第 84 章 探索性数据分析-企鹅的故事
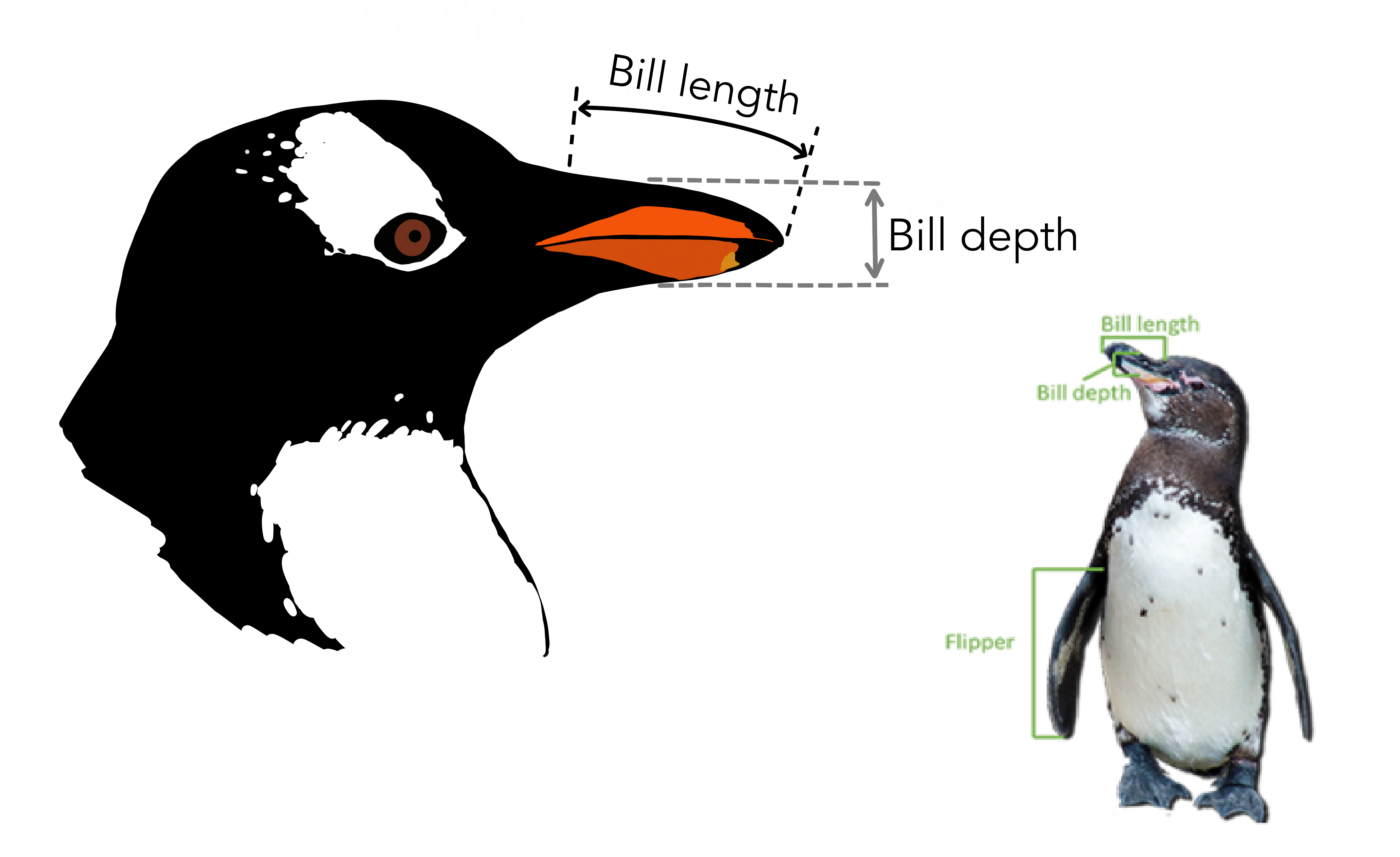
今天讲一个关于企鹅的数据故事。这个故事来源于科考人员记录的大量企鹅体征数据,图片来源这里.

84.1 数据
84.1.1 导入数据
可通过宏包palmerpenguins::penguins获取数据,也可以读取本地penguins.csv文件,
我们采取后面一种方法:
library(tidyverse)
penguins <- read_csv("./demo_data/penguins.csv") %>%
janitor::clean_names()
penguins %>%
head()## # A tibble: 6 × 8
## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
## <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Adelie Torgersen 39.1 18.7 181 3750
## 2 Adelie Torgersen 39.5 17.4 186 3800
## 3 Adelie Torgersen 40.3 18 195 3250
## 4 Adelie Torgersen NA NA NA NA
## 5 Adelie Torgersen 36.7 19.3 193 3450
## 6 Adelie Torgersen 39.3 20.6 190 3650
## # ℹ 2 more variables: sex <chr>, year <dbl>84.1.2 变量含义
| variable | class | description |
|---|---|---|
| species | integer | 企鹅种类 (Adelie, Gentoo, Chinstrap) |
| island | integer | 所在岛屿 (Biscoe, Dream, Torgersen) |
| bill_length_mm | double | 嘴峰长度 (单位毫米) |
| bill_depth_mm | double | 嘴峰深度 (单位毫米) |
| flipper_length_mm | integer | 鰭肢长度 (单位毫米) |
| body_mass_g | integer | 体重 (单位克) |
| sex | integer | 性别 |
| year | integer | 记录年份 |
84.1.3 数据清洗
检查缺失值(NA)这个很重要!
## # A tibble: 1 × 8
## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
## <int> <int> <int> <int> <int> <int>
## 1 0 0 2 2 2 2
## # ℹ 2 more variables: sex <int>, year <int>有缺失值的地方找出来看看
penguins %>% filter_all(
any_vars(is.na(.))
)## # A tibble: 11 × 8
## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
