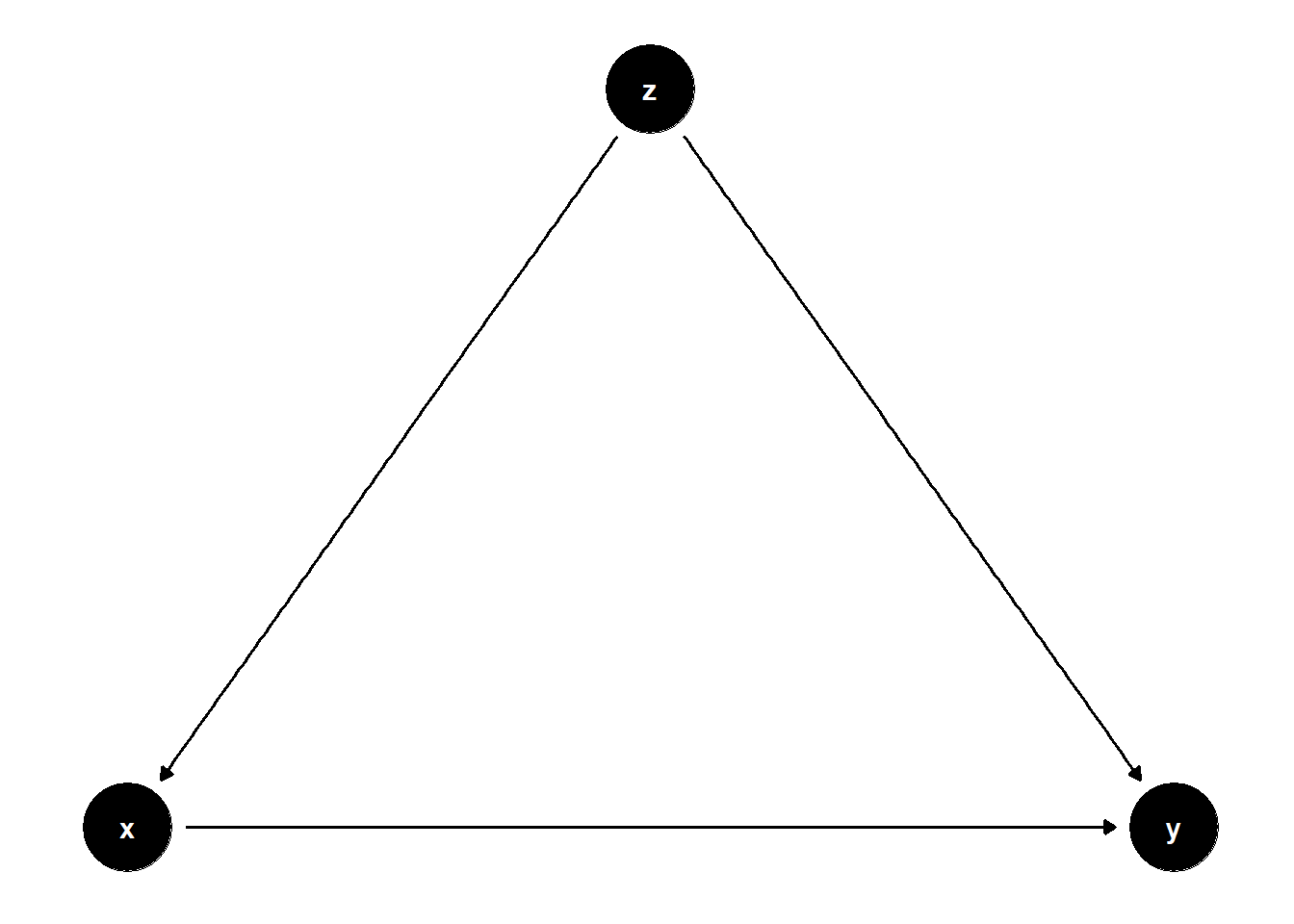
Unadjusted estimate is biased
adjusting for \(Z\) blocks the backdoor path
n <- 1e4
z <- rnorm(n)
causal_coef = 2
beta2 = 3
x <- z + rnorm(n)
y <- causal_coef * x + beta2 * z + rnorm(n)
jtools::export_summs(lm(y ~ x), lm(y ~ x + z))| Model 1 | Model 2 | |
|---|---|---|
| (Intercept) | 0.01 | -0.00 |
| (0.02) | (0.01) | |
| x | 3.51 *** | 2.00 *** |
| (0.02) | (0.01) | |
| z | 3.02 *** | |
| (0.01) | ||
| N | 10000 | 10000 |
| R2 | 0.82 | 0.97 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
# cleans workspace
rm(list = ls())
# Draw DAG
# specify edges
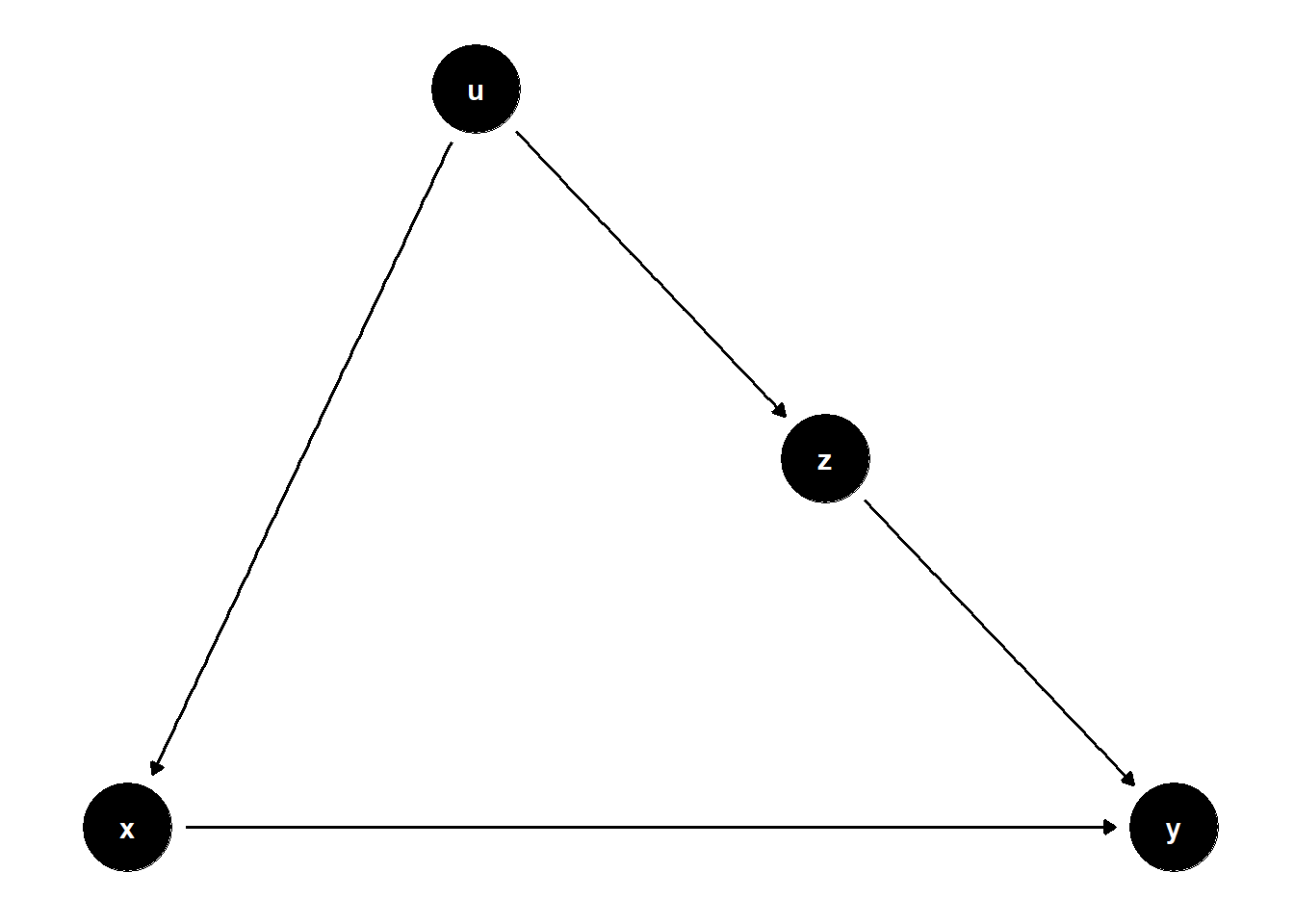
model <- dagitty("dag{x->y; u->z; z->x; u->y}")
# set u as latent
latents(model) <- "u"
## coordinates for plotting
coordinates(model) <- list(
