42.3 Model-based causal mediation analysis
Other resources:
Fit 2 models
mediator model: conditional distribution of the mediators \(M_i | T_i, X_i\)
Outcome model: conditional distribution of \(Y_i | T_i, M_i, X_i\)
mediation can accommodate almost all types of model for both mediator model and outcome model except Censored mediator model.
The update here is that estimation of ACME does not rely on product or difference of coefficients (see 40.2.1 ,
which requires very strict assumption: (1) linear regression models of mediator and outcome, (2) \(T_i\) and \(M_i\) effects are additive and no interaction
library(mediation)
set.seed(2014)
data("framing", package = "mediation")
med.fit <-
lm(emo ~ treat + age + educ + gender + income, data = framing)
out.fit <-
glm(
cong_mesg ~ emo + treat + age + educ + gender + income,
data = framing,
family = binomial("probit")
)
# Quasi-Bayesian Monte Carlo
med.out <-
mediate(
med.fit,
out.fit,
treat = "treat",
mediator = "emo",
robustSE = TRUE,
sims = 100 # should be 10000 in practice
)
summary(med.out)
#>
#> Causal Mediation Analysis
#>
#> Quasi-Bayesian Confidence Intervals
#>
#> Estimate 95% CI Lower 95% CI Upper p-value
#> ACME (control) 0.0791 0.0351 0.15 <2e-16 ***
#> ACME (treated) 0.0804 0.0367 0.16 <2e-16 ***
#> ADE (control) 0.0206 -0.0976 0.12 0.70
#> ADE (treated) 0.0218 -0.1053 0.12 0.70
#> Total Effect 0.1009 -0.0497 0.23 0.14
#> Prop. Mediated (control) 0.6946 -6.3109 3.68 0.14
#> Prop. Mediated (treated) 0.7118 -5.7936 3.50 0.14
#> ACME (average) 0.0798 0.0359 0.15 <2e-16 ***
#> ADE (average) 0.0212 -0.1014 0.12 0.70
#> Prop. Mediated (average) 0.7032 -6.0523 3.59 0.14
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Sample Size Used: 265
#>
#>
#> Simulations: 100Nonparametric bootstrap version
med.out <-
mediate(
med.fit,
out.fit,
boot = TRUE,
treat = "treat",
mediator = "emo",
sims = 100, # should be 10000 in practice
boot.ci.type = "bca" # bias-corrected and accelerated intervals
)
summary(med.out)
#>
#> Causal Mediation Analysis
#>
#> Nonparametric Bootstrap Confidence Intervals with the BCa Method
#>
#> Estimate 95% CI Lower 95% CI Upper p-value
#> ACME (control) 0.0848 0.0424 0.14 <2e-16 ***
#> ACME (treated) 0.0858 0.0410 0.14 <2e-16 ***
#> ADE (control) 0.0117 -0.0726 0.13 0.58
#> ADE (treated) 0.0127 -0.0784 0.14 0.58
#> Total Effect 0.0975 0.0122 0.25 0.06 .
#> Prop. Mediated (control) 0.8698 1.7460 151.20 0.06 .
#> Prop. Mediated (treated) 0.8804 1.6879 138.91 0.06 .
#> ACME (average) 0.0853 0.0434 0.14 <2e-16 ***
#> ADE (average) 0.0122 -0.0756 0.14 0.58
#> Prop. Mediated (average) 0.8751 1.7170 145.05 0.06 .