## <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Adelie Torgersen NA NA NA NA
## 2 Adelie Torgersen 34.1 18.1 193 3475
## 3 Adelie Torgersen 42 20.2 190 4250
## 4 Adelie Torgersen 37.8 17.1 186 3300
## 5 Adelie Torgersen 37.8 17.3 180 3700
## 6 Adelie Dream 37.5 18.9 179 2975
## 7 Gentoo Biscoe 44.5 14.3 216 4100
## 8 Gentoo Biscoe 46.2 14.4 214 4650
## 9 Gentoo Biscoe 47.3 13.8 216 4725
## 10 Gentoo Biscoe 44.5 15.7 217 4875
## 11 Gentoo Biscoe NA NA NA NA
## # ℹ 2 more variables: sex <chr>, year <dbl>发现共有11行至少有一处有缺失值,于是我们就删除这些行
## # A tibble: 333 × 8
## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
## <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Adelie Torgersen 39.1 18.7 181 3750
## 2 Adelie Torgersen 39.5 17.4 186 3800
## 3 Adelie Torgersen 40.3 18 195 3250
## 4 Adelie Torgersen 36.7 19.3 193 3450
## 5 Adelie Torgersen 39.3 20.6 190 3650
## 6 Adelie Torgersen 38.9 17.8 181 3625
## 7 Adelie Torgersen 39.2 19.6 195 4675
## 8 Adelie Torgersen 41.1 17.6 182 3200
## 9 Adelie Torgersen 38.6 21.2 191 3800
## 10 Adelie Torgersen 34.6 21.1 198 4400
## # ℹ 323 more rows
## # ℹ 2 more variables: sex <chr>, year <dbl>84.2 探索性分析
大家可以提出自己想探索的内容:
- 每种类型企鹅有多少只?
- 每种类型企鹅各种属性的均值和分布?
- 嘴峰长度和深度的关联?
- 体重与翅膀长度的关联?
- 嘴峰长度与嘴峰深度的比例?
- 不同种类的宝宝,体重具有显著性差异?
- 这体征中哪个因素对性别影响最大?
- …
84.2.1 每种类型企鹅有多少只
## # A tibble: 3 × 2
## species n
## <chr> <int>
## 1 Adelie 146
## 2 Gentoo 119
## 3 Chinstrap 6884.2.2 每个岛屿有多少企鹅?
## # A tibble: 3 × 2
## island n
## <chr> <int>
## 1 Biscoe 163
## 2 Dream 123
## 3 Torgersen 4784.2.3 每种类型企鹅各种体征属性的均值和分布
## # A tibble: 3 × 6
## species bill_length_mm bill_depth_mm flipper_length_mm body_mass_g year
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Adelie 38.8 18.3 190. 3706. 2008.