x = c(x=1, z=2, u=3, y = 4),
y = c(x=1, y=1, z=2, u = 3))
## ggplot
ggdag(model) + theme_dag()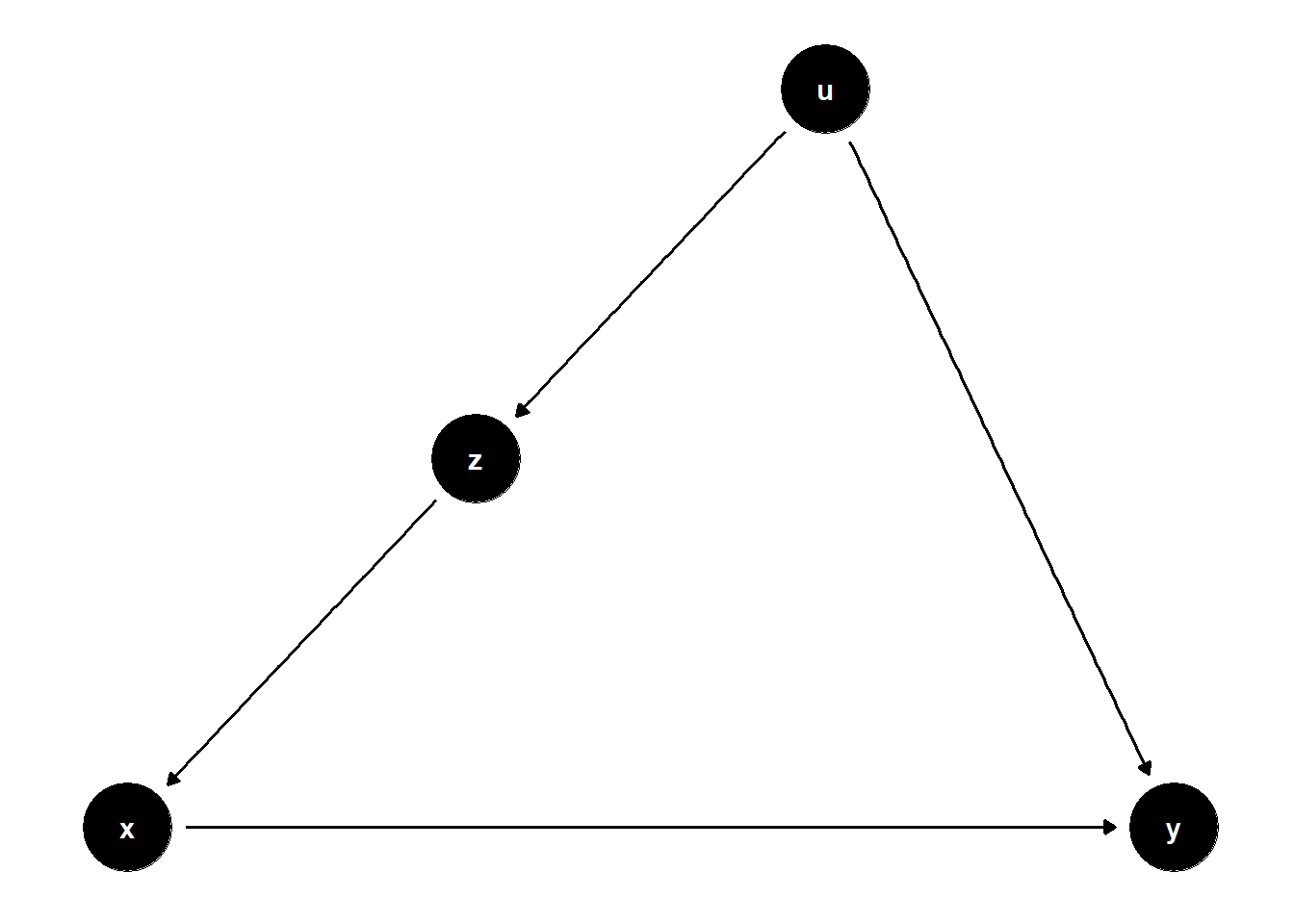
Unadjusted estimate is biased
adjusting for \(Z\) blocks the backdoor door path due to \(U\)
n <- 1e4
u <- rnorm(n)
z <- u + rnorm(n)
causal_coef = 2
x <- z + rnorm(n)
y <- causal_coef * x + u + rnorm(n)
jtools::export_summs(lm(y ~ x), lm(y ~ x + z))| Model 1 | Model 2 | |
|---|---|---|
| (Intercept) | 0.03 * | 0.03 * |
| (0.01) | (0.01) | |
| x | 2.34 *** | 2.01 *** |
| (0.01) | (0.01) | |
| z | 0.49 *** | |
| (0.01) | ||
| N | 10000 | 10000 |
| R2 | 0.91 | 0.92 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
Even though \(Z\) is significant, we cannot give it a causal interpretation.
