第 35 章 回望tidyverse之旅
前面几章先后介绍了tidyverse套餐的若干部件。感觉很难吗?如果是,那说明你认真听了。
本章做个小结,通过案例复习和串讲下tidyverse中常用的核心部件(事实上,tidyverse套餐比我们列出的要丰富)。
图 35.1: 图片来源Silvia Canelón在R-Ladies Chicago的报告
35.1 readr 宏包

读入数据是第一步,我们可以用readr导入数据
提示:
-
逗号(
,)分割的文件read_csv() -
制表符(
tab)分割的文件read_tsv() -
任意的分割符
read_delim() -
固定宽度的文件
read_fwf() -
空格分割的文件
read_table() -
网页log文件
read_log()
读取外部数据
penguins <- read_csv("./demo_data/penguins.csv") 保存到外部文件
35.2 tibble 宏包

tibble 是升级版的 dataframe, 之所以是升级版,是因为在tidyverse中tibble做很多优化。下面你可以看到两者的区别:
as_tibble(penguins)## # A tibble: 344 × 8
## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
## <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Adelie Torgersen 39.1 18.7 181 3750
## 2 Adelie Torgersen 39.5 17.4 186 3800
## 3 Adelie Torgersen 40.3 18 195 3250
## 4 Adelie Torgersen NA NA NA NA
## 5 Adelie Torgersen 36.7 19.3 193 3450
## 6 Adelie Torgersen 39.3 20.6 190 3650
## 7 Adelie Torgersen 38.9 17.8 181 3625
## 8 Adelie Torgersen 39.2 19.6 195 4675
## 9 Adelie Torgersen 34.1 18.1 193 3475
## 10 Adelie Torgersen 42 20.2 190 4250
## # ℹ 334 more rows
## # ℹ 2 more variables: sex <chr>, year <dbl>
as.data.frame(penguins) %>% head()## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
## 1 Adelie Torgersen 39.1 18.7 181 3750
## 2 Adelie Torgersen 39.5 17.4 186 3800
## 3 Adelie Torgersen 40.3 18.0 195 3250
## 4 Adelie Torgersen NA NA NA NA
## 5 Adelie Torgersen 36.7 19.3 193 3450
## 6 