## 2 Chinstrap 48.8 18.4 196. 3733. 2008.
## 3 Gentoo 47.6 15.0 217. 5092. 2008.84.2.4 每种类型企鹅的嘴峰长度的分布
penguins %>%
ggplot(aes(x = bill_length_mm)) +
geom_density() +
facet_wrap(vars(species), scales = "free")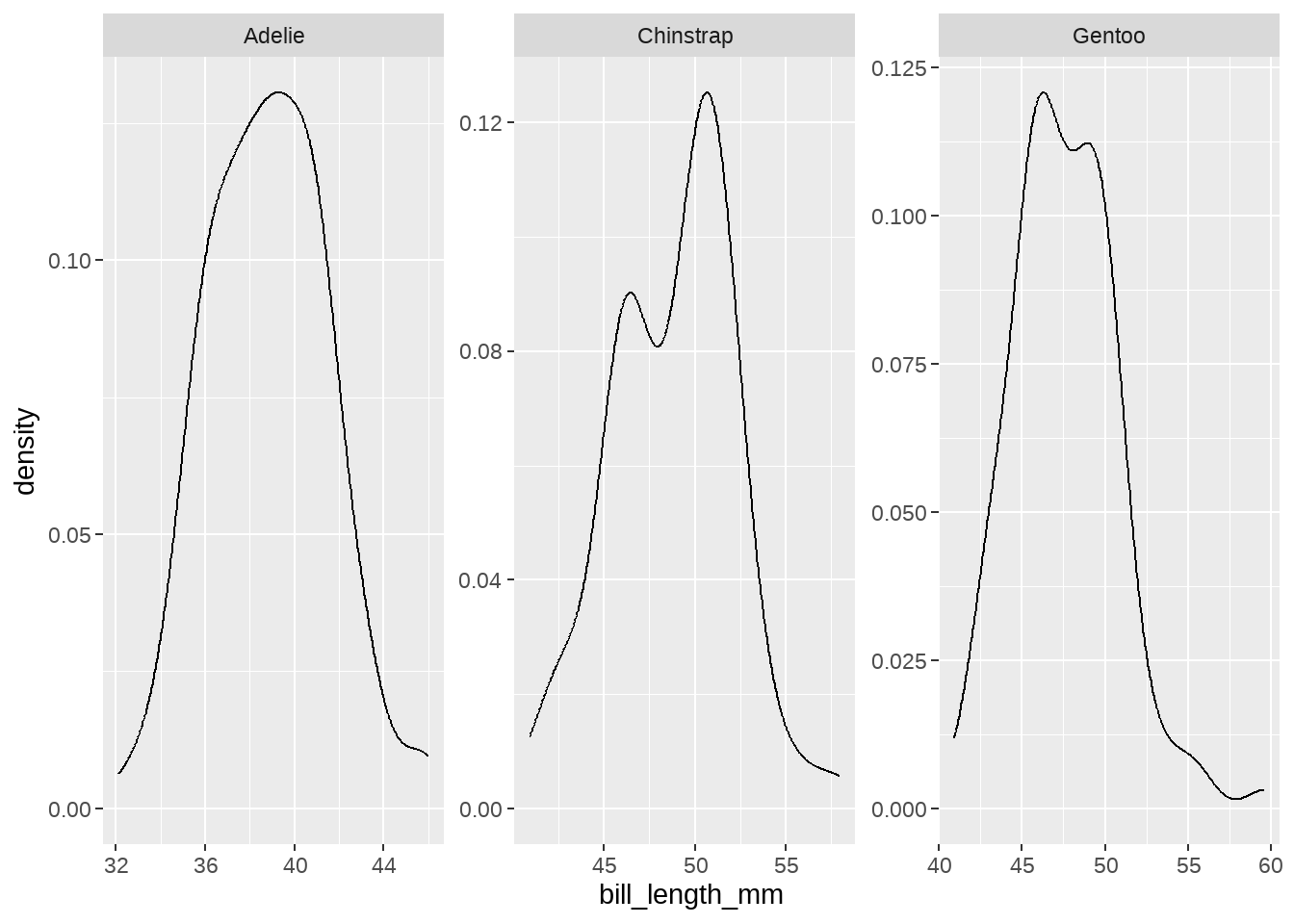
84.2.5 每种类型企鹅的嘴峰长度的分布(分性别)
penguins %>%
ggplot(aes(x = bill_length_mm)) +
geom_density(aes(fill = sex)) +
facet_wrap(vars(species), scales = "free")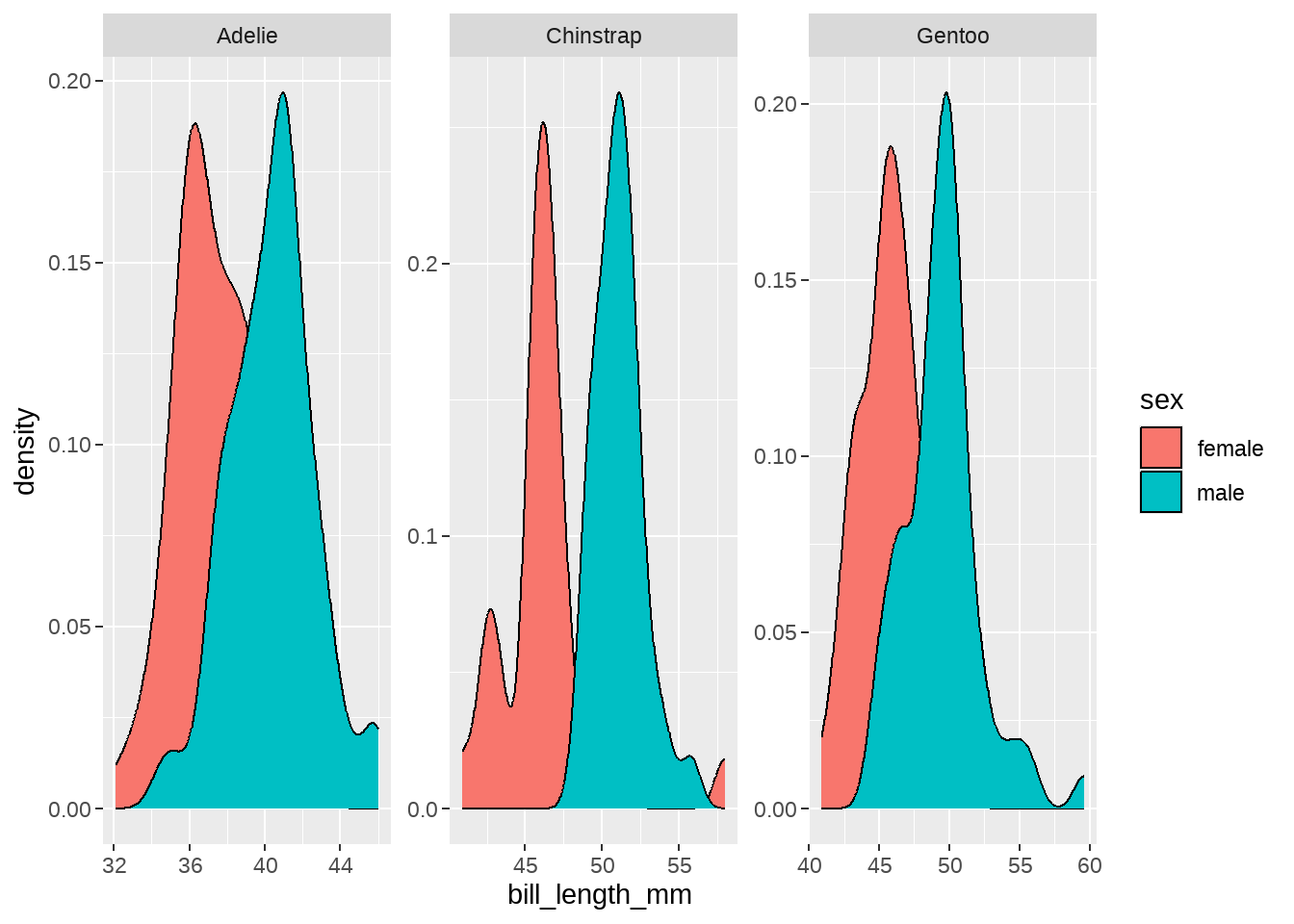
男宝宝的嘴巴要长些,哈哈。
来张更好看点的
penguins %>%
ggplot(aes(x = bill_length_mm, fill = sex)) +
geom_histogram(
position = "identity",
alpha = 0.7,
bins = 25
) +
scale_fill_manual(values = c("#66b3ff", "#8c8c8c")) +
ylab("number of penguins") +
xlab("length (mm)") +
theme_minimal() +
theme(
legend.position = "bottom",
legend.text = element_text(size = 11),
legend.title = element_blank(),
panel.grid.minor = element_blank(),
axis.title = element_text(color = "white", size = 10),
plot.title = element_text(size = 20),
plot.subtitle = element_text(size = 12, hjust = 1)
) +
facet_wrap(vars(species), scales = "free")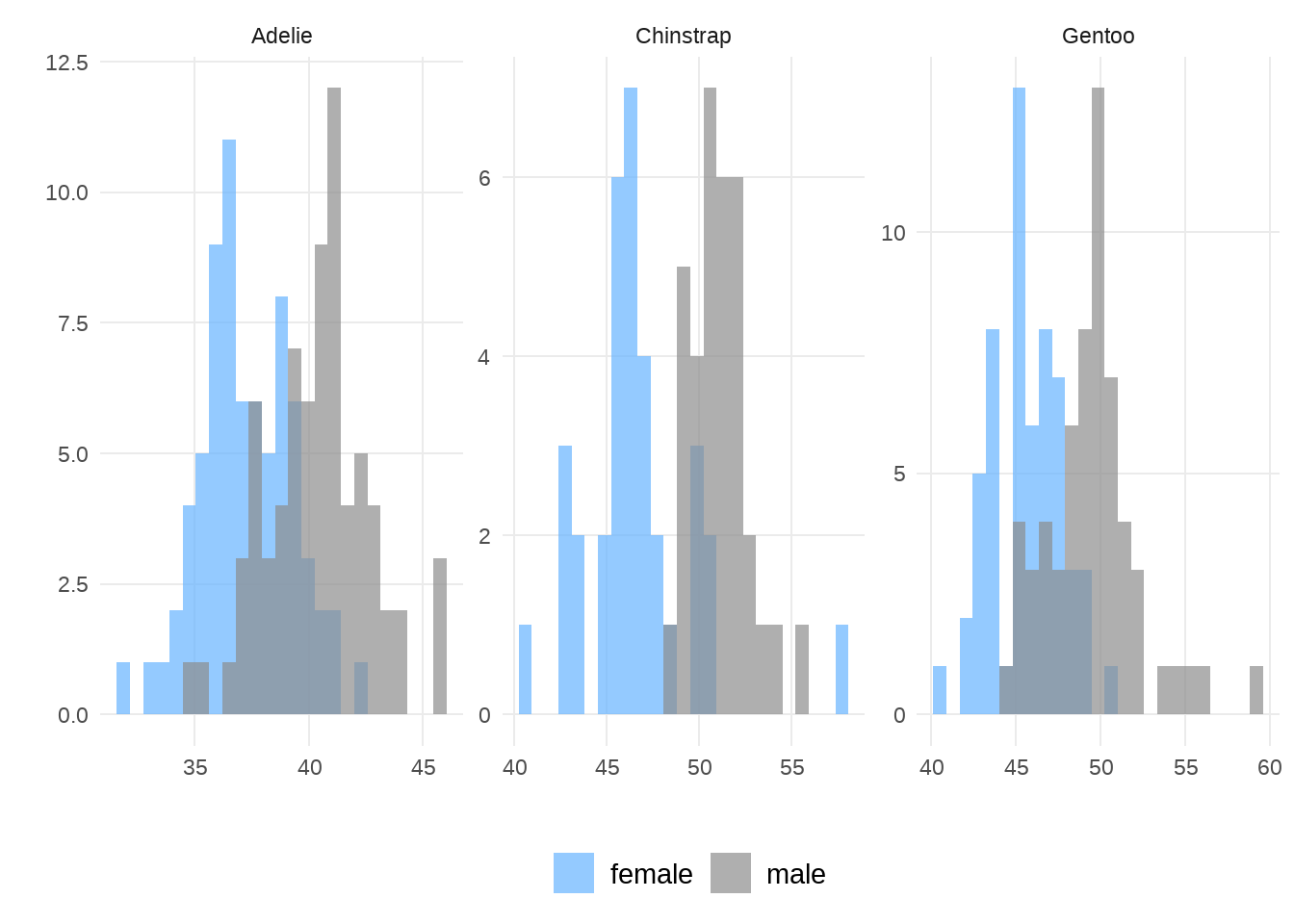
同理,可以画出其他属性的分布。当然,我更喜欢用山峦图来呈现不同分组的分布,因为竖直方向可以更方便比较
library(ggridges)
penguins %>%
ggplot(aes(x = bill_length_mm, y = species, fill = species)) +
ggridges::geom_density_ridges()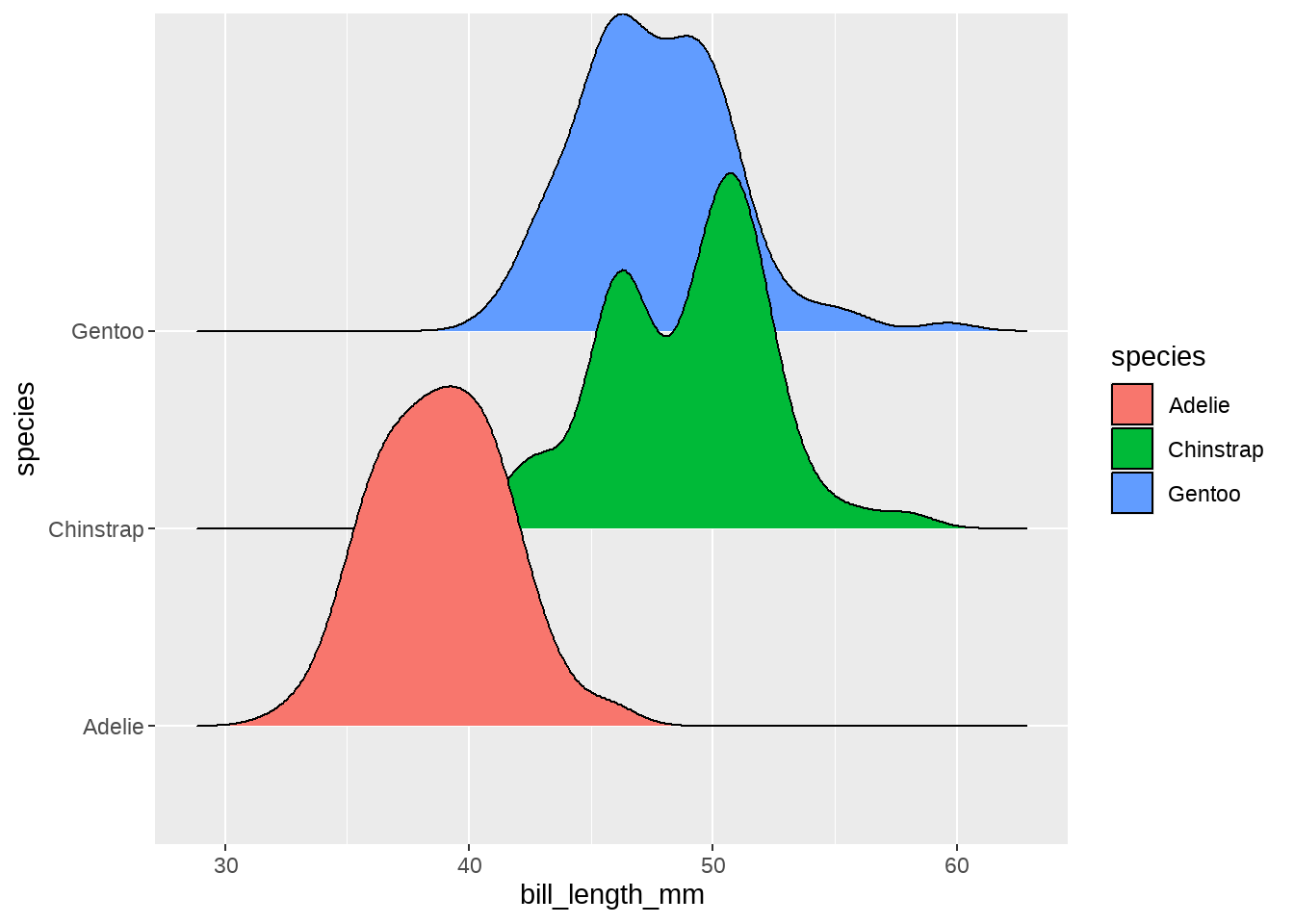
同样,我们也用颜色区分下性别,这样不同种类、不同性别企鹅的嘴峰长度分布一目了然
penguins %>%
ggplot(aes(x = bill_length_mm, y = species, fill = sex)) +
geom_density_ridges(alpha = 0.5)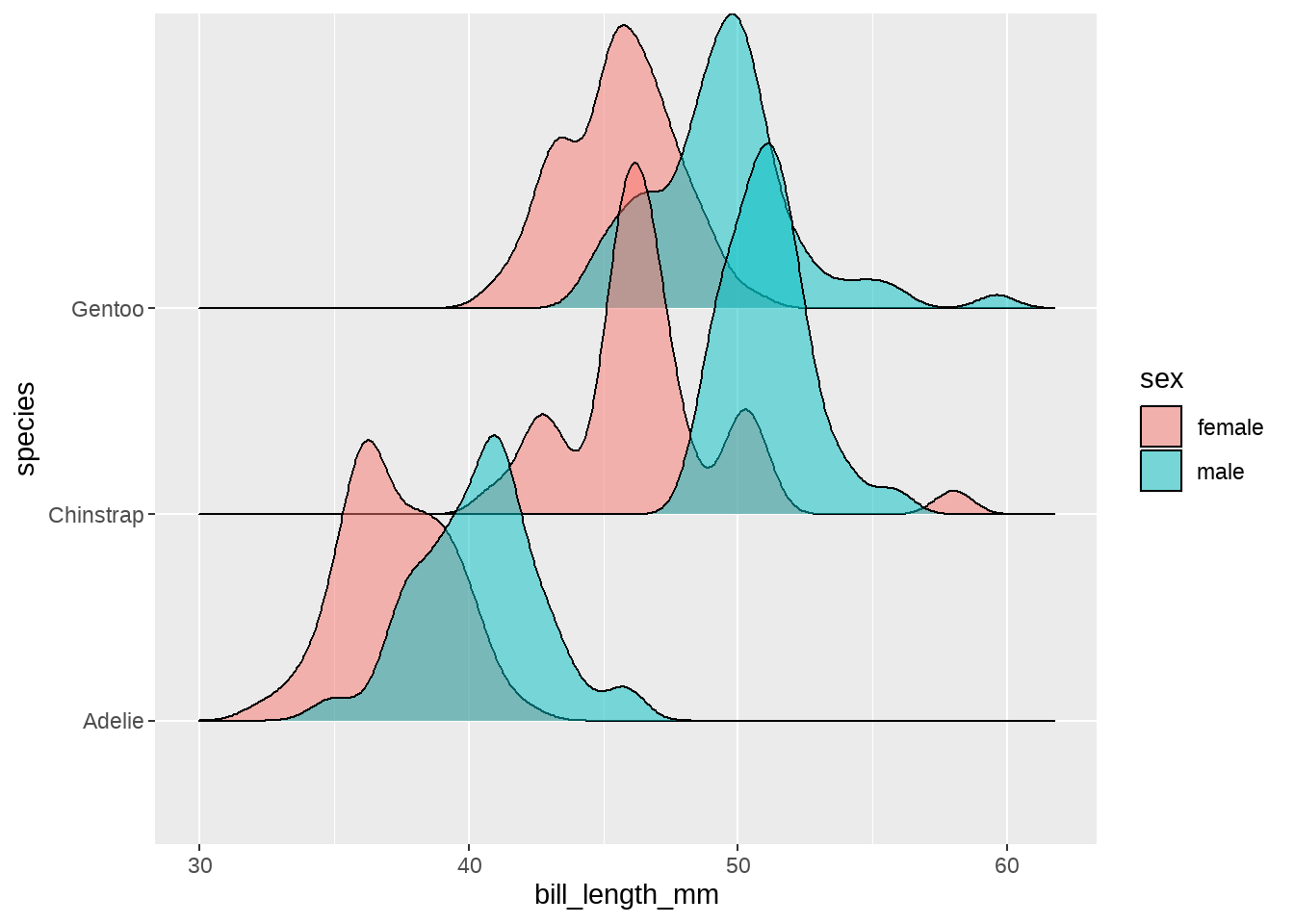
