29.1 MatchIt
Procedure typically involves (proposed by Noah Freifer using MatchIt)
- planning
- matching
- checking (balance)
- estimating the treatment effect
examine treat on re78
- Planning
select type of effect to be estimated (e.g., mediation effect, conditional effect, marginal effect)
select the target population
select variables to match/balance (Austin 2011) (T. J. VanderWeele 2019)
- Check Initial Imbalance
# No matching; constructing a pre-match matchit object
m.out0 <- matchit(
formula(treat ~ age + educ + race
+ married + nodegree + re74 + re75, env = lalonde),
data = data.frame(lalonde),
method = NULL,
# assess balance before matching
distance = "glm" # logistic regression
)
# Checking balance prior to matching
summary(m.out0)- Matching
# 1:1 NN PS matching w/o replacement
m.out1 <- matchit(treat ~ age + educ,
data = lalonde,
method = "nearest",
distance = "glm")
m.out1
#> A matchit object
#> - method: 1:1 nearest neighbor matching without replacement
#> - distance: Propensity score
#> - estimated with logistic regression
#> - number of obs.: 614 (original), 370 (matched)
#> - target estimand: ATT
#> - covariates: age, educ- Check balance
Sometimes you have to make trade-off between balance and sample size.
# Checking balance after NN matching
summary(m.out1, un = FALSE)
#>
#> Call:
#> matchit(formula = treat ~ age + educ, data = lalonde, method = "nearest",
#> distance = "glm")
#>
#> Summary of Balance for Matched Data:
#> Means Treated Means Control Std. Mean Diff. Var. Ratio eCDF Mean
#> distance 0.3080 0.3077 0.0094 0.9963 0.0033
#> age 25.8162 25.8649 -0.0068 1.0300 0.0050
#> educ 10.3459 10.2865 0.0296 0.5886 0.0253
#> eCDF Max Std. Pair Dist.
#> distance 0.0432 0.0146
#> age 0.0162 0.0597
#> educ 0.1189 0.8146
#>
#> Sample Sizes:
#> Control Treated
#> All 429 185
#> Matched 185 185
#> Unmatched 244 0
#> Discarded 0 0
# examine visually
plot(m.out1, type = "jitter", interactive = FALSE)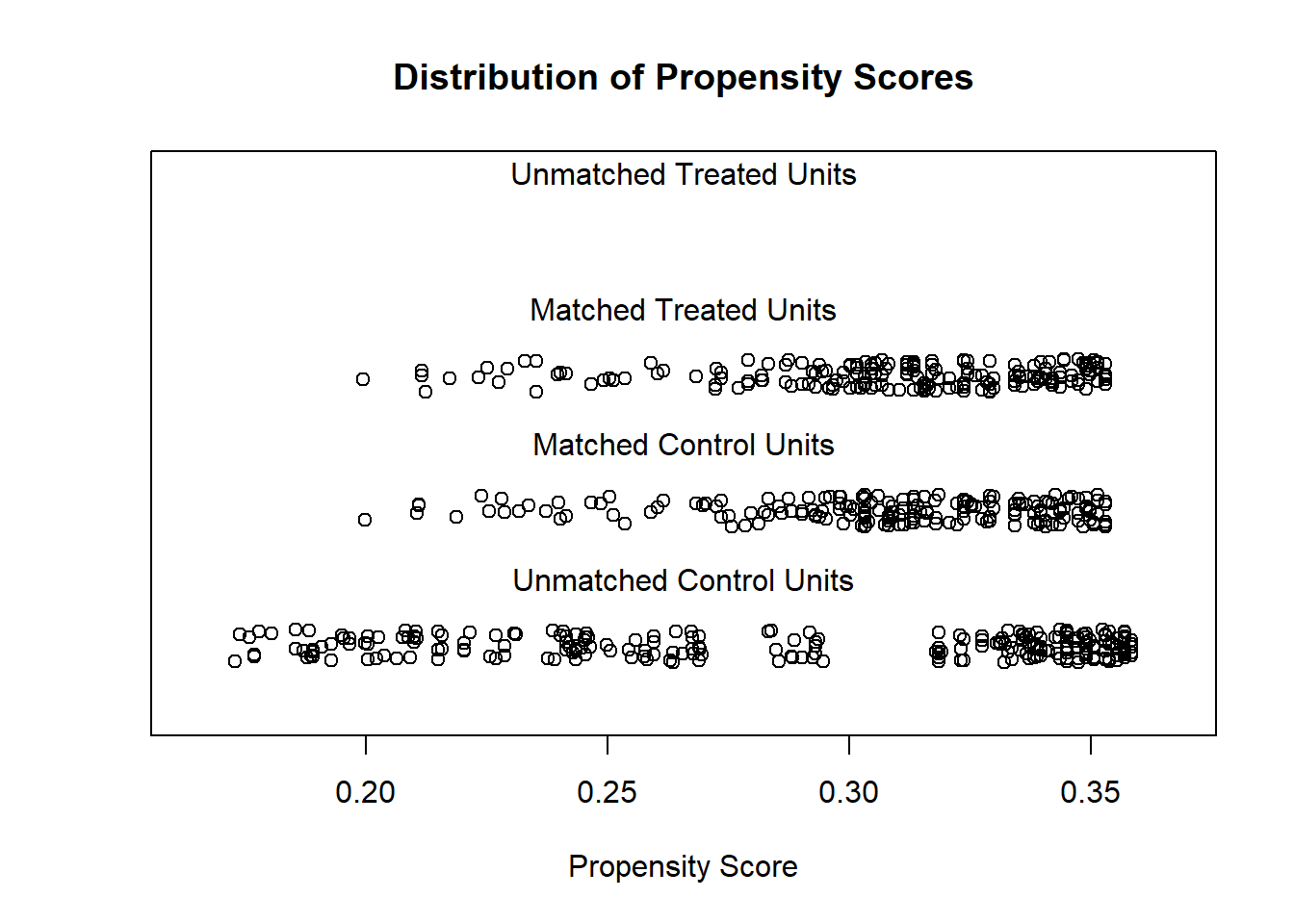