Adelie Torgersen 39.3 20.6 190 3650
## sex year
## 1 male 2007
## 2 female 2007
## 3 female 2007
## 4 <NA> 2007
## 5 female 2007
## 6 male 2007在R Markdown里两者区别不大,但在console中,区别很明显的。比如tibble不一样的地方有:
- 列出了变量的类型(这个很不错)
- 只列出10行
- 只列出有限的列数(与屏幕适应的)
- 高亮
NAs
35.3 ggplot2 宏包

35.3.1 查看数据
我们先查看下数据
glimpse(penguins)## Rows: 344
## Columns: 8
## $ species <chr> "Adelie", "Adelie", "Adelie", "Adelie", "Adelie", "A…
## $ island <chr> "Torgersen", "Torgersen", "Torgersen", "Torgersen", …
## $ bill_length_mm <dbl> 39.1, 39.5, 40.3, NA, 36.7, 39.3, 38.9, 39.2, 34.1, …
## $ bill_depth_mm <dbl> 18.7, 17.4, 18.0, NA, 19.3, 20.6, 17.8, 19.6, 18.1, …
## $ flipper_length_mm <dbl> 181, 186, 195, NA, 193, 190, 181, 195, 193, 190, 186…
## $ body_mass_g <dbl> 3750, 3800, 3250, NA, 3450, 3650, 3625, 4675, 3475, …
## $ sex <chr> "male", "female", "female", NA, "female", "male", "f…
## $ year <dbl> 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007…35.3.3 箱线图
ggplot(data = penguins, aes(x = sex, y = body_mass_g)) +
geom_boxplot()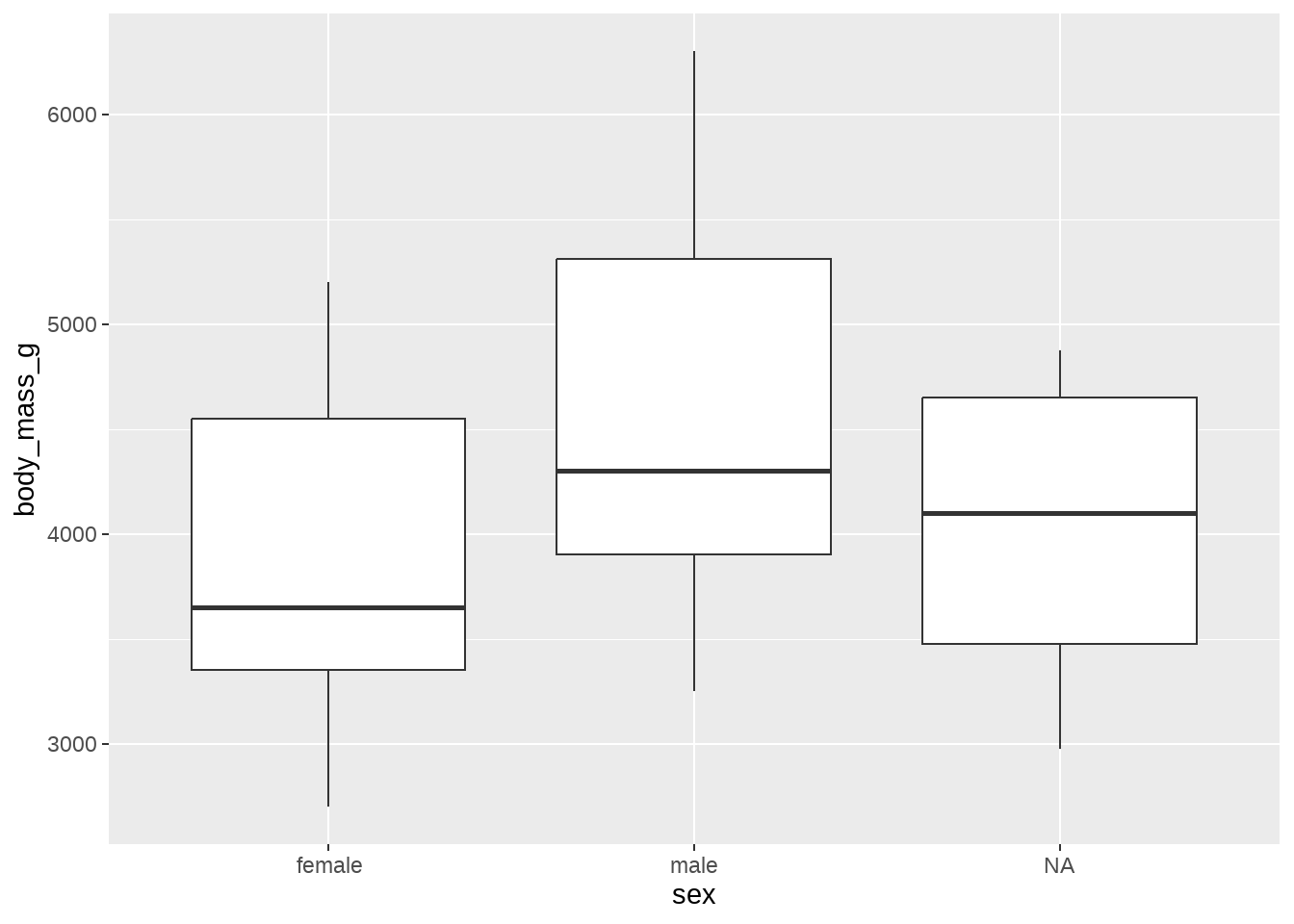
ggplot(data = penguins, aes(x = sex, y = body_mass_g)) +
geom_boxplot(aes(fill = species))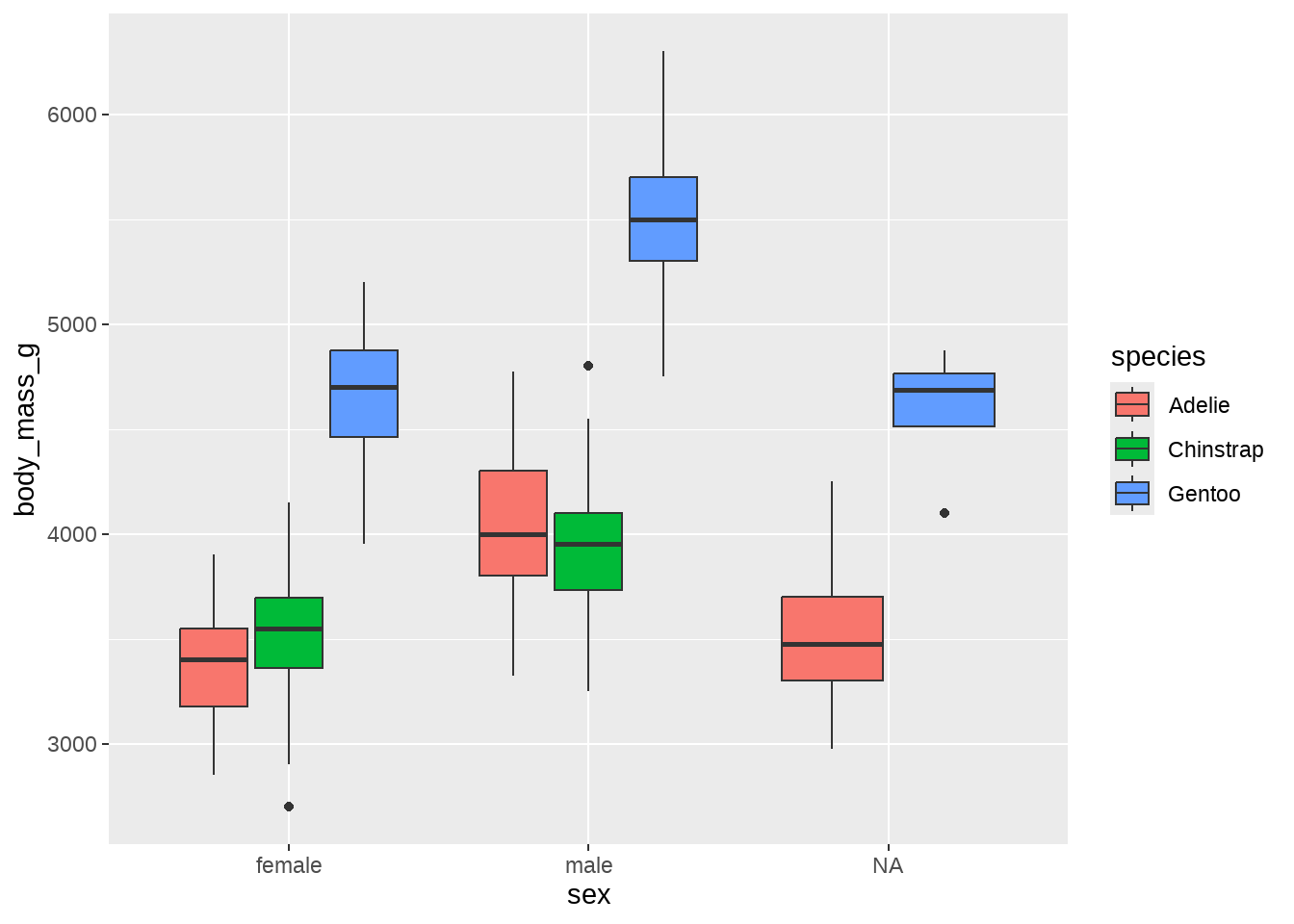
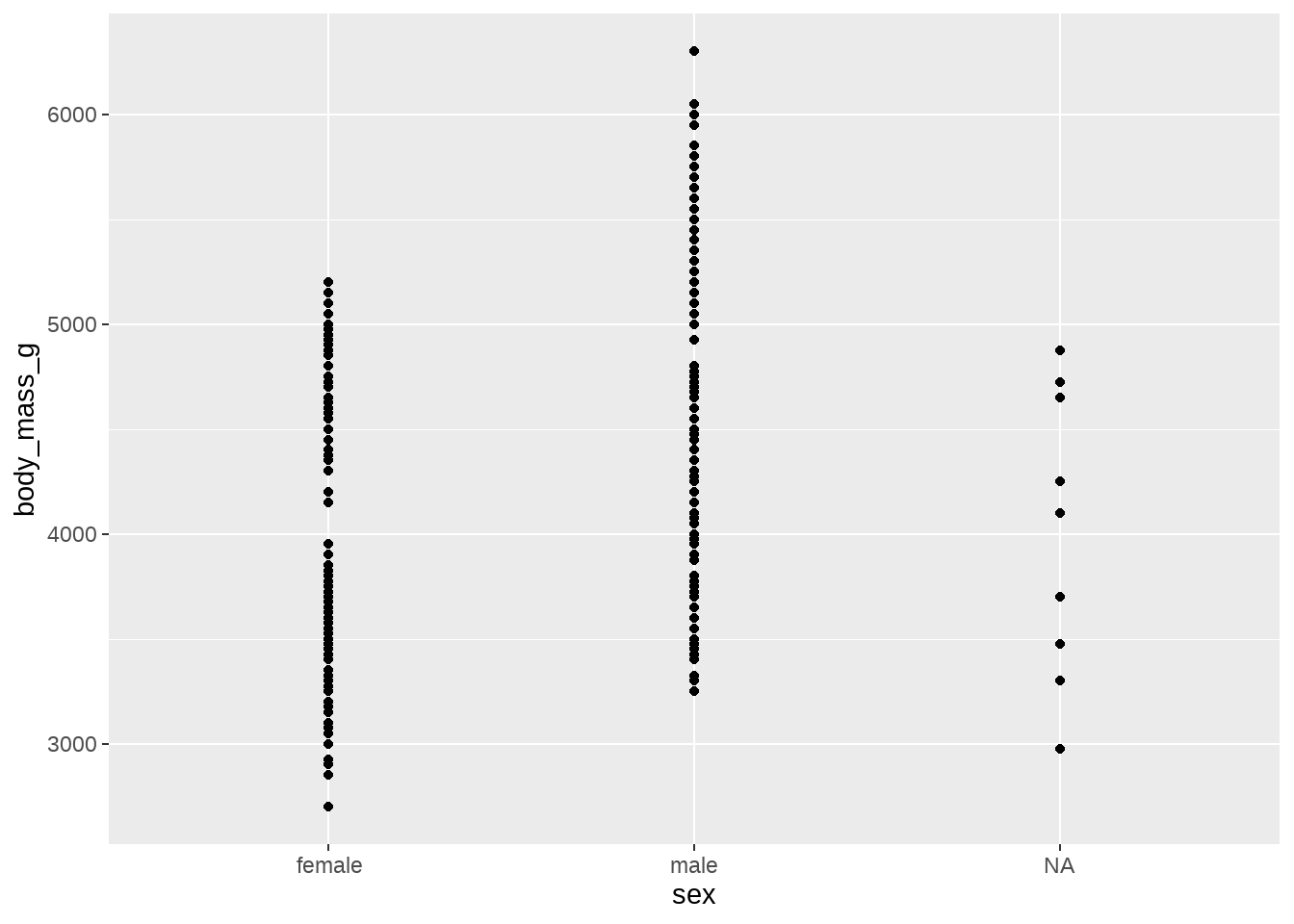
我们可能看到:
- Gentoo 类的企鹅 比 Adelie 和 Chinstrap 类的企鹅体重更重
- Gentoo 类型中,雄性企鹅比雌性企鹅体重更重
- Adelie 和 Chinstrap 两种类型的企鹅,区别不是很明显
- sex 这个变量有缺失值,主要集中在 Gentoo 和 Chinstrap 两种类型
那么每种类型的企鹅,数据中有多少是NA呢? 上dplyr吧!