同样的代码,类似地画个其他体征的分布,
penguins %>%
ggplot(aes(x = bill_depth_mm, fill = species)) +
ggridges::geom_density_ridges(aes(y = species))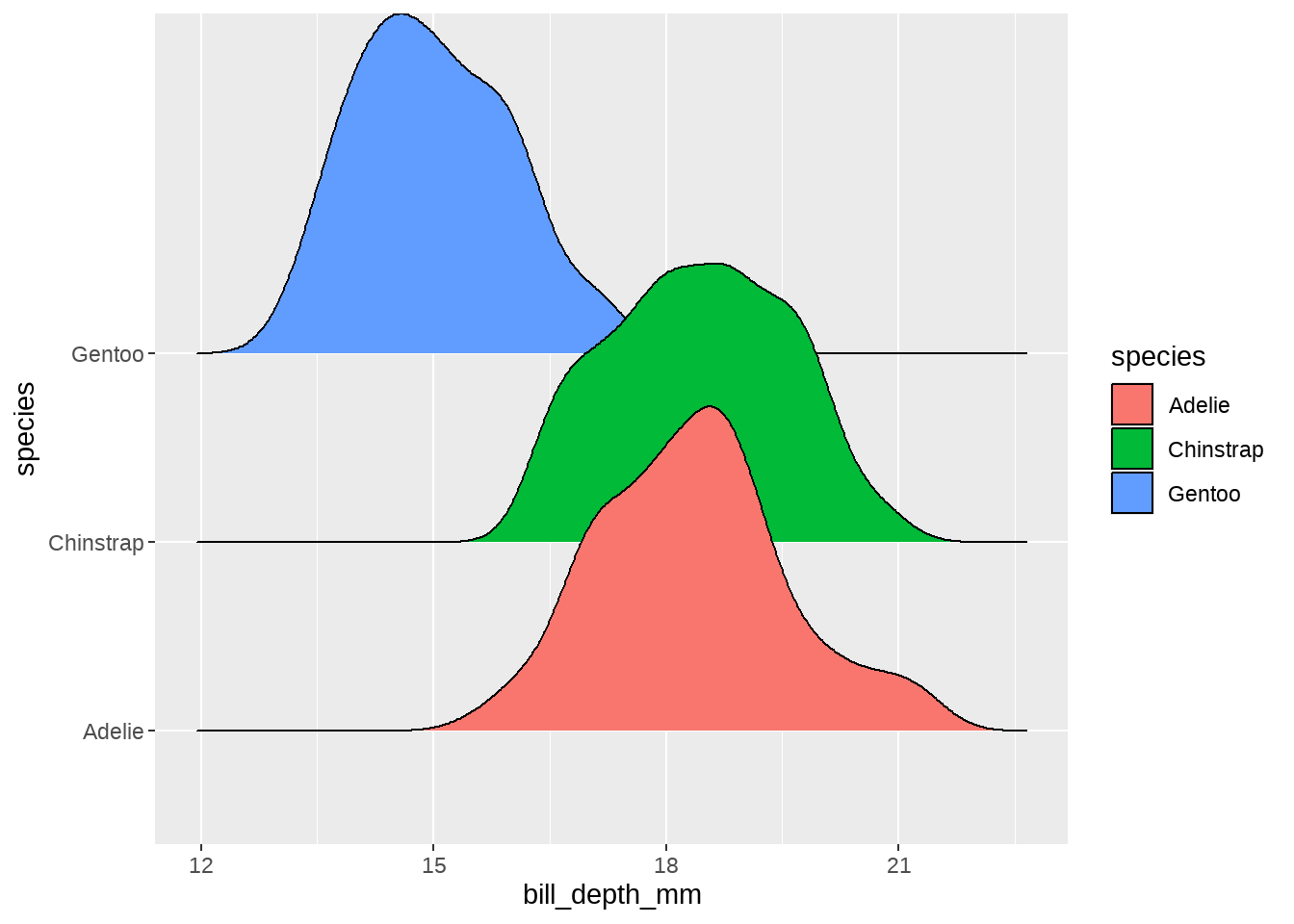
penguins %>%
ggplot(aes(x = bill_depth_mm, fill = sex)) +
ggridges::geom_density_ridges(aes(y = species))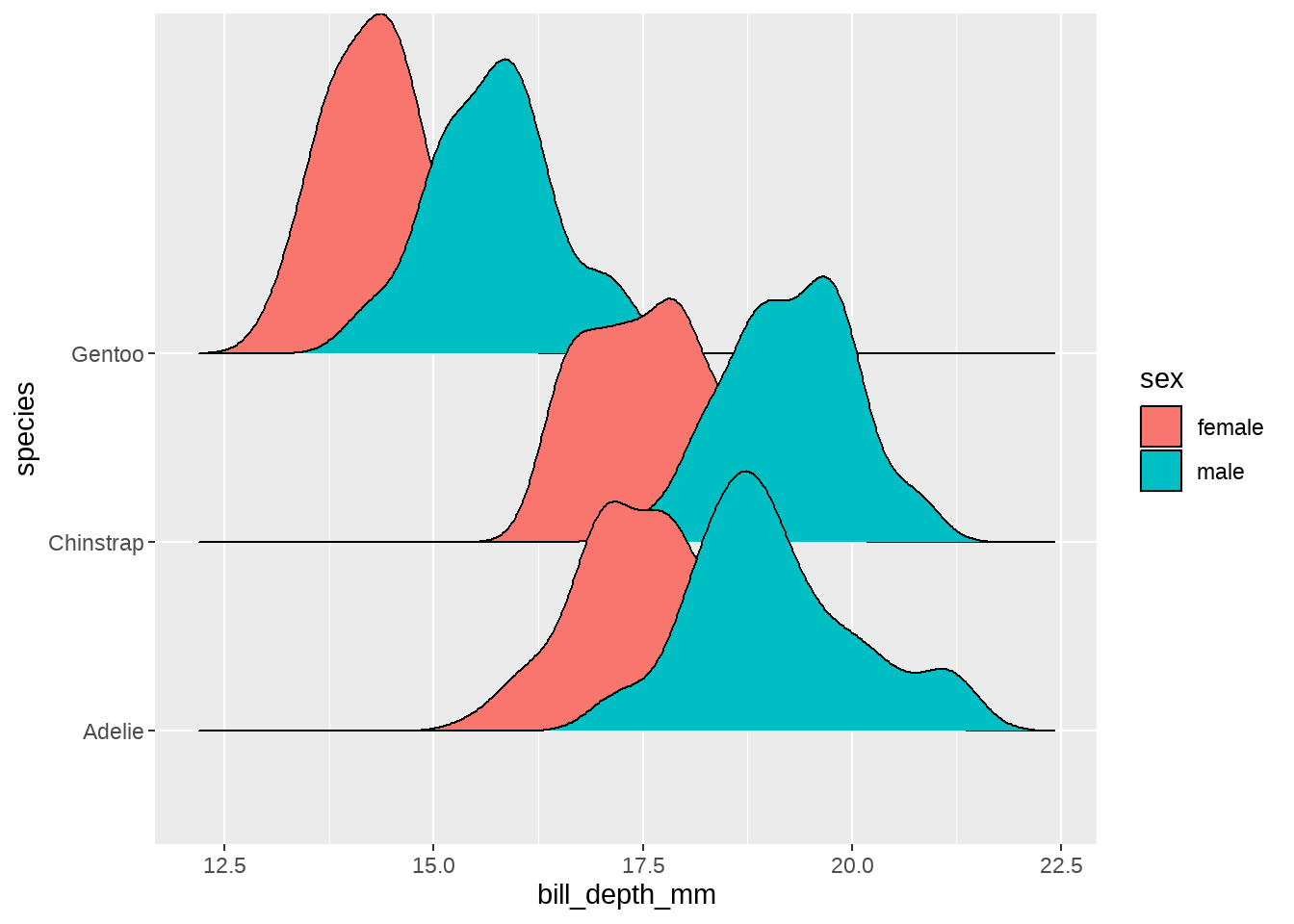
penguins %>%
ggplot(aes(x = body_mass_g, y = species, fill = sex)) +
ggridges::geom_density_ridges(alpha = 0.5)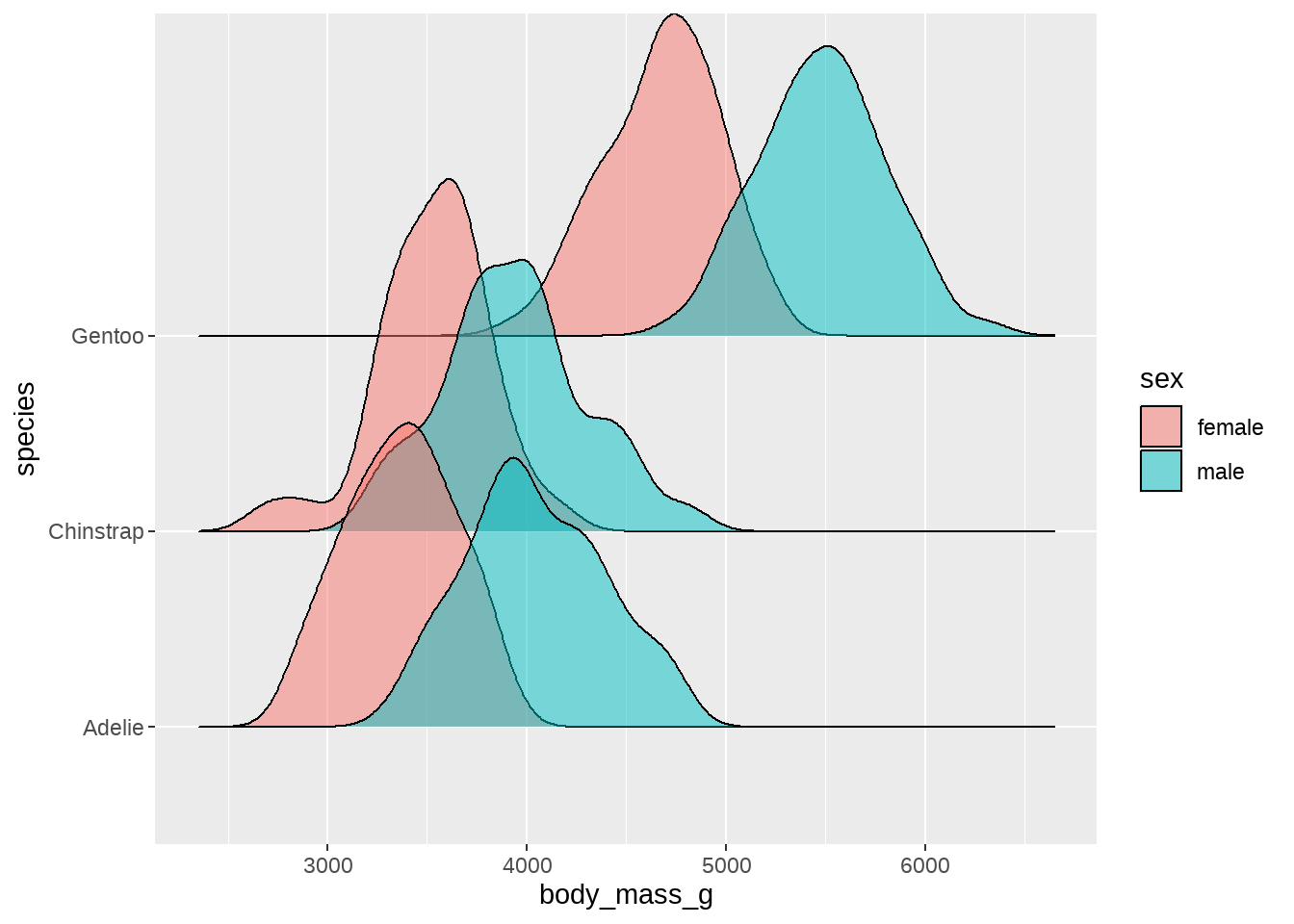
但这样一个特征一个特征的画,好麻烦。你知道程序员都是偷懒的,于是我们还有更骚的操作
penguins %>%
dplyr::select(species, bill_length_mm:body_mass_g) %>%
pivot_longer(-species, names_to = "measurement", values_to = "value") %>%
ggplot(aes(x = value)) +
geom_density(aes(color = species, fill = species), size = 1.2, alpha = 0.2) +
facet_wrap(vars(measurement), ncol = 2, scales = "free")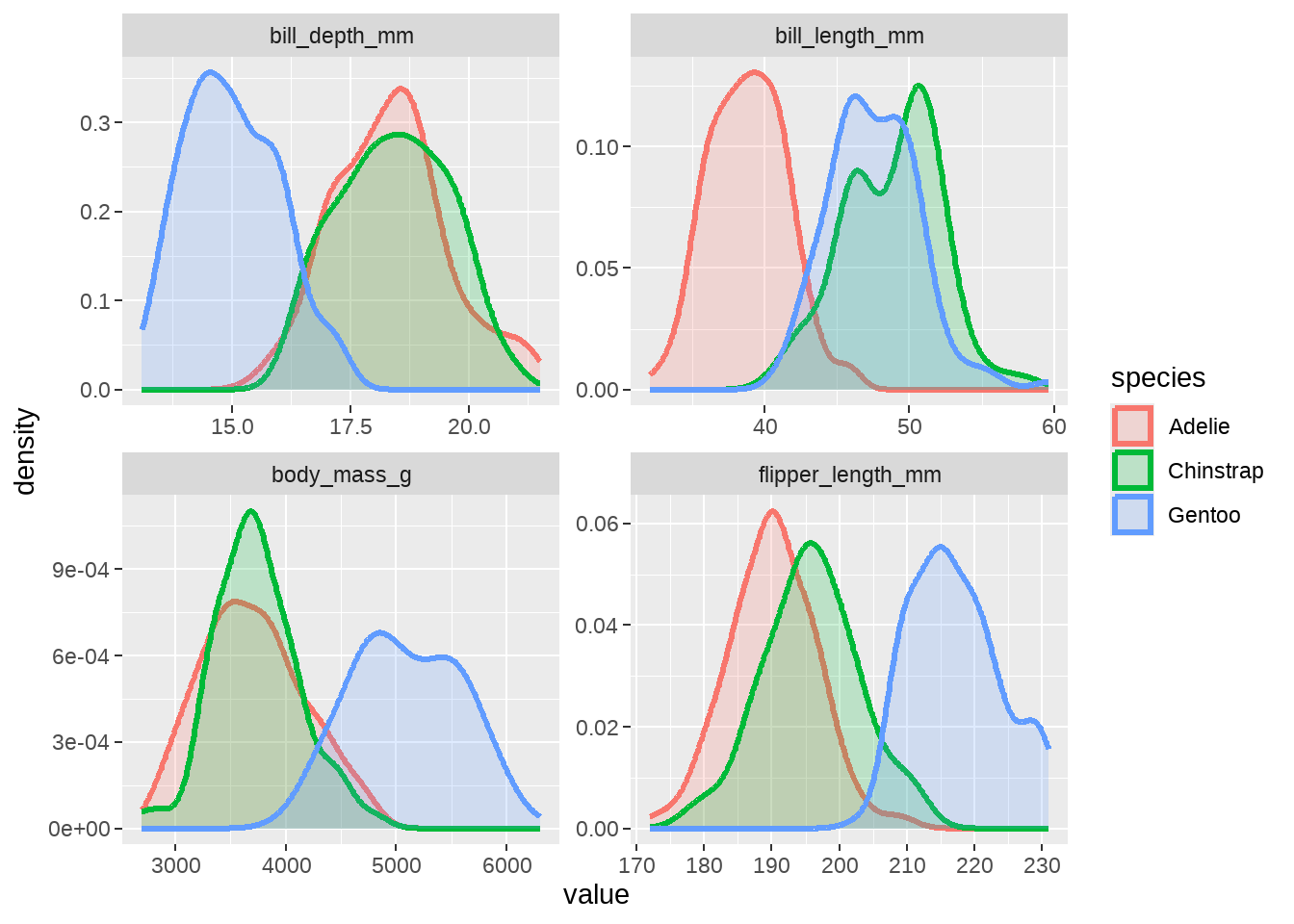
penguins %>%
dplyr::select(species, bill_length_mm:body_mass_g) %>%
pivot_longer(-species, names_to = "measurement", values_to = "value") %>%
ggplot(aes(x = species, y = value)) +
geom_boxplot(aes(color = species, fill = species), size = 1.2, alpha = 0.2) +
facet_wrap(vars(measurement), ncol = 2, scales = "free")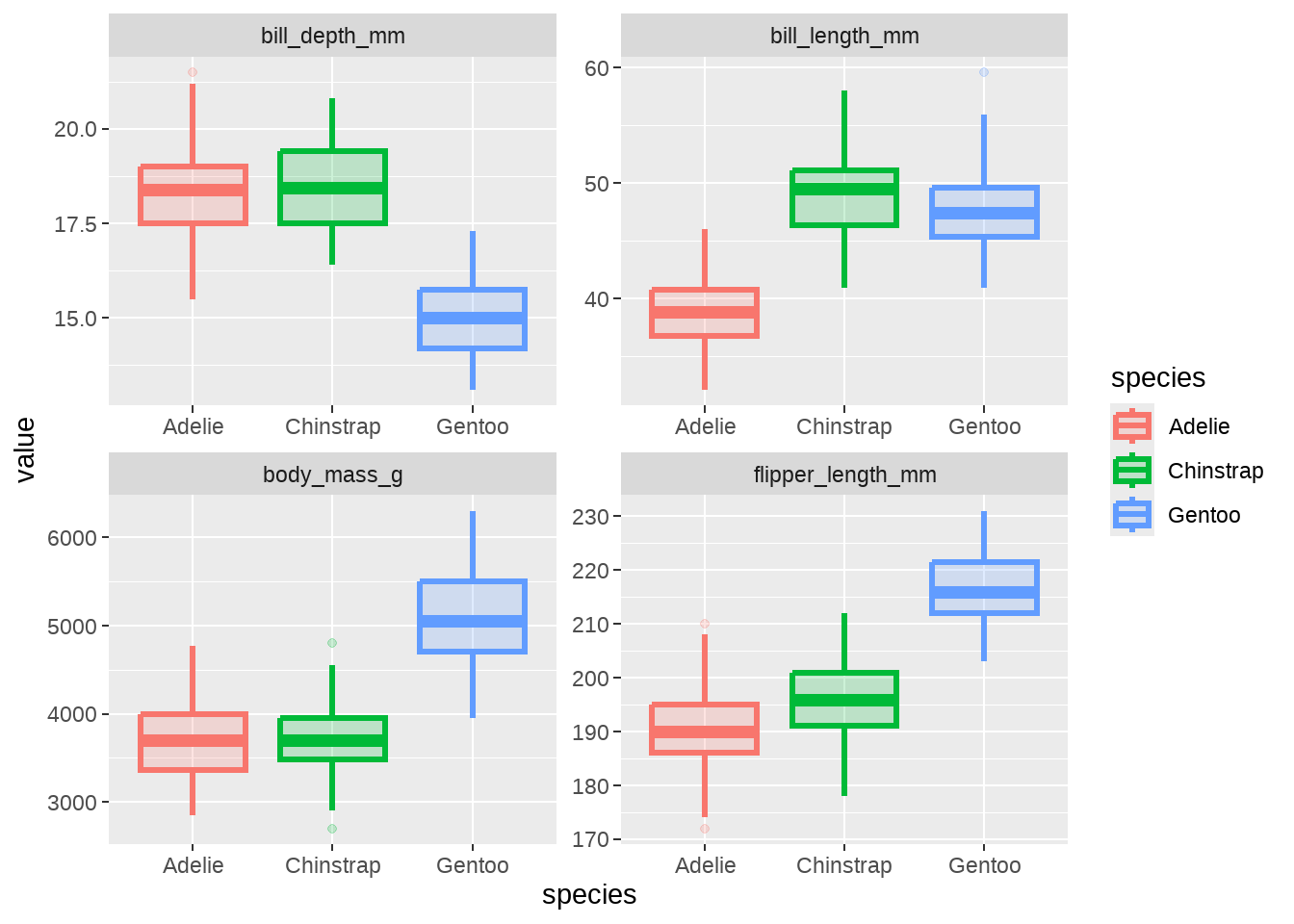
penguins %>%
dplyr::select(species, bill_length_mm:body_mass_g) %>%
pivot_longer(-species, names_to = "measurement", values_to = "value") %>%
ggplot(aes(x = value, y = species, fill = species)) +
ggridges::geom_density_ridges() +
facet_wrap(vars(measurement), scales = "free")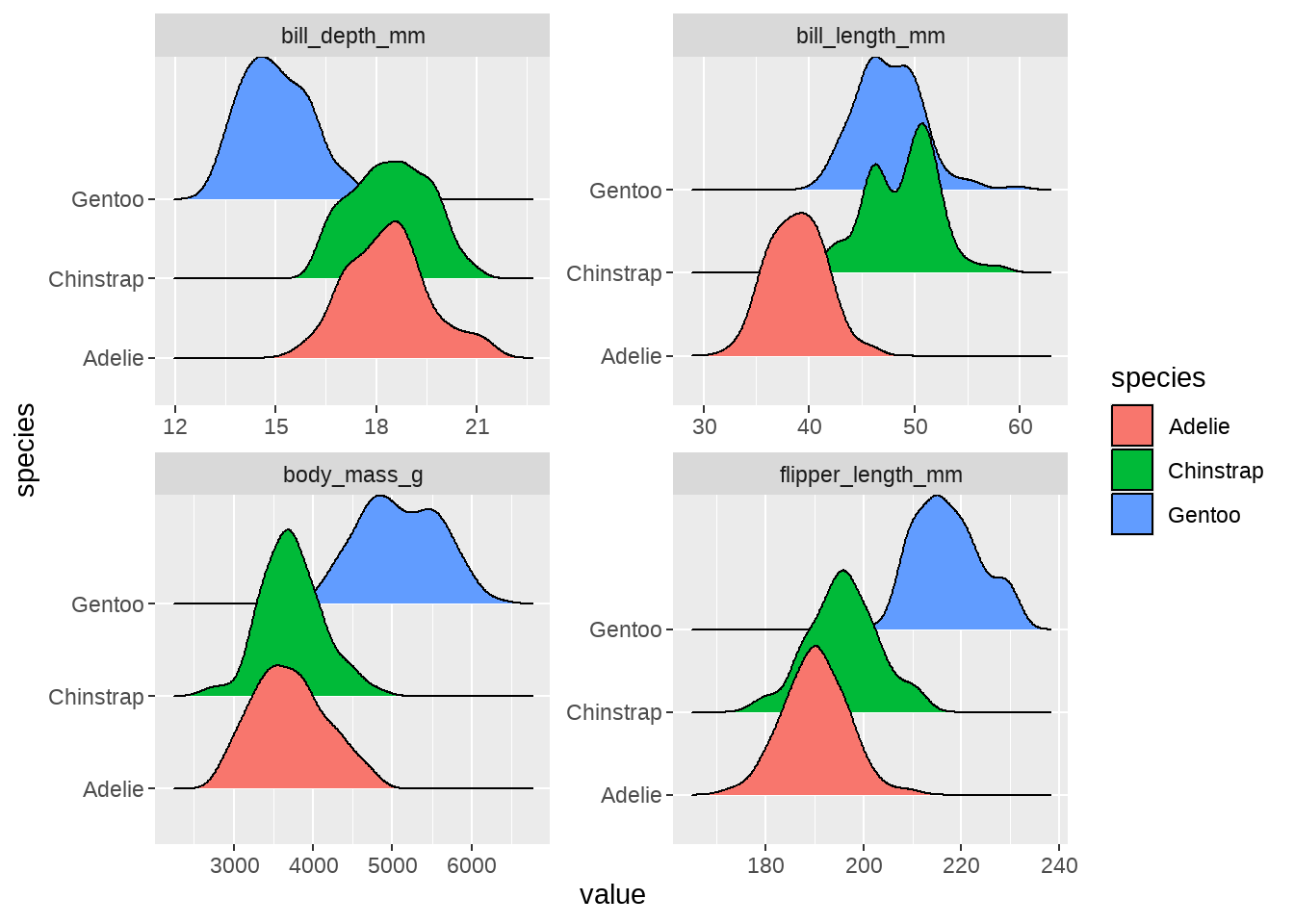
penguins %>%
dplyr::select(species,sex, bill_length_mm:body_mass_g) %>%