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Sample Size Used: 265
#>
#>
#> Simulations: 100If theoretically understanding suggests that there is treatment and mediator interaction
med.fit <-
lm(emo ~ treat + age + educ + gender + income, data = framing)
out.fit <-
glm(
cong_mesg ~ emo * treat + age + educ + gender + income,
data = framing,
family = binomial("probit")
)
med.out <-
mediate(
med.fit,
out.fit,
treat = "treat",
mediator = "emo",
robustSE = TRUE,
sims = 100
)
summary(med.out)
#>
#> Causal Mediation Analysis
#>
#> Quasi-Bayesian Confidence Intervals
#>
#> Estimate 95% CI Lower 95% CI Upper p-value
#> ACME (control) 0.07417 0.02401 0.14 <2e-16 ***
#> ACME (treated) 0.09496 0.02702 0.16 <2e-16 ***
#> ADE (control) -0.01353 -0.11855 0.11 0.76
#> ADE (treated) 0.00726 -0.11007 0.11 0.90
#> Total Effect 0.08143 -0.05646 0.19 0.26
#> Prop. Mediated (control) 0.64510 -14.31243 3.13 0.26
#> Prop. Mediated (treated) 0.98006 -17.83202 4.01 0.26
#> ACME (average) 0.08457 0.02738 0.15 <2e-16 ***
#> ADE (average) -0.00314 -0.11457 0.12 1.00
#> Prop. Mediated (average) 0.81258 -16.07223 3.55 0.26
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Sample Size Used: 265
#>
#>
#> Simulations: 100
test.TMint(med.out, conf.level = .95) # test treatment-mediator interaction effect
#>
#> Test of ACME(1) - ACME(0) = 0
#>
#> data: estimates from med.out
#> ACME(1) - ACME(0) = 0.020796, p-value = 0.3
#> alternative hypothesis: true ACME(1) - ACME(0) is not equal to 0
#> 95 percent confidence interval:
#> -0.01757310 0.07110837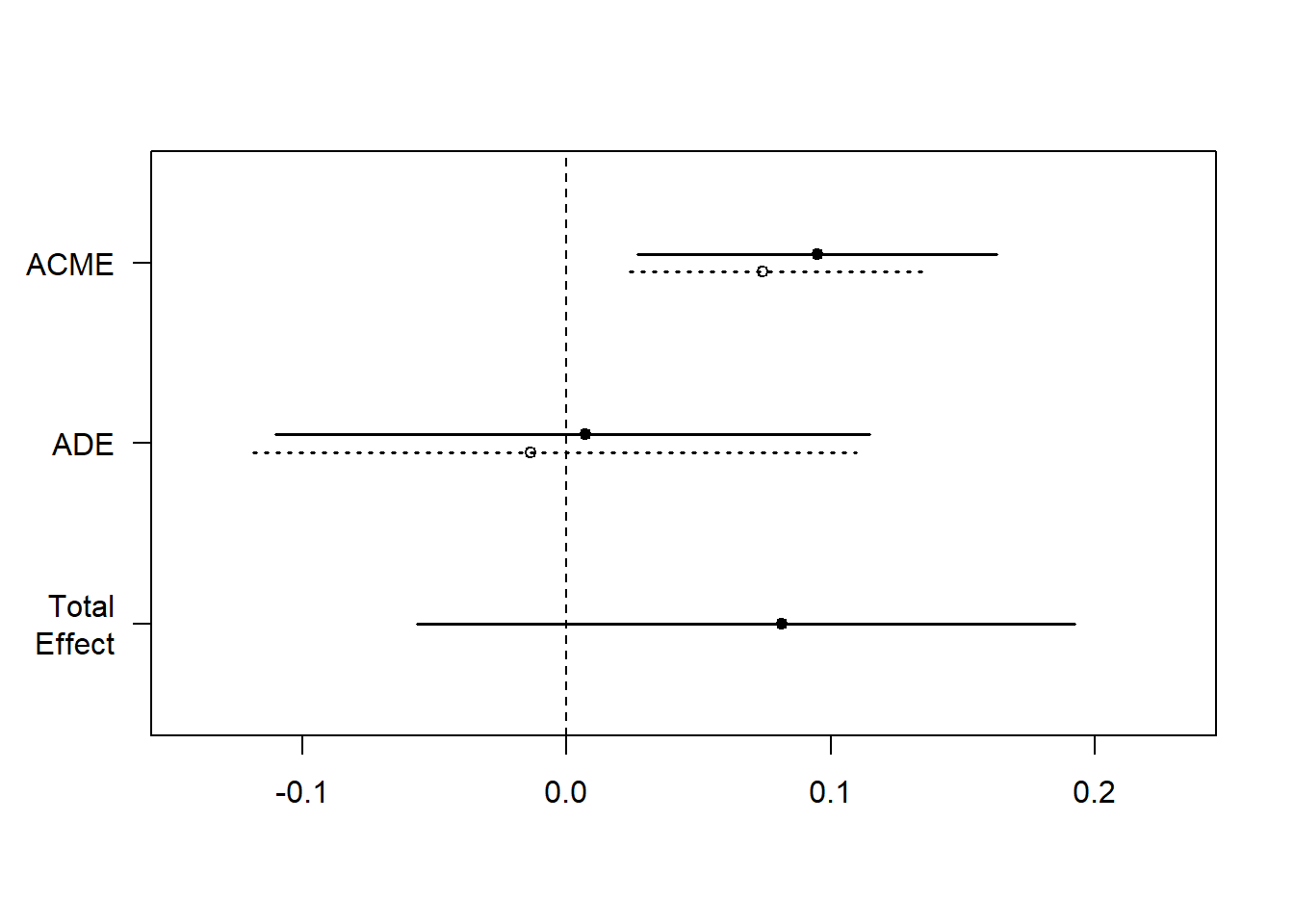
mediation can be used in conjunction with any of your imputation packages.
And it can also handle mediated moderation or non-binary treatment variables, or multi-level data
Sensitivity Analysis for sequential ignorability
test for unobserved pre-treatment covariates
\(\rho\) = correlation between the residuals of the mediator and outcome regressions.