35.4 dplyr 宏包

dplyr 宏包可以:
35.4.1 选取列
下面两个有什么区别?
select(penguins, species, sex, body_mass_g)## # A tibble: 344 × 3
## species sex body_mass_g
## <chr> <chr> <dbl>
## 1 Adelie male 3750
## 2 Adelie female 3800
## 3 Adelie female 3250
## 4 Adelie <NA> NA
## 5 Adelie female 3450
## 6 Adelie male 3650
## 7 Adelie female 3625
## 8 Adelie male 4675
## 9 Adelie <NA> 3475
## 10 Adelie <NA> 4250
## # ℹ 334 more rows## # A tibble: 344 × 3
## species sex body_mass_g
## <chr> <chr> <dbl>
## 1 Adelie male 3750
## 2 Adelie female 3800
## 3 Adelie female 3250
## 4 Adelie <NA> NA
## 5 Adelie female 3450
## 6 Adelie male 3650
## 7 Adelie female 3625
## 8 Adelie male 4675
## 9 Adelie <NA> 3475
## 10 Adelie <NA> 4250
## # ℹ 334 more rows35.4.2 行方向排序
glimpse(penguins)## Rows: 344
## Columns: 8
## $ species <chr> "Adelie", "Adelie", "Adelie", "Adelie", "Adelie", "A…
## $ island <chr> "Torgersen", "Torgersen", "Torgersen", "Torgersen", …
## $ bill_length_mm <dbl> 39.1, 39.5, 40.3, NA, 36.7, 39.3, 38.9, 39.2, 34.1, …
## $ bill_depth_mm <dbl> 18.7, 17.4, 18.0, NA, 19.3, 20.6, 17.8, 19.6, 18.1, …
## $ flipper_length_mm <dbl> 181, 186, 195, NA, 193, 190, 181, 195, 193, 190, 186…
## $ body_mass_g <dbl> 3750, 3800, 3250, NA, 3450, 3650, 3625, 4675, 3475, …
## $ sex <chr> "male", "female", "female", NA, "female", "male", "f…
## $ year <dbl> 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007…## # A tibble: 344 × 3
## species sex body_mass_g
## <chr> <chr> <dbl>
## 1 Gentoo male 6300
## 2 Gentoo male 6050
## 3 Gentoo male 6000
## 4 Gentoo male 6000
## 5 Gentoo male 5950
## 6 Gentoo male 5950
## 7 Gentoo male 5850
## 8 Gentoo male 5850
## 9 Gentoo male 5850
## 10 Gentoo male 5800
## # ℹ 334 more rows35.4.3 分组统计
## # A tibble: 8 × 3
## # Groups: species [3]
## species sex count
## <chr> <chr> <int>
## 1 Adelie female 73
## 2 Adelie male 73
## 3 Adelie <NA> 6
## 4 Chinstrap female 34
## 5 Chinstrap male 34
## 6 Gentoo female 58
## 7 Gentoo male 61
## 8 Gentoo <NA> 535.4.4 增加列
penguins %>%
group_by(species) %>%
mutate(count_species = n()) %>%
ungroup() %>%
group_by(species, sex, count_species) %>%
summarize(count = n()) %>%
mutate(prop = count/count_species*100)## # A tibble: 8 × 5
## # Groups: species, sex [8]
## species sex count_species count prop
## <chr> <chr> <int> <int> <dbl>
## 1 Adelie female 152 73 48.0
## 2 Adelie male 152 73 48.0
## 3 Adelie <NA> 152 6 3.95
## 4 Chinstrap female 68 34 50
## 5 Chinstrap male 68 34 50
## 6 Gentoo female 124 58 46.8
## 7 Gentoo male 124 61 49.2
## 8 Gentoo <NA> 124 5 4.0335.4.5 筛选
penguins %>%
group_by(species) %>%
mutate(count_species = n()) %>%
ungroup() %>%
group_by(species, sex, count_species) %>%
summarize(count = n()) %>%
mutate(percentage = count/count_species*100) %>%
filter(species == "Chinstrap")## # A tibble: 2 × 5
## # Groups: species, sex [2]
## species sex count_species count percentage
## <chr> <chr> <int> <int> <dbl>
## 1 Chinstrap female 68 34 50
## 2 Chinstrap male 68 34 5035.5 forcats 宏包

forcats 宏包主要用于分类变量和因子型变量,比如这里的 species, island, sex.