# cleans workspace
rm(list = ls())
# Draw DAG
# specify edges
model <- dagitty("dag{x->y; u->z; u->x; z->y}")
# set u as latent
latents(model) <- "u"
## coordinates for plotting
coordinates(model) <- list(
x = c(x=1, z=3, u=2, y = 4),
y = c(x=1, y=1, z=2, u = 3))
## ggplot
ggdag(model) + theme_dag()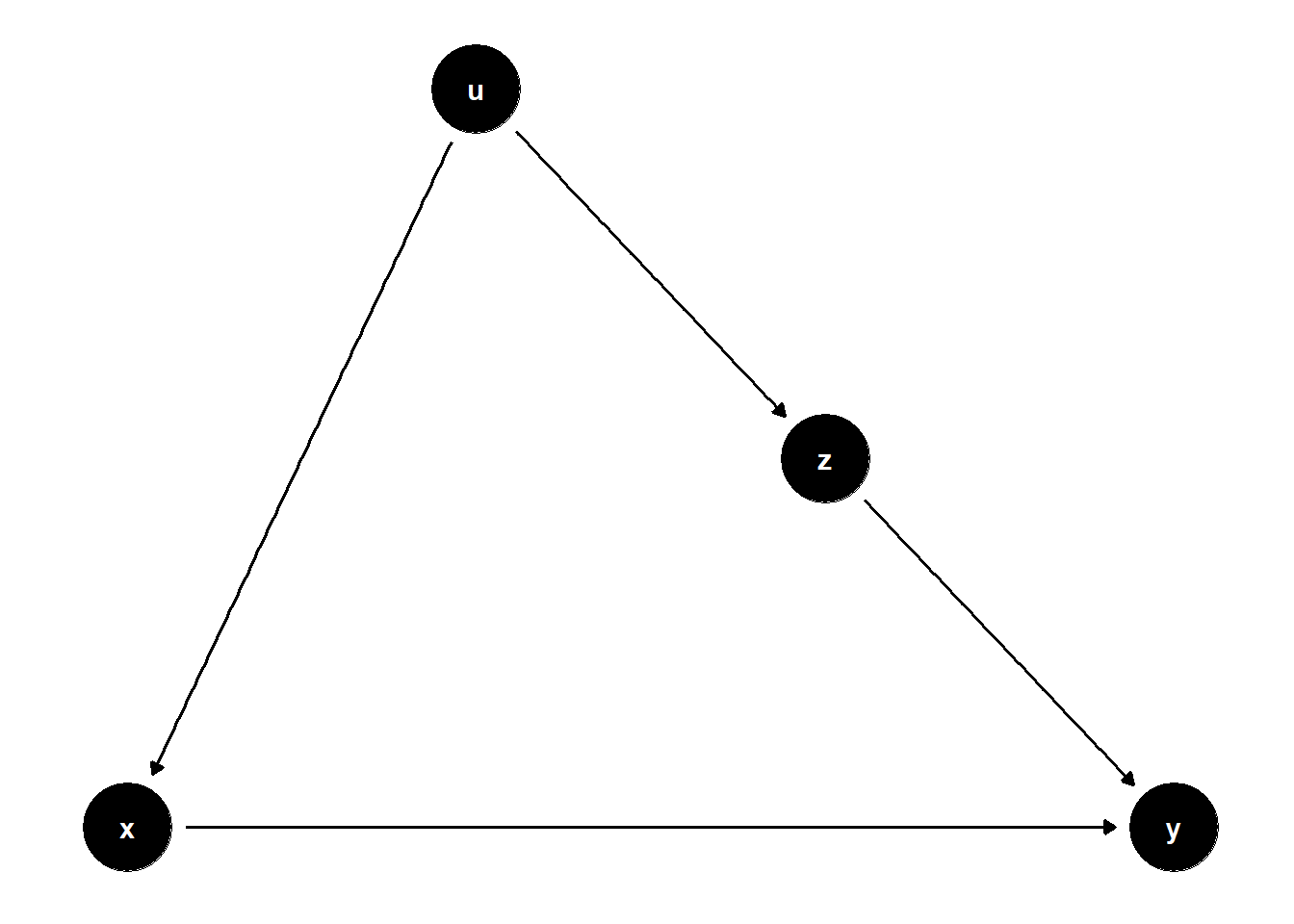
n <- 1e4
u <- rnorm(n)
z <- u + rnorm(n)
x <- u + rnorm(n)
causal_coef <- 2
y <- causal_coef * x + z + rnorm(n)
jtools::export_summs(lm(y ~ x), lm(y ~ x + z))| Model 1 | Model 2 | |
|---|---|---|
| (Intercept) | -0.03 | -0.01 |
| (0.02) | (0.01) | |
| x | 2.51 *** | 2.01 *** |
| (0.01) | (0.01) | |
| z | 1.01 *** | |
| (0.01) | ||
| N | 10000 | 10000 |
| R2 | 0.84 | 0.93 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
Even though \(Z\) is significant, we cannot give it a causal interpretation.