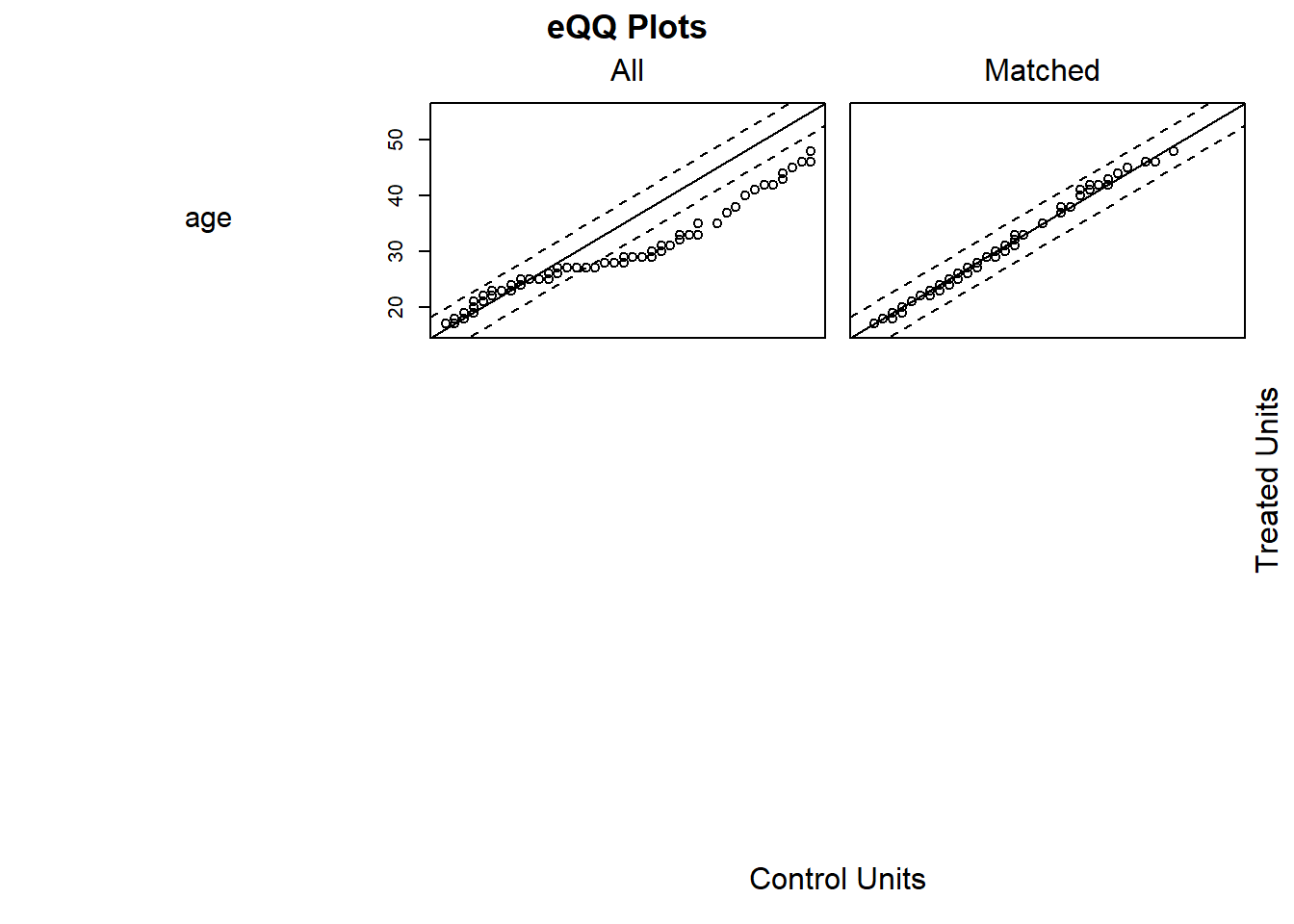
Try Full Match (i.e., every treated matches with one control, and every control with one treated).
# Full matching on a probit PS
m.out2 <- matchit(treat ~ age + educ,
data = lalonde,
method = "full",
distance = "glm",
link = "probit")
m.out2
#> A matchit object
#> - method: Optimal full matching
#> - distance: Propensity score
#> - estimated with probit regression
#> - number of obs.: 614 (original), 614 (matched)
#> - target estimand: ATT
#> - covariates: age, educChecking balance again
# Checking balance after full matching
summary(m.out2, un = FALSE)
#>
#> Call:
#> matchit(formula = treat ~ age + educ, data = lalonde, method = "full",
#> distance = "glm", link = "probit")
#>
#> Summary of Balance for Matched Data:
#> Means Treated Means Control Std. Mean Diff. Var. Ratio eCDF Mean
#> distance 0.3082 0.3081 0.0023 0.9815 0.0028
#> age 25.8162 25.8035 0.0018 0.9825 0.0062
#> educ 10.3459 10.2315 0.0569 0.4390 0.0481
#> eCDF Max Std. Pair Dist.