对于不是因子型的变量,比如这里 year 是数值型变量,我们也可以通过 factor() 函数 将它转换成因子型变量。
## # A tibble: 344 × 9
## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
## <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Adelie Torgersen 39.1 18.7 181 3750
## 2 Adelie Torgersen 39.5 17.4 186 3800
## 3 Adelie Torgersen 40.3 18 195 3250
## 4 Adelie Torgersen NA NA NA NA
## 5 Adelie Torgersen 36.7 19.3 193 3450
## 6 Adelie Torgersen 39.3 20.6 190 3650
## 7 Adelie Torgersen 38.9 17.8 181 3625
## 8 Adelie Torgersen 39.2 19.6 195 4675
## 9 Adelie Torgersen 34.1 18.1 193 3475
## 10 Adelie Torgersen 42 20.2 190 4250
## # ℹ 334 more rows
## # ℹ 3 more variables: sex <chr>, year <dbl>, year_factor <fct>我们保存到新的数据集中,再看看有什么变化
## # A tibble: 344 × 9
## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
## <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Adelie Torgersen 39.1 18.7 181 3750
## 2 Adelie Torgersen 39.5 17.4 186 3800
## 3 Adelie Torgersen 40.3 18 195 3250
## 4 Adelie Torgersen NA NA NA NA
## 5 Adelie Torgersen 36.7 19.3 193 3450
## 6 Adelie Torgersen 39.3 20.6 190 3650
## 7 Adelie Torgersen 38.9 17.8 181 3625
## 8 Adelie Torgersen 39.2 19.6 195 4675
## 9 Adelie Torgersen 34.1 18.1 193 3475
## 10 Adelie Torgersen 42 20.2 190 4250
## # ℹ 334 more rows
## # ℹ 3 more variables: sex <chr>, year <dbl>, year_factor <fct>
class(penguins_new$year_factor)## [1] "factor"
levels(penguins_new$year_factor)## [1] "2007" "2008" "2009"大家回想下,弄成因子型变量有什么好处呢?
35.6 stringr 宏包

stringr宏包包含了非常丰富的处理字符串的函数,比如
- 匹配
- 字符串子集
- 字符串长度
- 字符串合并
- 字符串分割
- 更多
35.6.1 字符串转换
penguins %>%
select(species, island) %>%
mutate(ISLAND = str_to_upper(island))## # A tibble: 344 × 3
## species island ISLAND
## <chr> <chr> <chr>
## 1 Adelie Torgersen TORGERSEN
## 2 Adelie Torgersen TORGERSEN
## 3 Adelie Torgersen TORGERSEN
## 4 Adelie Torgersen TORGERSEN
## 5 Adelie Torgersen TORGERSEN
## 6 Adelie Torgersen TORGERSEN
## 7 Adelie Torgersen TORGERSEN
## 8 Adelie Torgersen TORGERSEN
## 9 Adelie Torgersen TORGERSEN
## 10 Adelie Torgersen TORGERSEN
## # ℹ 334 more rows35.6.2 字符串合并
penguins %>%
select(species, island) %>%
mutate(ISLAND = str_to_upper(island)) %>%
mutate(species_island = str_c(species, ISLAND, sep = "_"))## # A tibble: 344 × 4
## species island ISLAND species_island
## <chr> <chr> <chr> <chr>
## 1 Adelie Torgersen TORGERSEN Adelie_TORGERSEN
## 2 Adelie Torgersen TORGERSEN Adelie_TORGERSEN
## 3 Adelie Torgersen TORGERSEN Adelie_TORGERSEN
## 4 Adelie Torgersen TORGERSEN Adelie_TORGERSEN
## 5 Adelie Torgersen TORGERSEN Adelie_TORGERSEN
## 6 Adelie Torgersen TORGERSEN Adelie_TORGERSEN
## 7 Adelie Torgersen TORGERSEN Adelie_TORGERSEN
## 8 Adelie Torgersen TORGERSEN Adelie_TORGERSEN
## 9 Adelie Torgersen TORGERSEN Adelie_TORGERSEN
## 10 Adelie Torgersen TORGERSEN Adelie_TORGERSEN
## # ℹ 334 more rows35.7 tidyr 宏包

想想什么叫tidy data?