Summary
# cleans workspace
rm(list = ls())
# Model 1
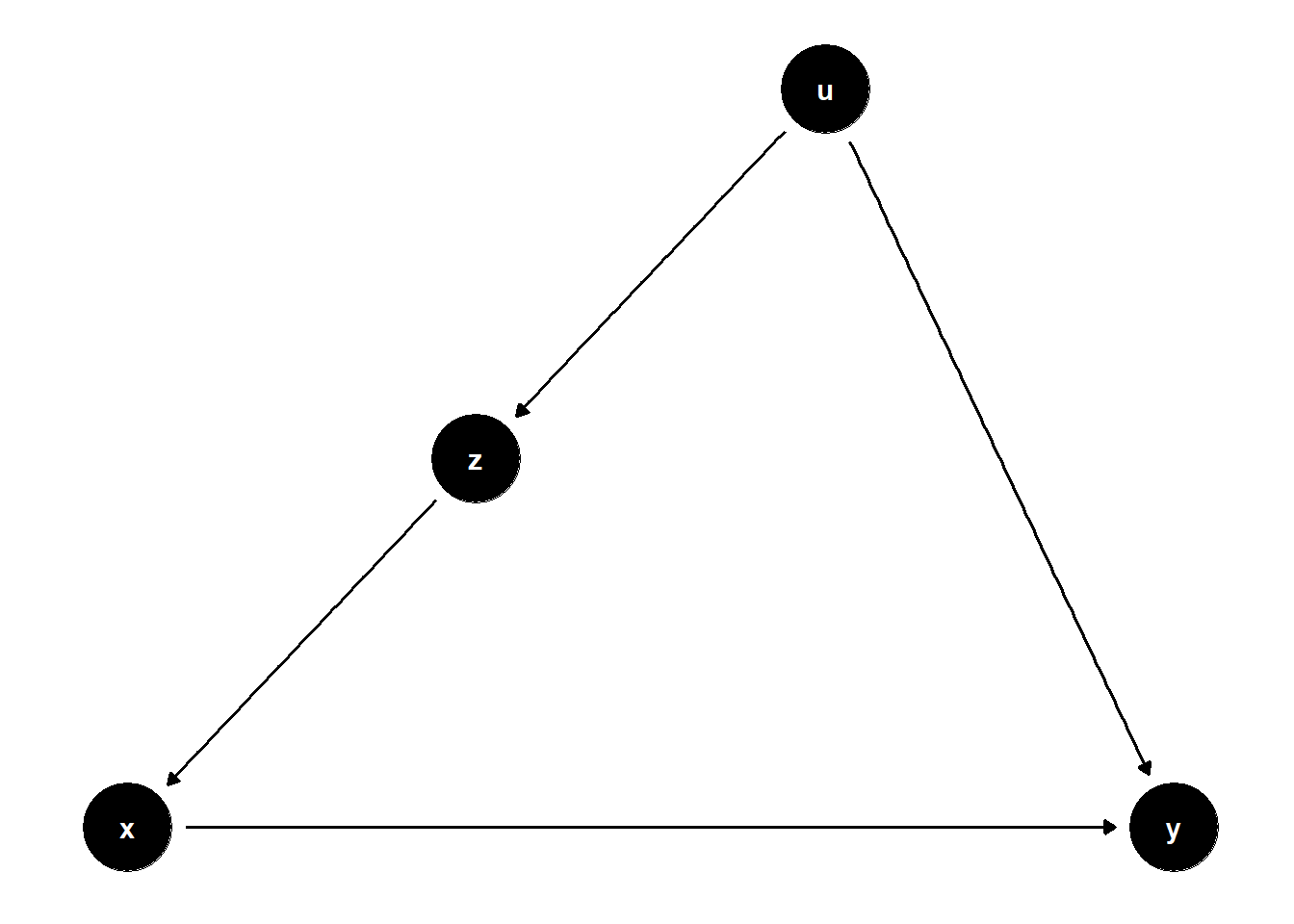
model1 <- dagitty("dag{x->y; z->x; z->y}")
## coordinates for plotting
coordinates(model1) <- list(
x = c(x=1, y=3, z=2),
y = c(x=1, y=1, z=2))
# Model 2
# specify edges
model2 <- dagitty("dag{x->y; u->z; z->x; u->y}")
# set u as latent
latents(model2) <- "u"
## coordinates for plotting
coordinates(model2) <- list(
x = c(x=1, z=2, u=3, y = 4),
y = c(x=1, y=1, z=2, u = 3))
# Model 3
# specify edges
model3 <- dagitty("dag{x->y; u->z; u->x; z->y}")
# set u as latent
latents(model3) <- "u"
## coordinates for plotting
coordinates(model3) <- list(
x = c(x=1, z=3, u=2, y = 4),
y = c(x=1, y=1, z=2, u = 3))
par(mfrow=c(1,3))
## ggplot
ggdag(model1) + theme_dag()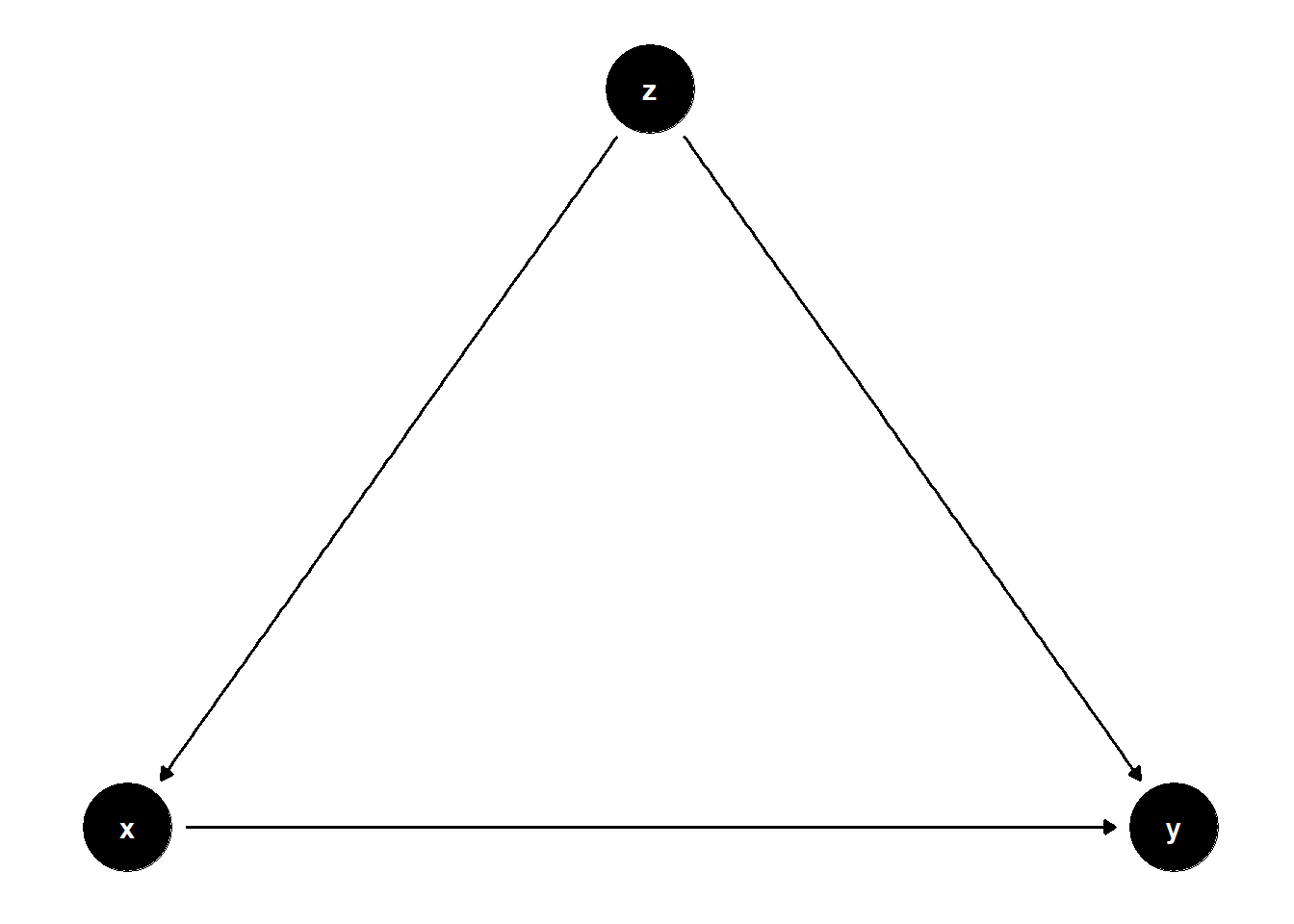
39.2.2 Omitted Variable Bias in Mediation Correction