#> distance 0.0270 0.0382
#> age 0.0249 0.1110
#> educ 0.1300 0.9805
#>
#> Sample Sizes:
#> Control Treated
#> All 429. 185
#> Matched (ESS) 145.23 185
#> Matched 429. 185
#> Unmatched 0. 0
#> Discarded 0. 0
plot(summary(m.out2))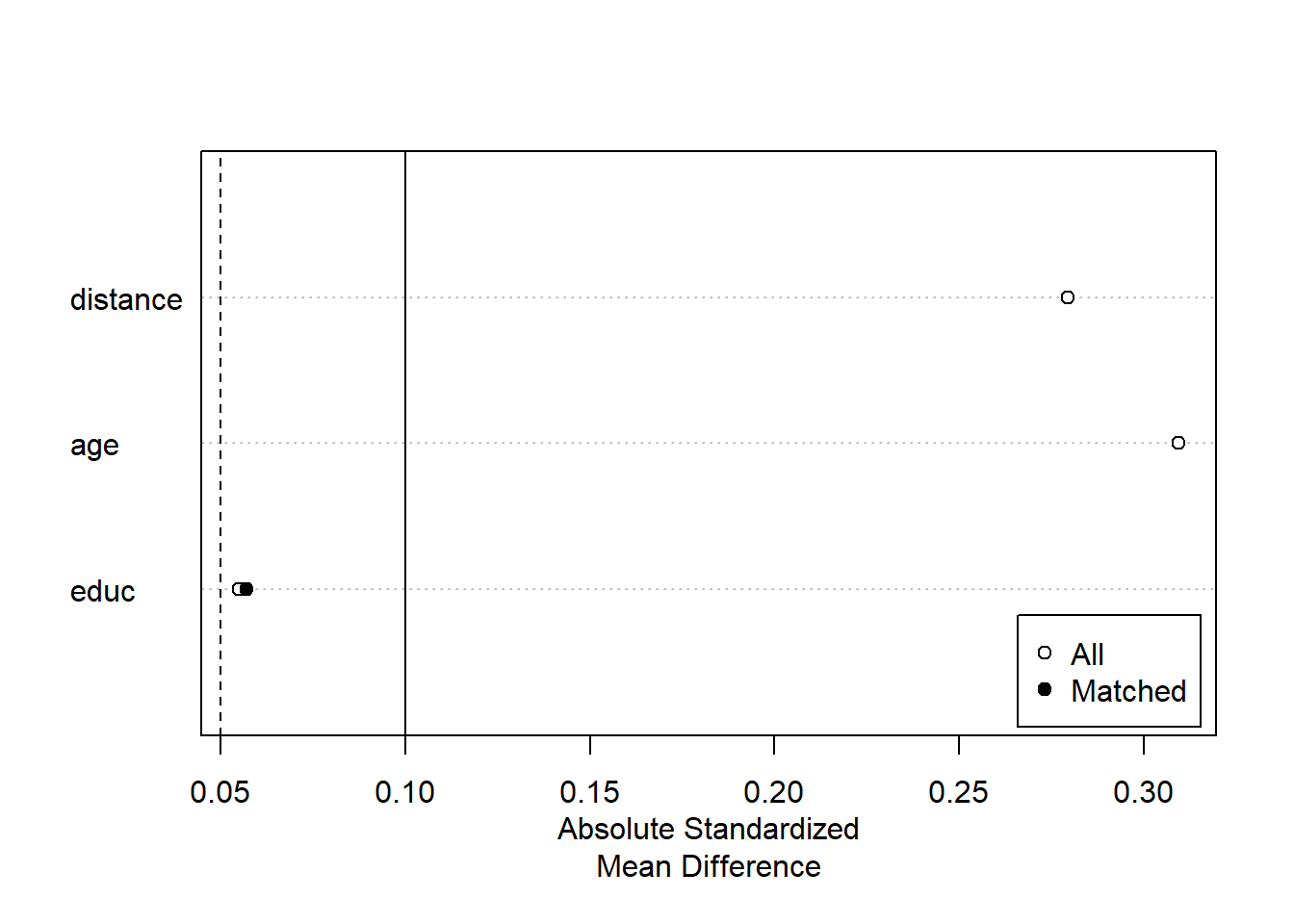
Exact Matching
# Full matching on a probit PS
m.out3 <-
matchit(
treat ~ age + educ,
data = lalonde,
method = "exact"
)
m.out3
#> A matchit object
#> - method: Exact matching
#> - number of obs.: 614 (original), 332 (matched)
#> - target estimand: ATT
#> - covariates: age, educSubclassfication
m.out4 <- matchit(
treat ~ age + educ,
data = lalonde,
method = "subclass"
)
m.out4
#> A matchit object
#> - method: Subclassification (6 subclasses)
#> - distance: Propensity score
#> - estimated with logistic regression
#> - number of obs.: 614 (original), 614 (matched)
#> - target estimand: ATT
#> - covariates: age, educ
# Or you can use in conjunction with "nearest"
m.out4 <- matchit(
treat ~ age + educ,
data = lalonde,
method = "nearest",
option = "subclass"
)
m.out4
#> A matchit object
#> - method: 1:1 nearest neighbor matching without replacement
#> - distance: Propensity score
#> - estimated with logistic regression
#> - number of obs.: 614 (original), 370 (matched)
#> - target estimand: ATT
#> - covariates: age, educOptimal Matching
m.out5 <- matchit(
treat ~ age + educ,
data = lalonde,
method = "optimal",
ratio = 2
)
m.out5
#> A matchit object
#> - method: 2:1 optimal pair matching
#> - distance: Propensity score
#> - estimated with logistic regression
#> - number of obs.: 614 (original), 555 (matched)
#> - target estimand: ATT
#> - covariates: age, educGenetic Matching
m.out6 <- matchit(
treat ~ age + educ,
data = lalonde,
method = "genetic"
)
m.out6
#> A matchit object
#> - method: 1:1 genetic matching without replacement
#> - distance: Propensity score
#> - estimated with logistic regression
#> - number of obs.: 614 (original), 370 (matched)
#> - target estimand: ATT
#> - covariates: age, educ- Estimating the Treatment Effect
# get matched data
m.data1 <- match.data(m.out1)
head(m.data1)
#> treat age educ race married nodegree re74 re75 re78 distance