35.7.1 长表格变宽表格
untidy_penguins <-
penguins %>%
pivot_wider(names_from = sex,
values_from = body_mass_g)
untidy_penguins## # A tibble: 344 × 9
## species island bill_length_mm bill_depth_mm flipper_length_mm year male
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Adelie Torgersen 39.1 18.7 181 2007 3750
## 2 Adelie Torgersen 39.5 17.4 186 2007 NA
## 3 Adelie Torgersen 40.3 18 195 2007 NA
## 4 Adelie Torgersen NA NA NA 2007 NA
## 5 Adelie Torgersen 36.7 19.3 193 2007 NA
## 6 Adelie Torgersen 39.3 20.6 190 2007 3650
## 7 Adelie Torgersen 38.9 17.8 181 2007 NA
## 8 Adelie Torgersen 39.2 19.6 195 2007 4675
## 9 Adelie Torgersen 34.1 18.1 193 2007 NA
## 10 Adelie Torgersen 42 20.2 190 2007 NA
## # ℹ 334 more rows
## # ℹ 2 more variables: female <dbl>, `NA` <dbl>35.7.2 宽表格变长表格
untidy_penguins %>%
pivot_longer(cols = male:`NA`,
names_to = "sex",
values_to = "body_mass_g")## # A tibble: 1,032 × 8
## species island bill_length_mm bill_depth_mm flipper_length_mm year sex
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr>
## 1 Adelie Torgersen 39.1 18.7 181 2007 male
## 2 Adelie Torgersen 39.1 18.7 181 2007 female
## 3 Adelie Torgersen 39.1 18.7 181 2007 NA
## 4 Adelie Torgersen 39.5 17.4 186 2007 male
## 5 Adelie Torgersen 39.5 17.4 186 2007 female
## 6 Adelie Torgersen 39.5 17.4 186 2007 NA
## 7 Adelie Torgersen 40.3 18 195 2007 male
## 8 Adelie Torgersen 40.3 18 195 2007 female
## 9 Adelie Torgersen 40.3 18 195 2007 NA
## 10 Adelie Torgersen NA NA NA 2007 male
## # ℹ 1,022 more rows
## # ℹ 1 more variable: body_mass_g <dbl>35.8 purrr 宏包

purrr 宏包提供了map()等一系列函数,取代 for 和 while循环方式,实现高效迭代,保持语法一致性,同时增强了代码的可读性。
## $species
## [1] 0
##
## $island
## [1] 0
##
## $bill_length_mm
## [1] 2
##
## $bill_depth_mm
## [1] 2
##
## $flipper_length_mm
## [1] 2
##
## $body_mass_g
## [1] 2
##
## $sex
## [1] 11
##
## $year
## [1] 0
penguins %>%
group_nest(species) %>%
mutate(model = purrr::map(data, ~ lm(bill_depth_mm ~ bill_length_mm, data = .))) %>%
mutate(result = purrr::map(model, ~ broom::tidy(.))) %>%
tidyr::unnest(result)## # A tibble: 6 × 8
## species data model term estimate std.error statistic p.value
## <chr> <list<tibble[,7]>> <lis> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Adelie [152 × 7] <lm> (Int… 11.4 1.34 8.52 1.61e-14
## 2 Adelie [152 × 7] <lm> bill… 0.179 0.0344 5.19 6.67e- 7
## 3 Chinstrap [68 × 7] <lm> (Int… 7.57 1.55 4.88 6.99e- 6
## 4 Chinstrap [68 × 7] <lm> bill… 0.222 0.0317 7.01 1.53e- 9
## 5 Gentoo [124 × 7] <lm> (Int… 5.25 1.05 4.98 2.15e- 6
## 6 Gentoo [124 × 7] <lm> bill… 0.205 0.0222 9.24 1.02e-15