Common causes of \(X\) and any mediator (between \(X\) and \(Y\)) confound the effect of \(X\) on \(Y\)
# cleans workspace
rm(list = ls())
# DAG
## specify edges
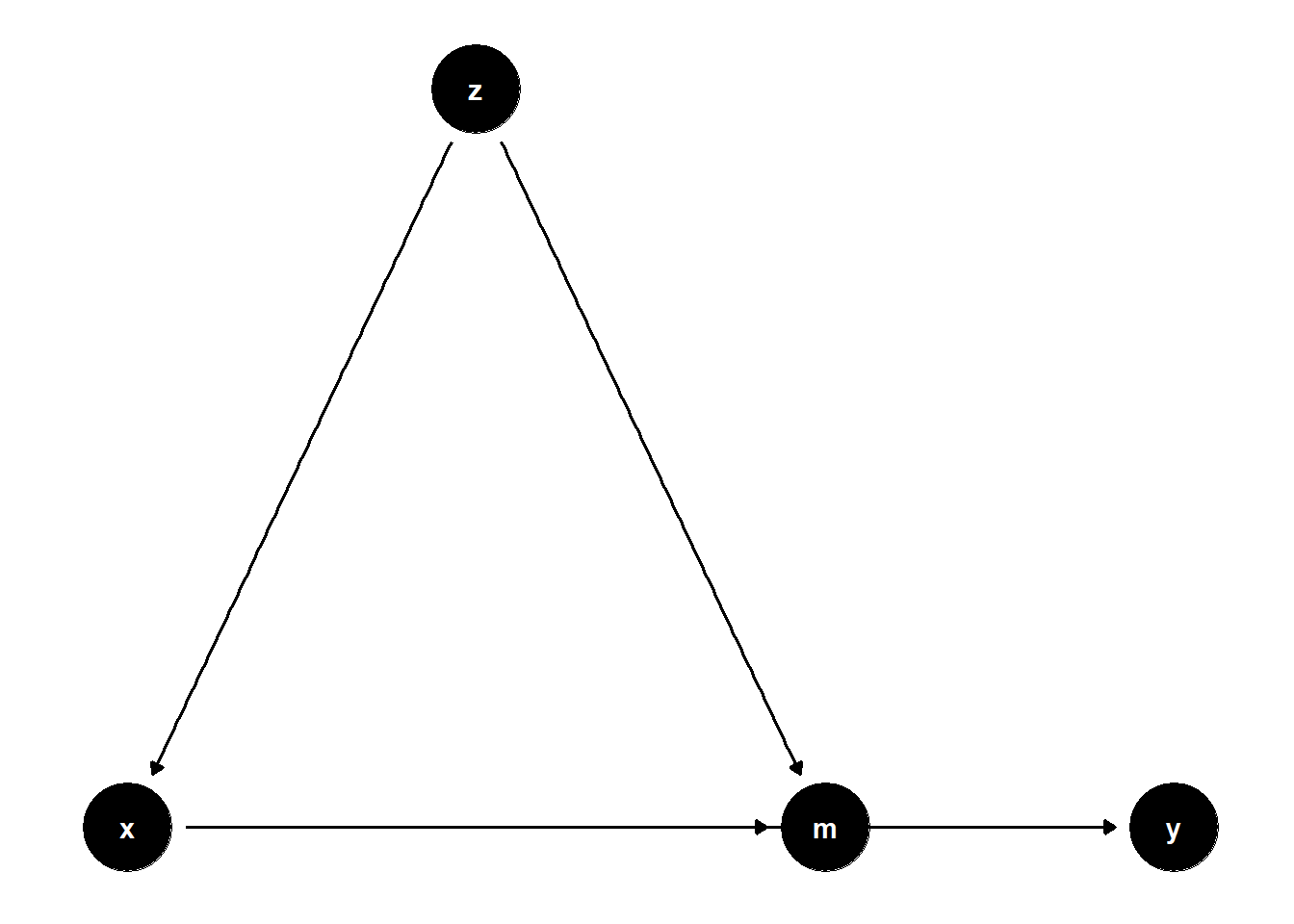
model <- dagitty("dag{x->y; z->x; x->m; z->m; m->y}")
## coordinates for plotting
coordinates(model) <- list(
x = c(x=1, z=2, m=3, y=4),
y = c(x=1, z=2, m=1, y=1))
## ggplot
ggdag(model) + theme_dag()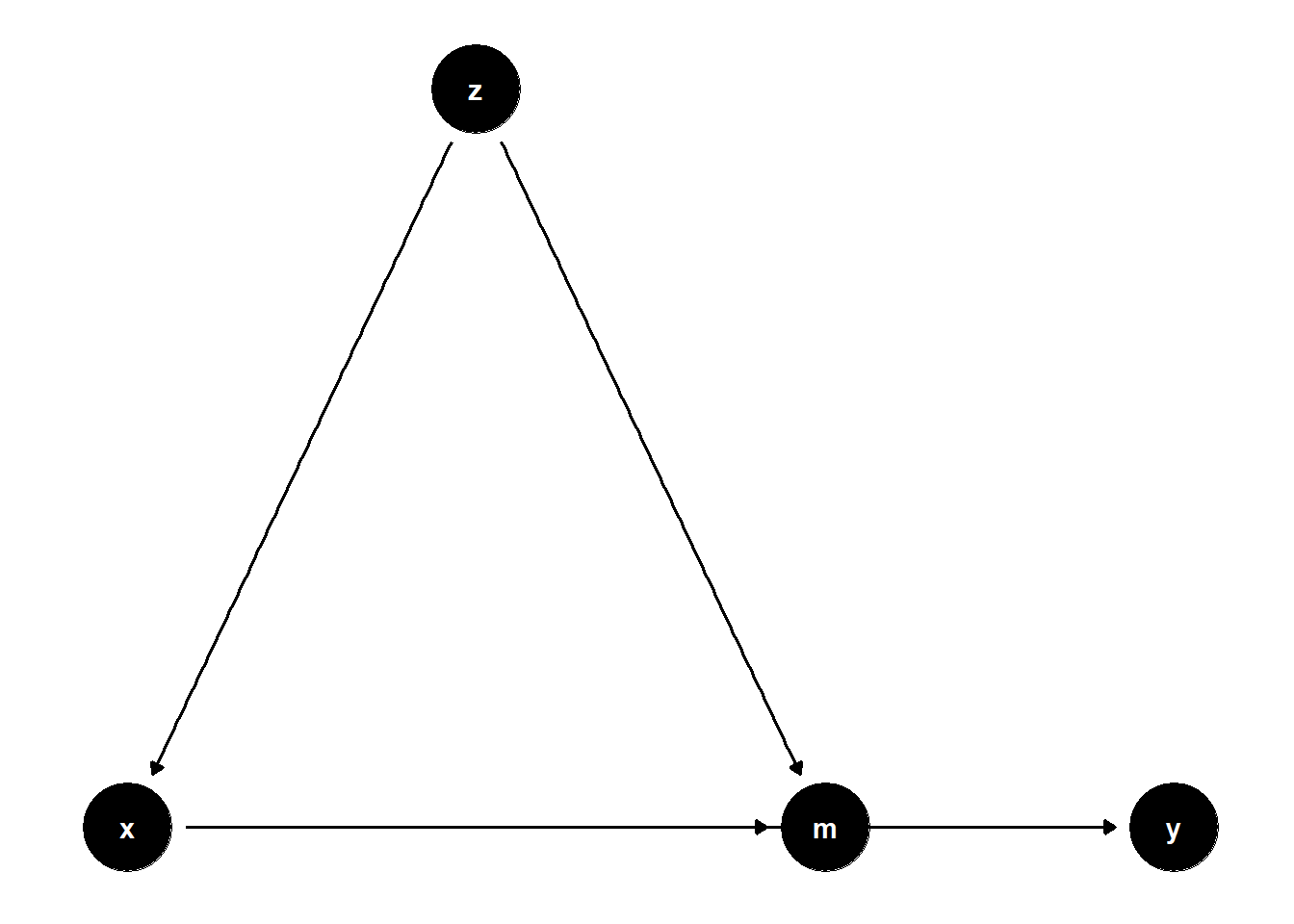
\(Z\) is a confounder of both the mediator \(M\) and \(X\)
n <- 1e4
z <- rnorm(n)
x <- z + rnorm(n)
causal_coef <- 2
m <- causal_coef * x + z + rnorm(n)
y <- m + rnorm(n)
jtools::export_summs(lm(y ~ x), lm(y ~ x + z))| Model 1 | Model 2 | |
|---|---|---|
| (Intercept) | -0.02 | -0.01 |
| (0.02) | (0.01) | |
| x | 2.49 *** | 1.97 *** |
| (0.01) | (0.01) | |