pivot_longer(
-c(species, sex),
names_to = "measurement",
values_to = "value"
) %>%
ggplot(aes(x = value, y = species, fill = sex)) +
ggridges::geom_density_ridges() +
facet_wrap(vars(measurement), scales = "free")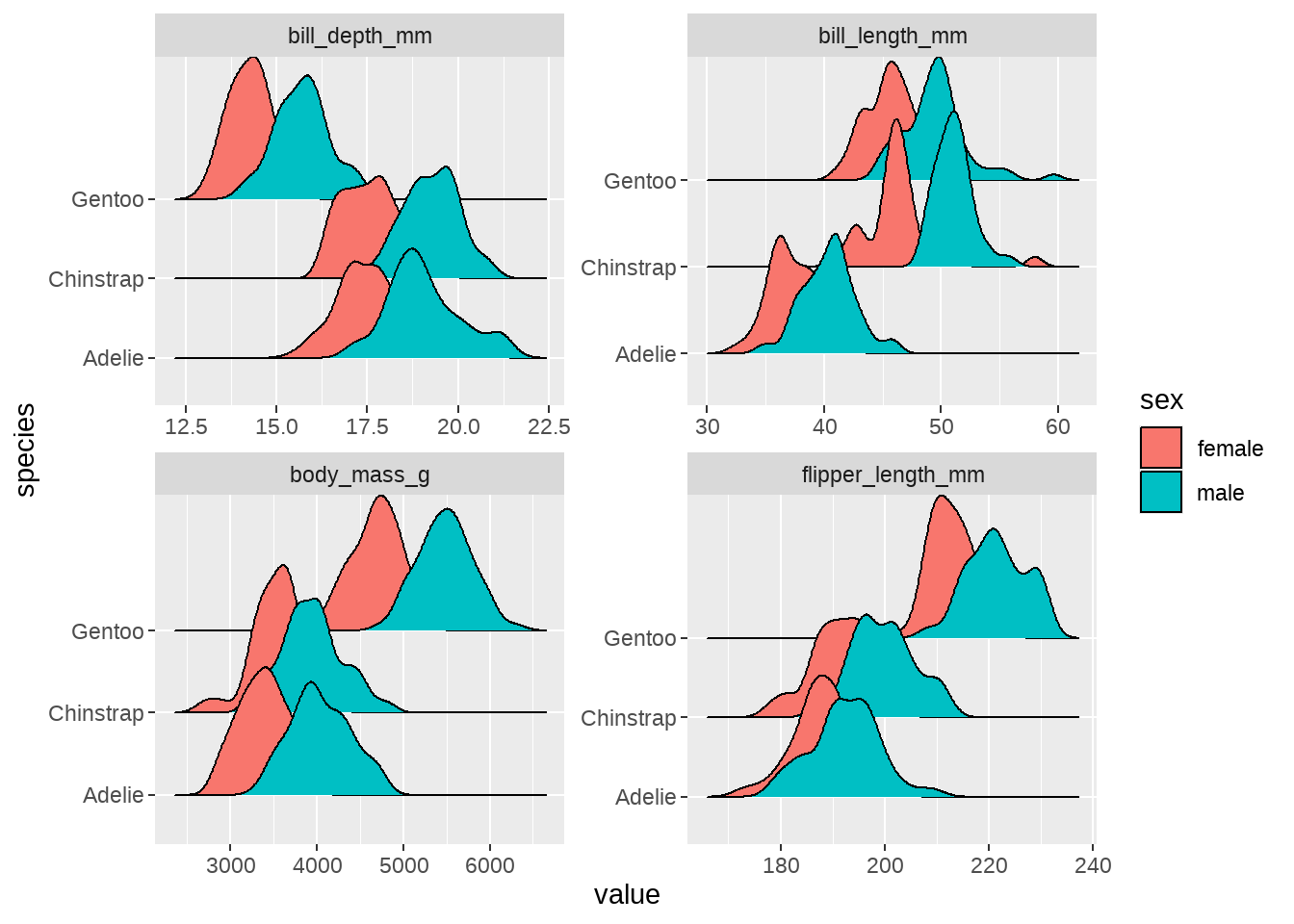
我若有所思的看着这张图,似乎看到了一些特征(pattern)了。
84.2.6 嘴峰长度和深度的关联
嘴巴越长,嘴巴也会越厚?
penguins %>%
ggplot(aes(
x = bill_length_mm, y = bill_depth_mm,
shape = species, color = species
)) +
geom_point()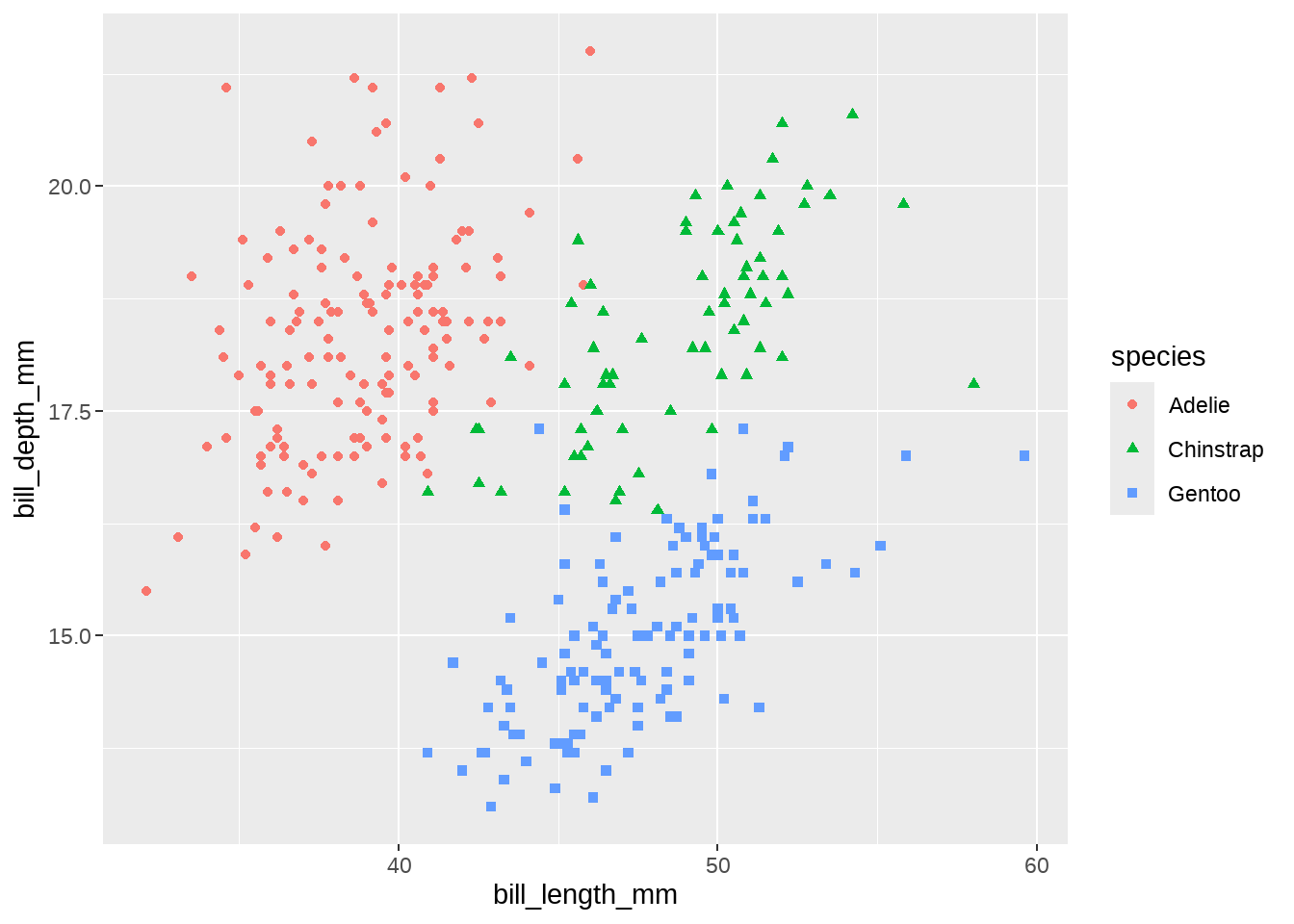
我们把不同的种类,用不同的颜色区分看看
penguins %>%
ggplot(aes(
x = bill_length_mm, y = bill_depth_mm,
shape = species, color = species
)) +
geom_point(aes(size = body_mass_g))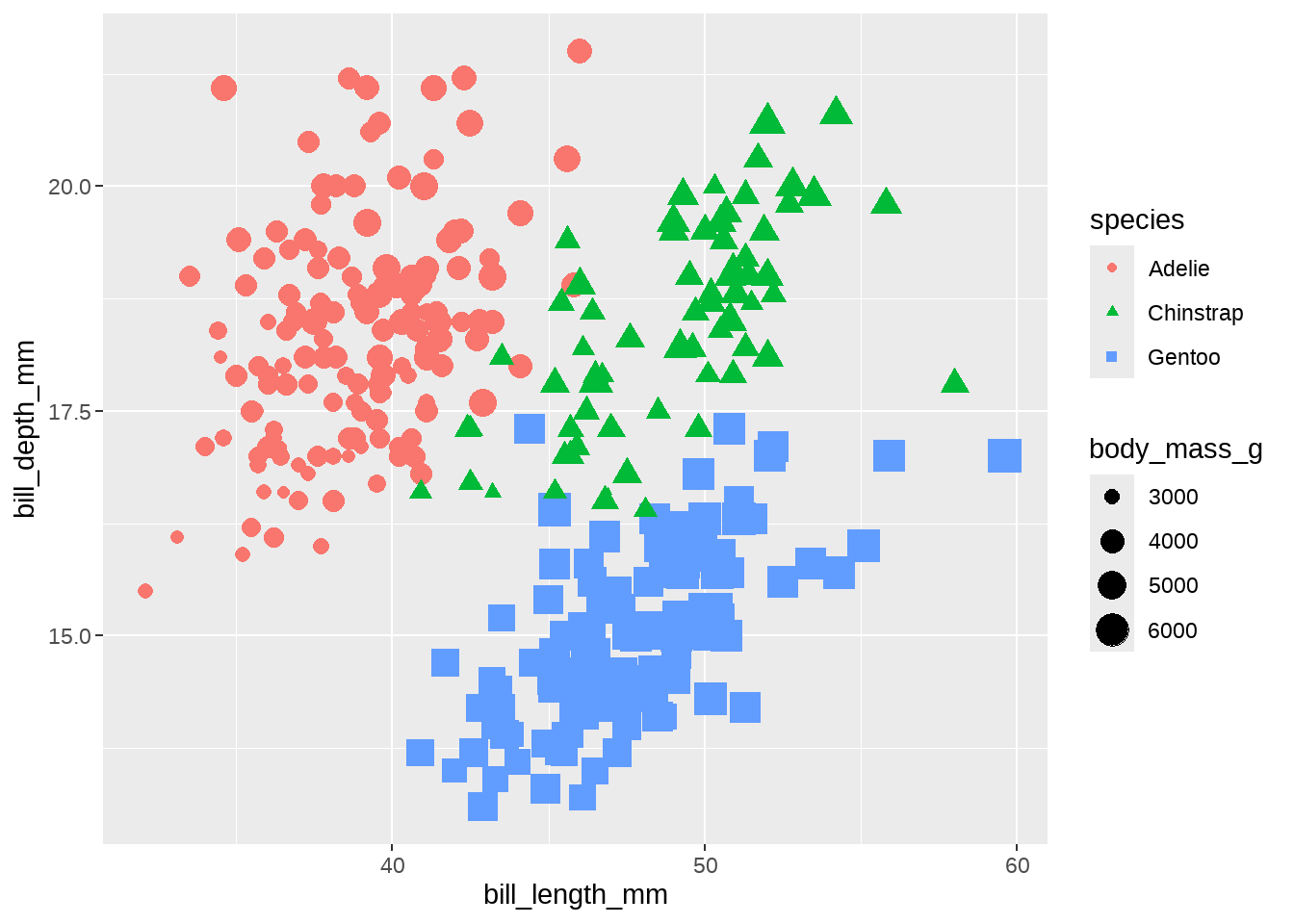
感觉这是一个辛普森佯谬, 我们画图看看
penguins %>%
ggplot(aes(x = bill_length_mm, y = bill_depth_mm)) +
geom_point(aes(color = species, shape = species)) +
geom_smooth(method = lm) +
geom_smooth(method = lm, aes(color = species))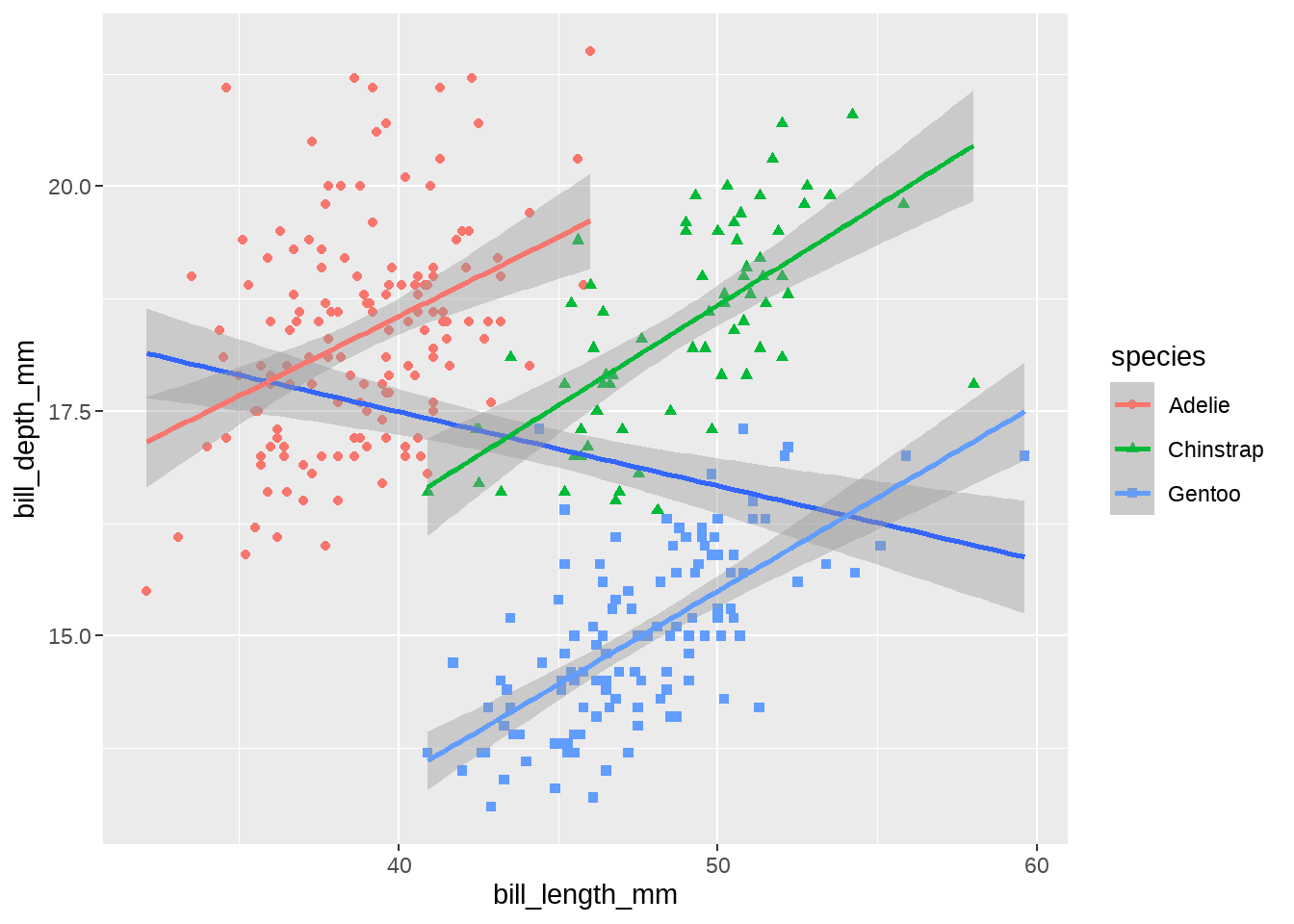
84.2.7 体重与翅膀长度的关联
翅膀越长,体重越大?
penguins %>%
group_by(species, island, sex) %>%
ggplot(aes(
x = body_mass_g, y = reorder(species, -body_mass_g),
color = species
)) +
geom_jitter(position = position_jitter(seed = 2020, width = 0.2), alpha = 0.4, size = 2) +
stat_summary(fun = mean, geom = "point", size = 5, alpha = 1)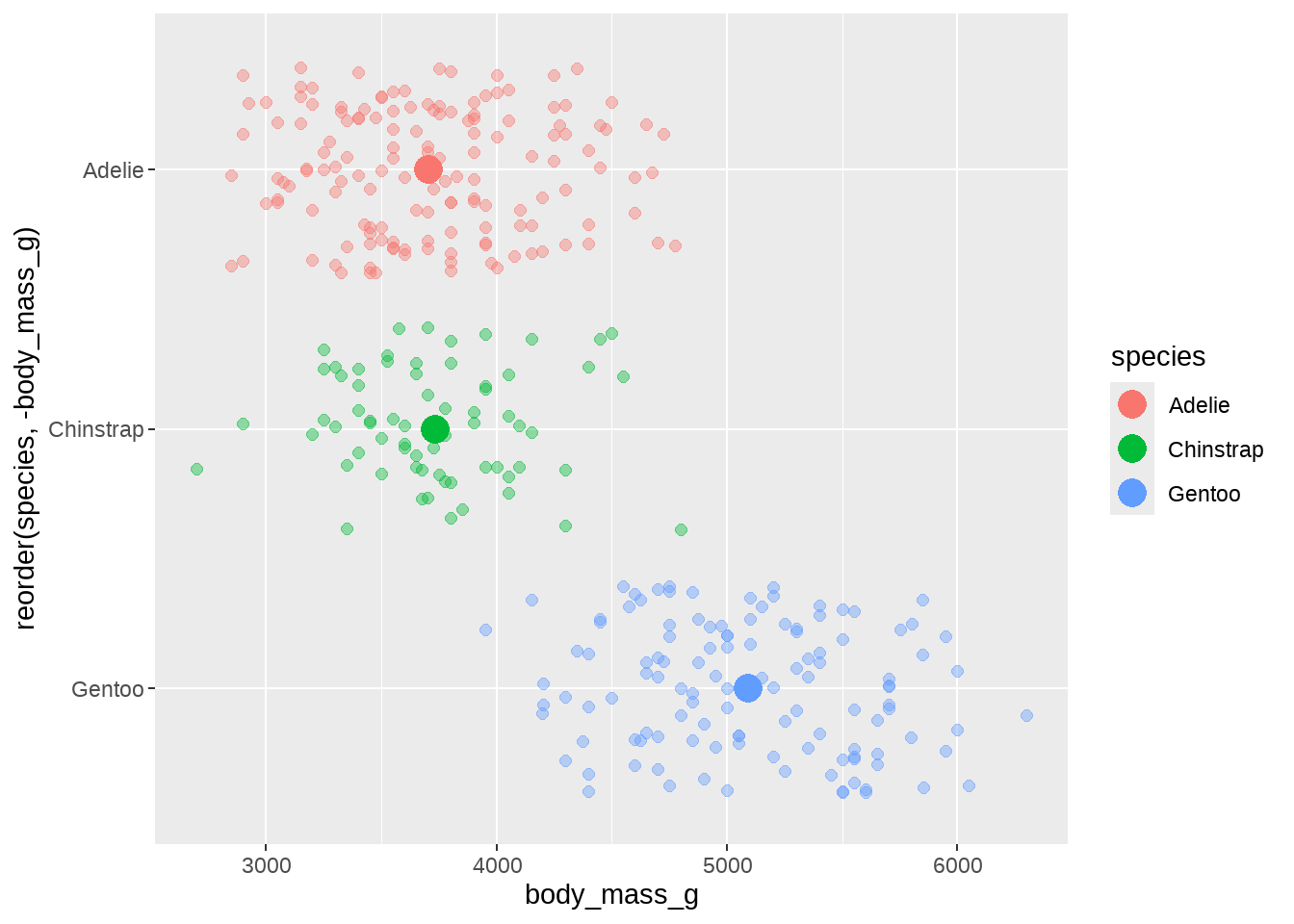
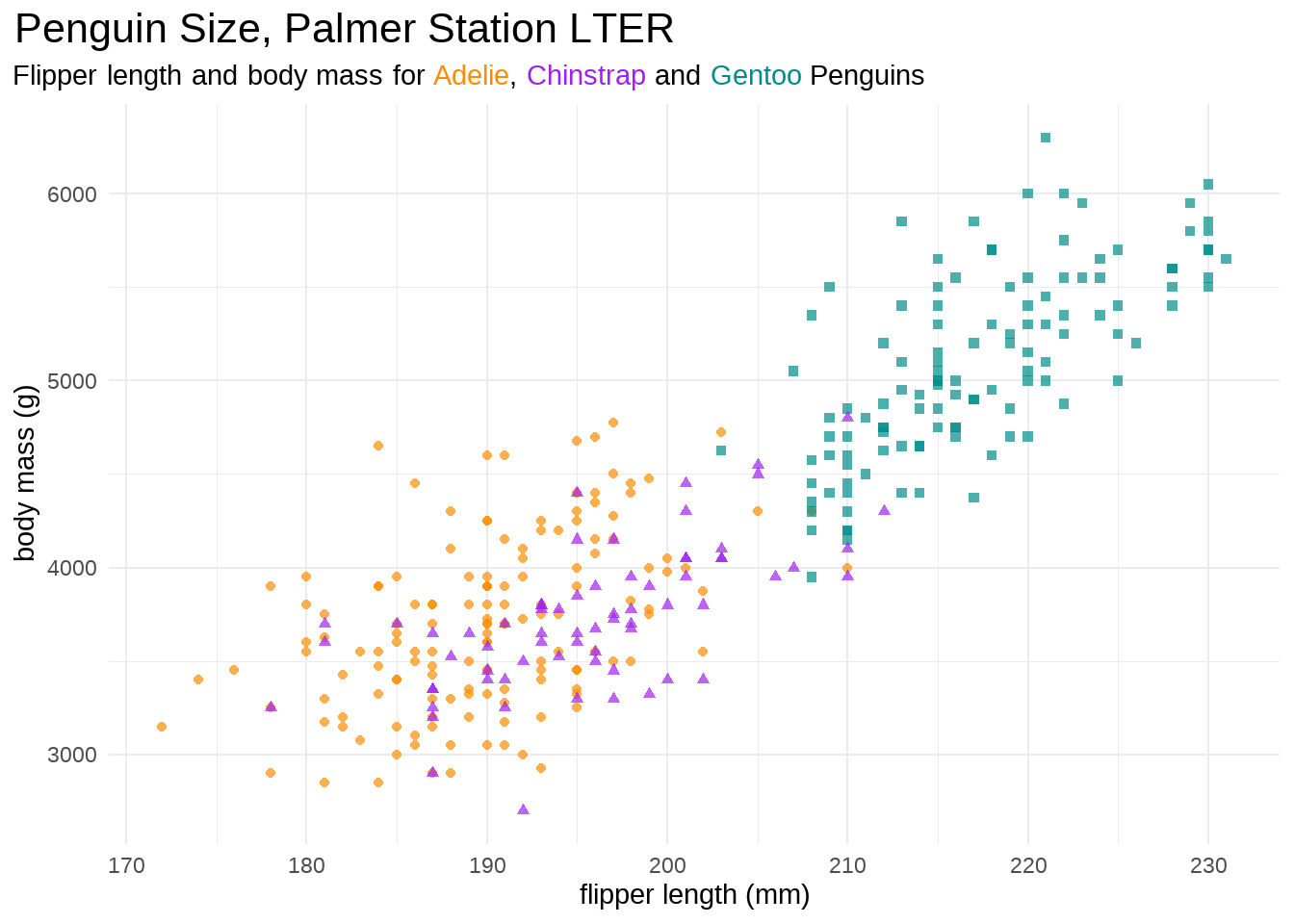
library(ggtext)
penguins %>%
ggplot(aes(flipper_length_mm, body_mass_g, group = species)) +
geom_point(aes(colour = species, shape = species), alpha = 0.7) +
scale_color_manual(values = c("darkorange", "purple", "cyan4")) +