#> NSW1 1 37 11 black 1 1 0 0 9930.0460 0.2536942
#> NSW2 1 22 9 hispan 0 1 0 0 3595.8940 0.3245468
#> NSW3 1 30 12 black 0 0 0 0 24909.4500 0.2881139
#> NSW4 1 27 11 black 0 1 0 0 7506.1460 0.3016672
#> NSW5 1 33 8 black 0 1 0 0 289.7899 0.2683025
#> NSW6 1 22 9 black 0 1 0 0 4056.4940 0.3245468
#> weights subclass
#> NSW1 1 1
#> NSW2 1 98
#> NSW3 1 109
#> NSW4 1 120
#> NSW5 1 131
#> NSW6 1 142library("lmtest") #coeftest
library("sandwich") #vcovCL
# imbalance matched dataset
fit1 <- lm(re78 ~ treat + age + educ ,
data = m.data1,
weights = weights)
coeftest(fit1, vcov. = vcovCL, cluster = ~subclass)
#>
#> t test of coefficients:
#>
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) -174.902 2445.013 -0.0715 0.943012
#> treat -1139.085 780.399 -1.4596 0.145253
#> age 153.133 55.317 2.7683 0.005922 **
#> educ 358.577 163.860 2.1883 0.029278 *
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1treat coefficient = estimated ATT
# balance matched dataset
m.data2 <- match.data(m.out2)
fit2 <- lm(re78 ~ treat + age + educ ,
data = m.data2, weights = weights)
coeftest(fit2, vcov. = vcovCL, cluster = ~subclass)
#>
#> t test of coefficients:
#>
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 2151.952 3141.152 0.6851 0.49355
#> treat -725.184 703.297 -1.0311 0.30289
#> age 120.260 53.933 2.2298 0.02612 *
#> educ 175.693 241.694 0.7269 0.46755
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1When reporting, remember to mention
- the matching specification (method, and additional options)
- the distance measure (e.g., propensity score)
- other methods, and rationale for the final chosen method.
- balance statistics of the matched dataset.
- number of matched, unmatched, discarded
- estimation method for treatment effect.
References
Austin, Peter C. 2011. “Optimal Caliper Widths for Propensity-Score Matching When Estimating Differences in Means and Differences in Proportions in Observational Studies.” Pharmaceutical Statistics 10 (2): 150–61.
VanderWeele, Tyler J. 2019. “Principles of Confounder Selection.” European Journal of Epidemiology 34: 211–19.