| z | 1.02 *** | |
| (0.02) | ||
| N | 10000 | 10000 |
| R2 | 0.83 | 0.86 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
# cleans workspace
rm(list = ls())
# DAG
## specify edges
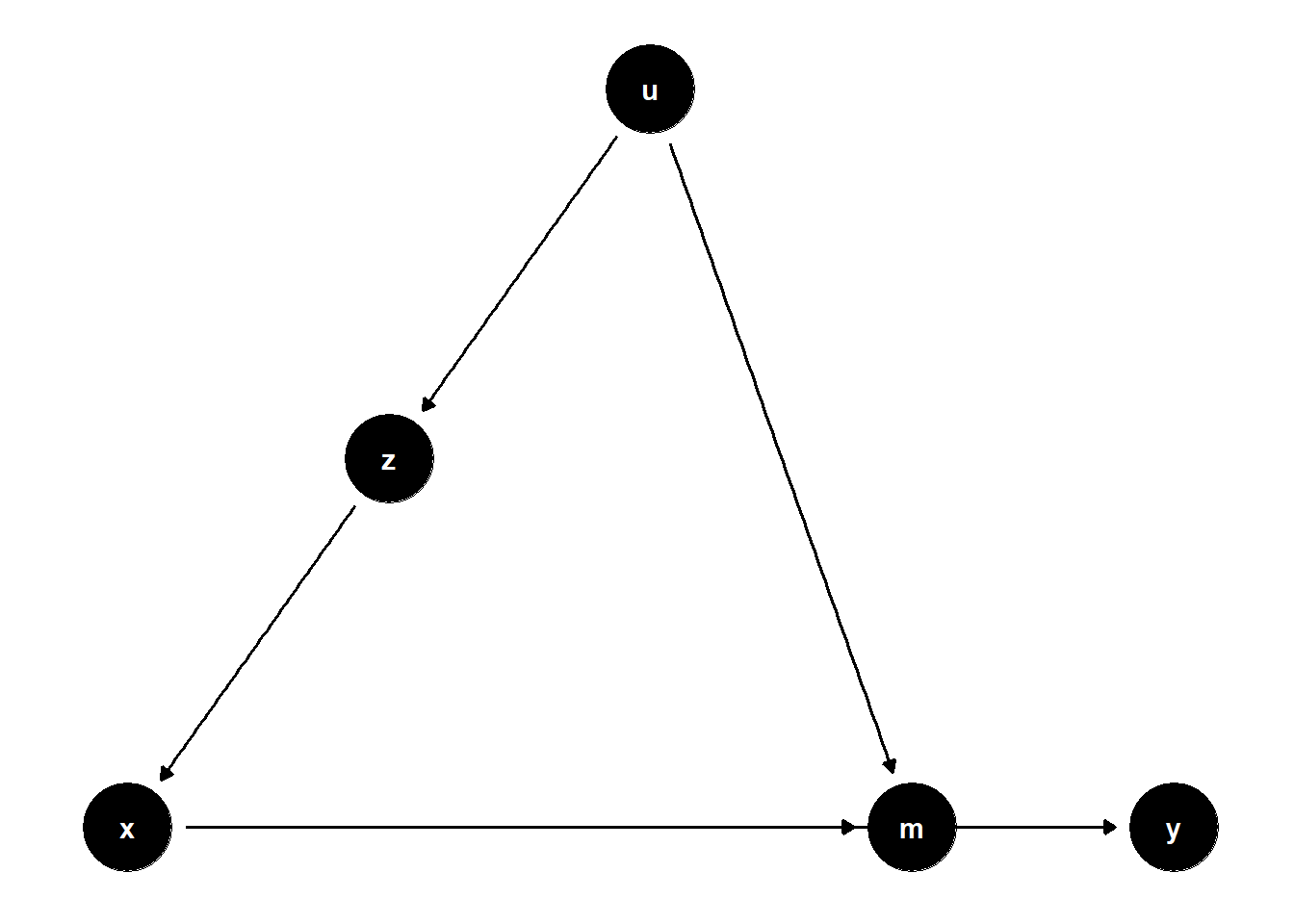
model <- dagitty("dag{x->y; u->z; z->x; x->m; u->m; m->y}")
# set u as latent
latents(model) <- "u"
## coordinates for plotting
coordinates(model) <- list(
x = c(x=1, z=2, u=3, m=4, y=5),
y = c(x=1, z=2, u=3, m=1, y=1))
## ggplot
ggdag(model) + theme_dag()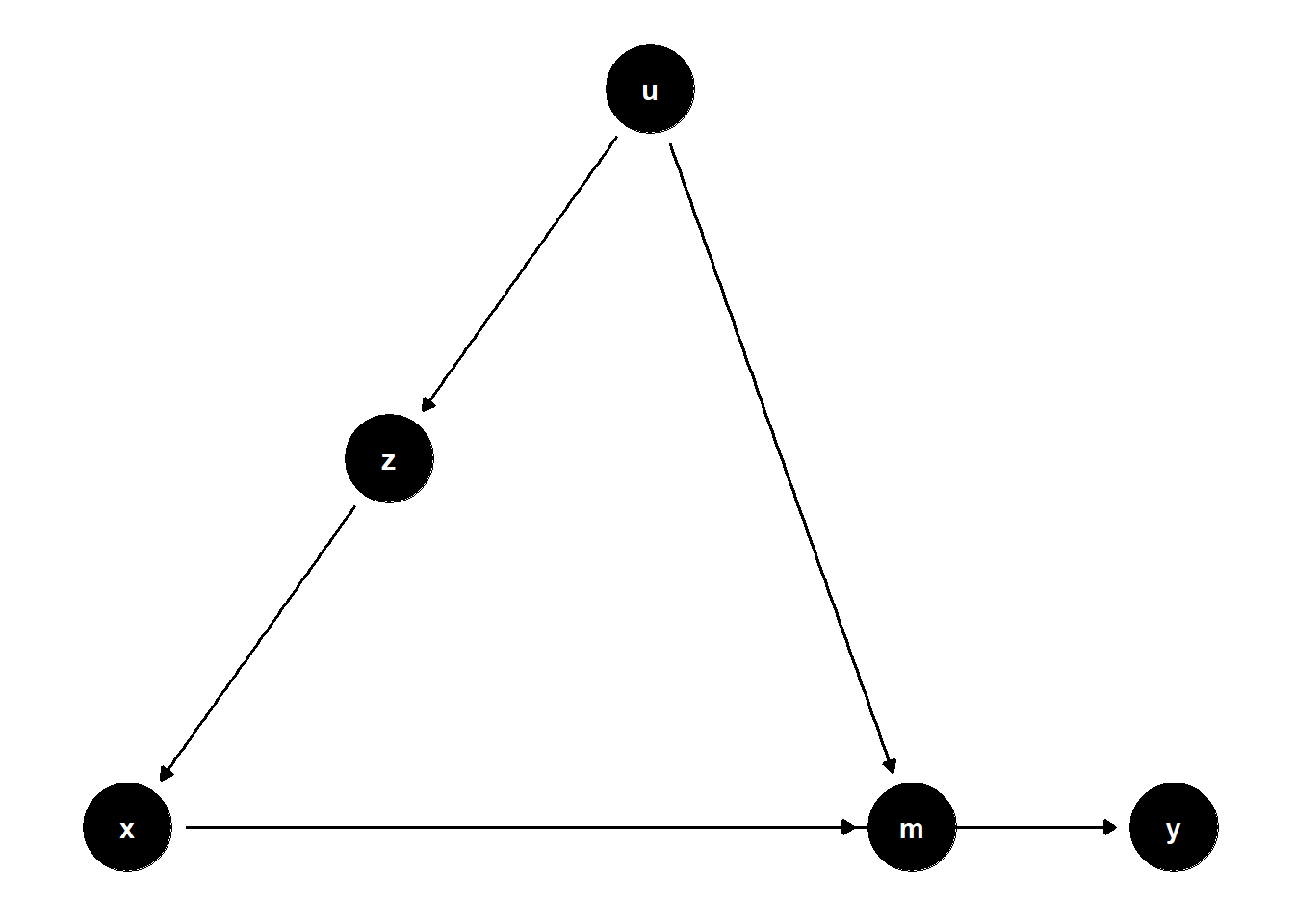
n <- 1e4
u <- rnorm(n)
z <- u + rnorm(n)
x <- z + rnorm(n)
causal_coef <- 2
m <- causal_coef * x + u + rnorm(n)
y <- m + rnorm(n)
jtools::export_summs(lm(y ~ x), lm(y ~ x + z))| Model 1 | Model 2 | |
|---|---|---|
| (Intercept) | -0.01 | -0.01 |
| (0.02) | (0.02) | |
| x | 2.31 *** | 2.00 *** |
| (0.01) | (0.02) | |