labs(
title = "Penguin Size, Palmer Station LTER",
subtitle = "Flipper length and body mass for <span style = 'color:darkorange;'>Adelie</span>, <span style = 'color:purple;'>Chinstrap</span> and <span style = 'color:cyan4;'>Gentoo</span> Penguins",
x = "flipper length (mm)",
y = "body mass (g)"
) +
theme_minimal() +
theme(
legend.position = "none",
# text = element_text(family = "Futura"),
# (I only have 'Light' )
plot.title = element_text(size = 16),
plot.subtitle = element_markdown(), # element_markdown from `ggtext` to parse the css in the subtitle
plot.title.position = "plot",
plot.caption = element_text(size = 8, colour = "grey50"),
plot.caption.position = "plot"
)
84.2.8 不同种类的宝宝,体重具有显著性差异?
先分组计算体重的均值和标准差
penguins %>%
group_by(species) %>%
summarise(
count = n(),
mean_body_mass = mean(body_mass_g),
sd_body_mass = sd(body_mass_g)
)## # A tibble: 3 × 4
## species count mean_body_mass sd_body_mass
## <chr> <int> <dbl> <dbl>
## 1 Adelie 146 3706. 459.
## 2 Chinstrap 68 3733. 384.
## 3 Gentoo 119 5092. 501.
penguins %>%
ggplot(aes(x = species, y = body_mass_g)) +
geom_boxplot() +
geom_jitter()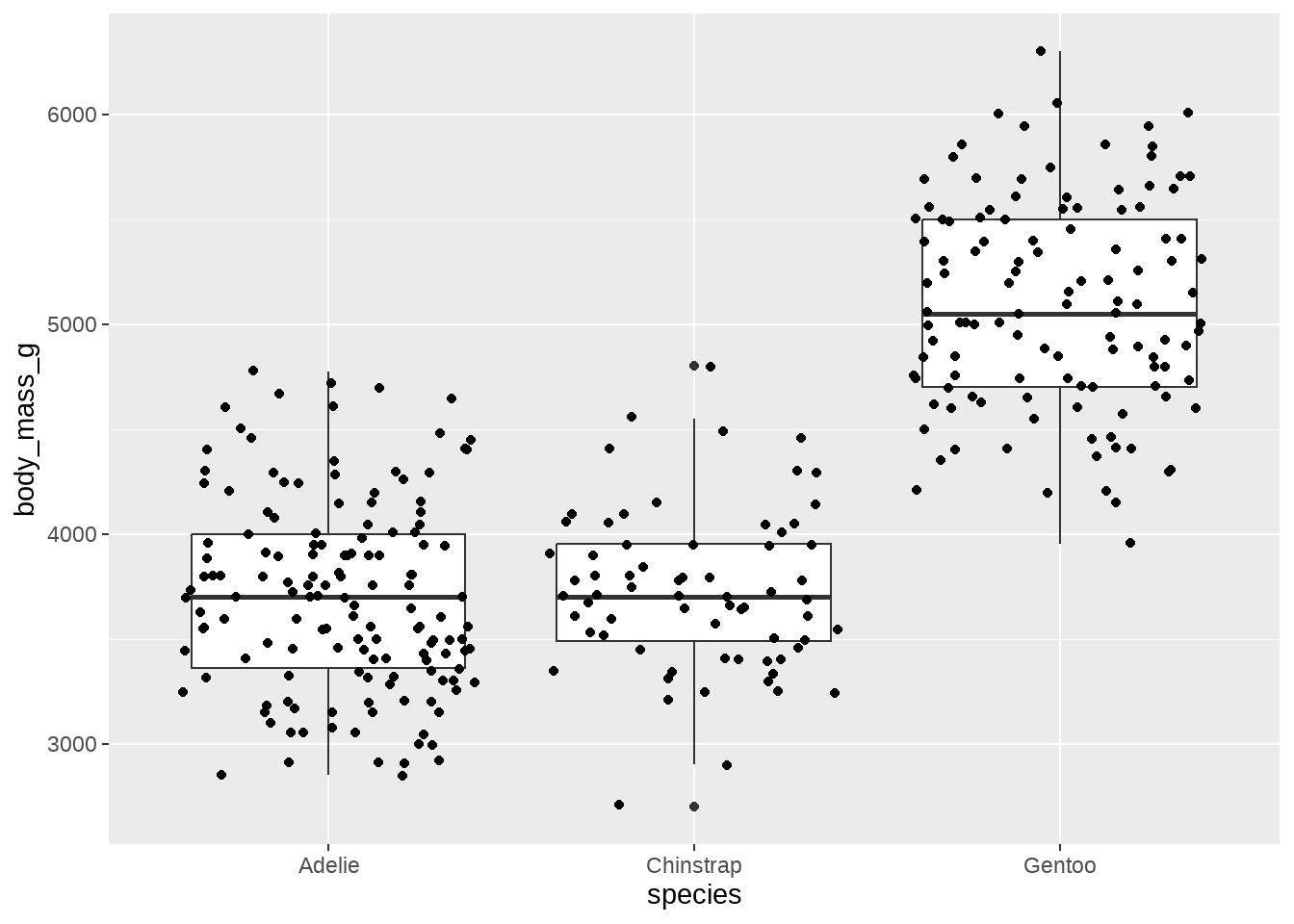
用统计方法验证下我们的猜测吧。记住,我们是有科学精神的的人!
84.2.8.1 参数检验
- one-way ANOVA(要求等方差)
## Df Sum Sq Mean Sq F value Pr(>F)
## species 2 145190219 72595110 341.9 <2e-16 ***
## Residuals 330 70069447 212332
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1p-value 很小,说明不同种类企鹅之间体重是有显著差异的,但aov只给出了species在整体上引起了体重差异(只要有任意两组之间有显著差异,aov给出的p-value都很小),如果想知道不同种类两两之间是否有显著差异,这就需要用到TukeyHSD().
- one-way ANOVA(不要求等方差),相关介绍看here