If \(\rho\) is significant, we have evidence for violation of sequential ignorability (i.e., there is unobserved pre-treatment confounders).
med.fit <-
lm(emo ~ treat + age + educ + gender + income, data = framing)
out.fit <-
glm(
cong_mesg ~ emo + treat + age + educ + gender + income,
data = framing,
family = binomial("probit")
)
med.out <-
mediate(
med.fit,
out.fit,
treat = "treat",
mediator = "emo",
robustSE = TRUE,
sims = 100
)
sens.out <-
medsens(med.out,
rho.by = 0.1, # \rho varies from -0.9 to 0.9 by 0.1
effect.type = "indirect", # sensitivity on ACME
# effect.type = "direct", # sensitivity on ADE
# effect.type = "both", # sensitivity on ACME and ADE
sims = 100)
summary(sens.out)
#>
#> Mediation Sensitivity Analysis: Average Mediation Effect
#>
#> Sensitivity Region: ACME for Control Group
#>
#> Rho ACME(control) 95% CI Lower 95% CI Upper R^2_M*R^2_Y* R^2_M~R^2_Y~
#> [1,] 0.3 0.0062 -0.0073 0.0188 0.09 0.0493
#> [2,] 0.4 -0.0084 -0.0238 0.0017 0.16 0.0877
#>
#> Rho at which ACME for Control Group = 0: 0.3
#> R^2_M*R^2_Y* at which ACME for Control Group = 0: 0.09
#> R^2_M~R^2_Y~ at which ACME for Control Group = 0: 0.0493
#>
#>
#> Sensitivity Region: ACME for Treatment Group
#>
#> Rho ACME(treated) 95% CI Lower 95% CI Upper R^2_M*R^2_Y* R^2_M~R^2_Y~
#> [1,] 0.3 0.0071 -0.0092 0.0213 0.09 0.0493
#> [2,] 0.4 -0.0101 -0.0295 0.0023 0.16 0.0877
#>
#> Rho at which ACME for Treatment Group = 0: 0.3
#> R^2_M*R^2_Y* at which ACME for Treatment Group = 0: 0.09
#> R^2_M~R^2_Y~ at which ACME for Treatment Group = 0: 0.0493ACME confidence intervals contains 0 when \(\rho \in (0.3,0.4)\)
Alternatively, using \(R^2\) interpretation, we need to specify the direction of confounder that affects the mediator and outcome variables in plot using sign.prod = "positive" (i.e., same direction) or sign.prod = "negative" (i.e., opposite direction).