| z | 0.49 *** | |
| (0.02) | ||
| N | 10000 | 10000 |
| R2 | 0.86 | 0.86 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
# cleans workspace
rm(list = ls())
# DAG
## specify edges
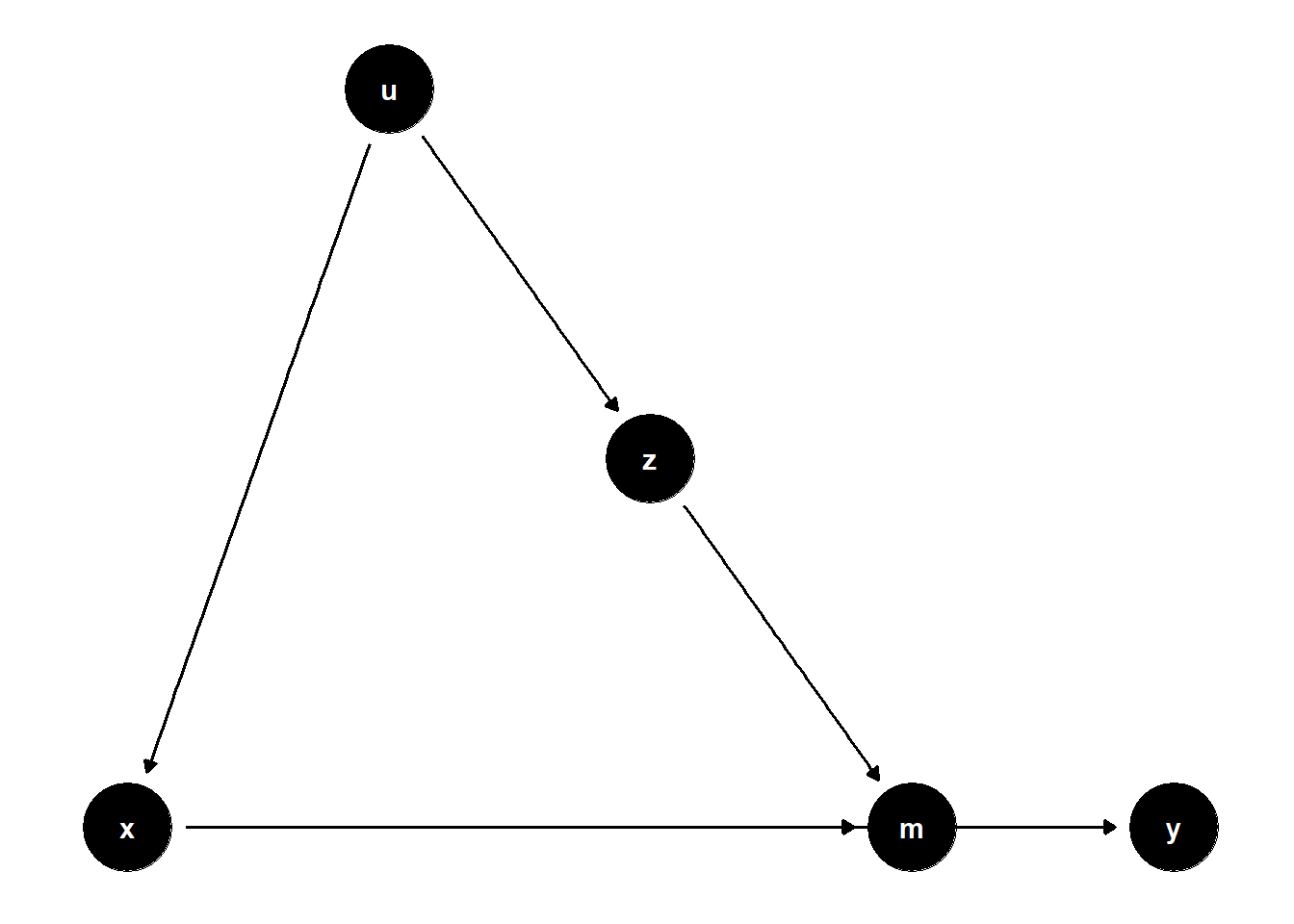
model <- dagitty("dag{x->y; u->z; z->m; x->m; u->x; m->y}")
# set u as latent
latents(model) <- "u"
## coordinates for plotting
coordinates(model) <- list(
x = c(x=1, z=3, u=2, m=4, y=5),
y = c(x=1, z=2, u=3, m=1, y=1))
## ggplot
ggdag(model) + theme_dag()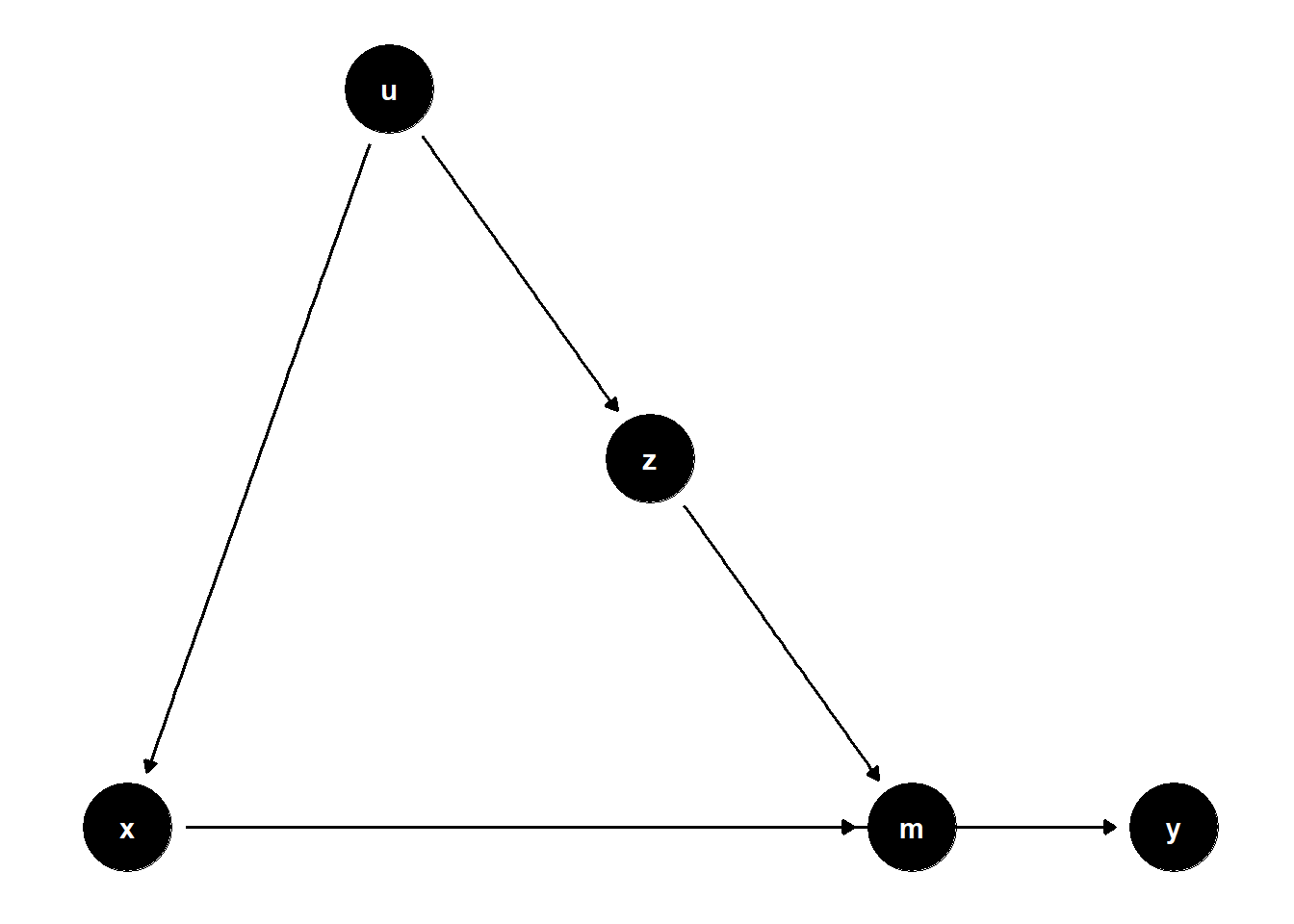
n <- 1e4
u <- rnorm(n)
z <- u + rnorm(n)
x <- u + rnorm(n)
causal_coef <- 2
m <- causal_coef * x + z + rnorm(n)
y <- m + rnorm(n)
jtools::export_summs(lm(y ~ x), lm(y ~ x + z))| Model 1 | Model 2 | |
|---|---|---|
| (Intercept) | 0.01 | -0.00 |
| (0.02) | (0.01) | |
| x | 2.50 *** | 1.99 *** |
| (0.01) | (0.01) | |
| z | 1.02 *** | |
| (0.01) | ||
| N | 10000 | 10000 |
| R2 | 0.78 | 0.87 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
Summary
# model 4
model4 <- dagitty("dag{x->y; z->x; x->m; z->m; m->y}")
## coordinates for plotting
coordinates(model4) <- list(
x = c(x=1, z=2, m=3, y=4),
y = c(x=1, z=2, m=1, y=1))
# model 5
model5 <- dagitty("dag{x->y; u->z; z->x; x->m; u->m; m->y}")
# set u as latent
latents(model5) <- "u"
## coordinates for plotting
coordinates(model5) <- list(
x = c(x=1, z=2, u=3, m=4, y=5),
y = c(x=1, z=2, u=3, m=1, y=1))
# model 6
model6 <- dagitty("dag{x->y; u->z; z->m; x->m; u->x; m->y}")
# set u as latent
latents(model6) <- "u"
## coordinates for plotting
coordinates(model6) <- list(
x = c(x=1, z=3, u=2, m=4, y=5),
y = c(x=1, z=2, u=3, m=1, y=1))
par(mfrow=c(1,3))
## ggplot
ggdag(model4) + theme_dag()