oneway.test(body_mass_g ~ species, data = penguins)##
## One-way analysis of means (not assuming equal variances)
##
## data: body_mass_g and species
## F = 316.5, num df = 2.00, denom df = 187.68, p-value < 2.2e-16
stats::aov(formula = body_mass_g ~ species, data = penguins) %>%
TukeyHSD(which = "species") %>%
broom::tidy()## # A tibble: 3 × 7
## term contrast null.value estimate conf.low conf.high adj.p.value
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 species Chinstrap-Adelie 0 26.9 -132. 186. 9.16e- 1
## 2 species Gentoo-Adelie 0 1386. 1252. 1520. 5.82e-13
## 3 species Gentoo-Chinstrap 0 1359. 1194. 1524. 5.82e-13表格第一行instrap-Adelie 的 p-value = 0.916,没通过显著性检验;而Gentoo-Adelie 和 Gentoo-Chinstrap 他们的p-value都接近0,通过显著性检验,这和图中的结果是一致的。
作为统计出生的R语言,有很多宏包可以帮助我们验证我们的结论,我这里推荐可视化学统计的宏包ggstatsplot宏包将统计分析的结果写在图片里,统计结果和图形融合在一起,让统计结果更容易懂了。(使用这个宏包辅助我们学习统计)
library(ggstatsplot)
penguins %>%
ggstatsplot::ggbetweenstats(
x = species, # > 2 groups
y = body_mass_g,
type = "parametric",
pairwise.comparisons = TRUE,
pairwise.display = "all",
messages = FALSE,
var.equal = FALSE
)84.2.8.2 非参数检验
相关介绍看here
kruskal.test(body_mass_g ~ species, data = penguins)##
## Kruskal-Wallis rank sum test
##
## data: body_mass_g by species
## Kruskal-Wallis chi-squared = 212.09, df = 2, p-value < 2.2e-16
penguins %>%
ggstatsplot::ggbetweenstats(
x = species,
y = body_mass_g,
type = "nonparametric",
mean.ci = TRUE,
pairwise.comparisons = TRUE, # <<
pairwise.display = "all", # ns = only non-significant
p.adjust.method = "fdr", # <<
messages = FALSE
)哇,原来统计可以这样学!
84.2.9 嘴峰长度与嘴峰深度的比例
penguins %>%
mutate(ratio = bill_length_mm / bill_depth_mm) %>%
group_by(species) %>%
summarise(mean = mean(ratio))## # A tibble: 3 × 2
## species mean
## <chr> <dbl>
## 1 Adelie 2.12
## 2 Chinstrap 2.65
## 3 Gentoo 3.18
penguins %>%
mutate(ratio = bill_length_mm / bill_depth_mm) %>%
ggplot(aes(x = ratio, fill = species)) +
ggridges::geom_density_ridges(aes(y = species))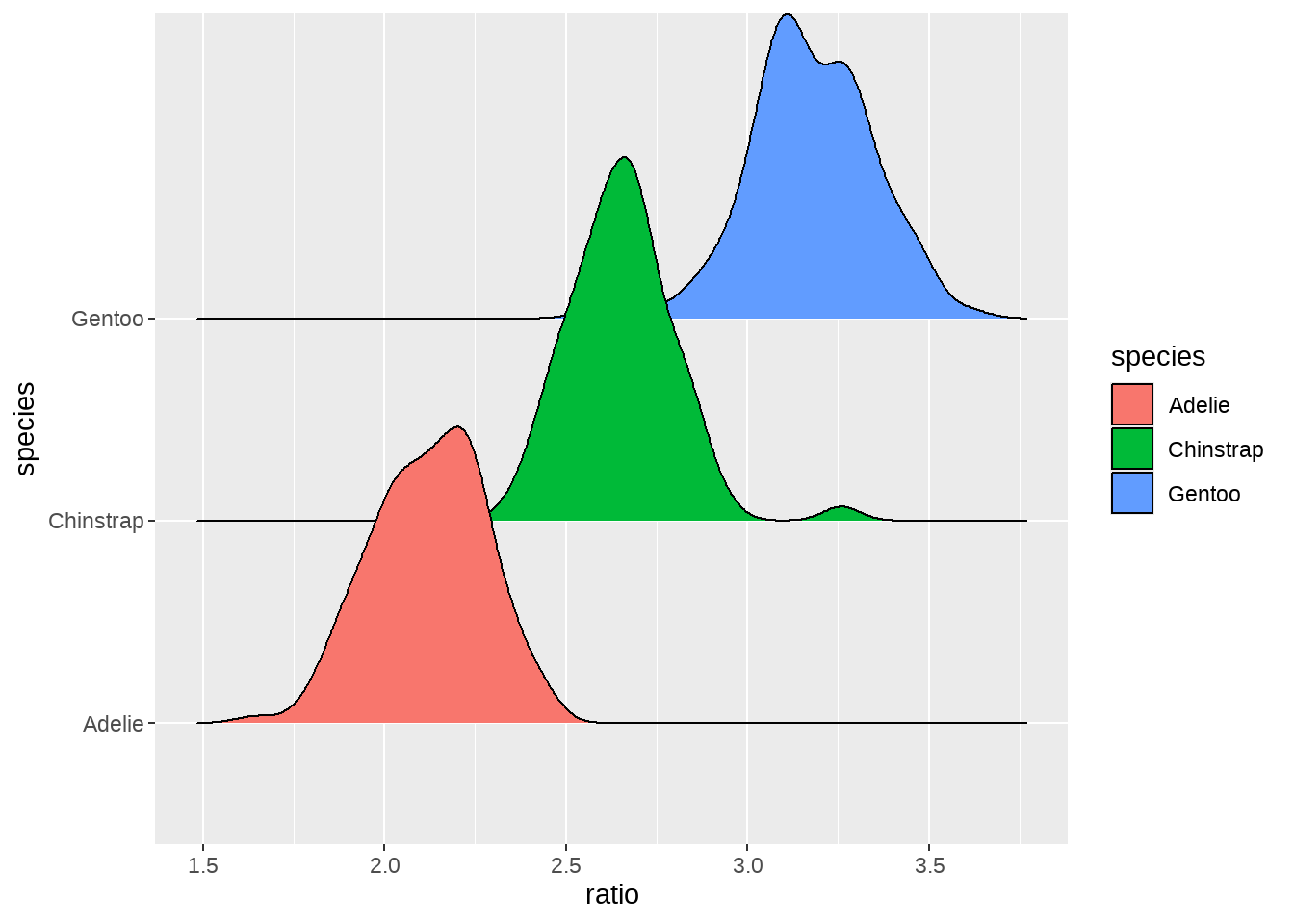
男宝宝和女宝宝颜色区分下,代码只需要修改一个地方,留给大家自己实践下吧。
84.2.10 建立模型
建模需要标准化数据,并对分类变量(比如sex)编码为 1 和 0; (这是第二个好习惯)
scale_fun <- function(x) {
(x - mean(x)) / sd(x)
}
d <- penguins %>%
select(sex, species, bill_length_mm:body_mass_g) %>%
mutate(
across(where(is.numeric), scale_fun)
) %>%
mutate(male = if_else(sex == "male", 1, 0))
d## # A tibble: 333 × 7
## sex species bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
## <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 male Adelie -0.895 0.780 -1.42 -0.568
## 2 female Adelie -0.822 0.119 -1.07 -0.506
## 3 female Adelie -0.675 0.424 -0.426 -1.19
## 4 female Adelie -1.33 1.08 -0.568 -0.940
## 5 male Adelie -0.858 1.74 -0.782 -0.692
## 6 female Adelie -0.931 0.323 -1.42 -0.723
## 7 male Adelie -0.876 1.24 -0.426 0.581
## 8 female Adelie -0.529 0.221 -1.35 -1.25
## 9 male Adelie -0.986 2.05 -0.711 -0.506
## 10 male Adelie -1.72 2.00 -0.212 0.240
## # ℹ 323 more rows
## # ℹ 1 more variable: male <dbl>按照species分组后,对flipper_length_mm标准化?这样数据会聚拢到一起了喔, 还是不要了
penguins %>%
select(sex, species, bill_length_mm:body_mass_g) %>%
group_by(species) %>%
mutate(
across(where(is.numeric), scale_fun)
) %>%
ungroup()84.2.10.1 model_01
我们将性别sex视为响应变量,其他变量为预测变量。这里性别变量是二元的(0 或者 1),所以我们用logistic回归
logit_mod1 <- glm(
male ~ 1 + species + bill_length_mm + bill_depth_mm +
flipper_length_mm + body_mass_g,
data = d,
family = binomial(link = "logit")
)
summary(logit_mod1)##
## Call:
## glm(formula = male ~ 1 + species + bill_length_mm + bill_depth_mm +
## flipper_length_mm + body_mass_g, family = binomial(link = "logit"),
## data = d)
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) 4.6835 1.1865 3.947 7.90e-05 ***
## speciesChinstrap -6.9803 1.5743 -4.434 9.25e-06 ***
## speciesGentoo -8.3539 2.5235 -3.310 0.000932 ***
## bill_length_mm 3.3566 0.7164 4.685 2.80e-06 ***
## bill_depth_mm 3.1958 0.6546 4.882 1.05e-06 ***
## flipper_length_mm 0.2912 0.6704 0.434 0.664046
## body_mass_g 4.7228 0.8723 5.414 6.15e-08 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 461.61 on 332 degrees of freedom
## Residual deviance: 127.11 on 326 degrees of freedom
## AIC: 141.11
##
## Number of Fisher Scoring iterations: 7计算每个变量的平均边际效应
library(margins)
logit_mod1_m <- logit_mod1 %>%
margins() %>%
summary() %>%
as_tibble()
logit_mod1_m## # A tibble: 6 × 7
## factor AME SE z p lower upper
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 bill_depth_mm 0.185 0.0290 6.38 1.82e-10 0.128 0.242
## 2 bill_length_mm 0.194 0.0339 5.72 1.04e- 8 0.128 0.261
## 3 body_mass_g 0.273 0.0378 7.22 5.08e-13 0.199 0.347
## 4 flipper_length_mm 0.0169 0.0388 0.434 6.64e- 1 -0.0592 0.0929
## 5 speciesChinstrap -0.373 0.0513 -7.27 3.67e-13 -0.473 -0.272
## 6 speciesGentoo -0.434 0.0740 -5.86 4.66e- 9 -0.579 -0.289
logit_mod1_m %>%
ggplot(aes(
x = reorder(factor, AME),
y = AME, ymin = lower, ymax = upper
)) +
geom_hline(yintercept = 0, color = "gray80") +
geom_pointrange() +
coord_flip() +
labs(x = NULL, y = "Average Marginal Effect")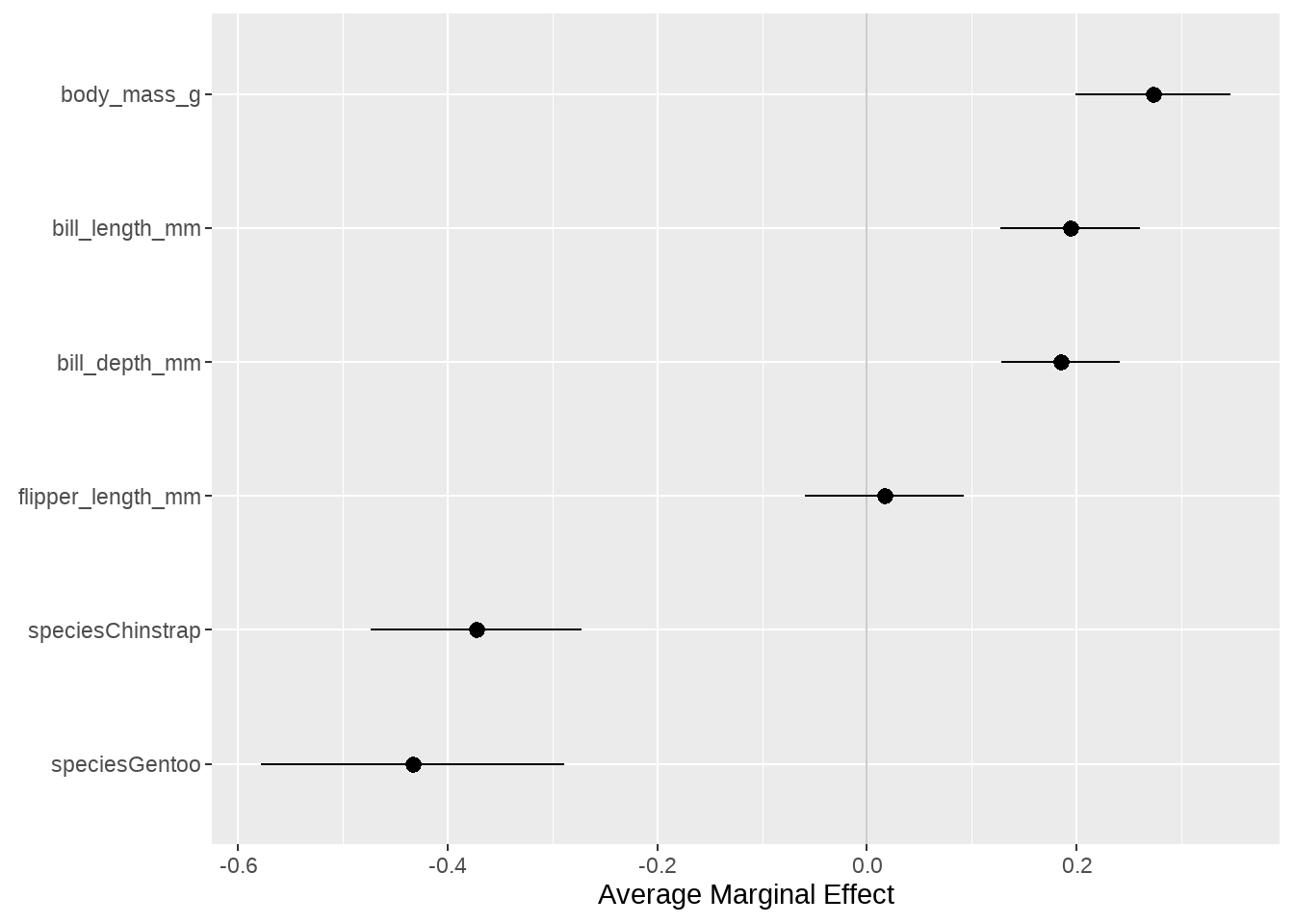
84.2.10.2 model_02
library(brms)
brms_mod2 <- brm(
male ~ 1 + bill_length_mm + bill_depth_mm + flipper_length_mm + body_mass_g + (1 | species),
data = d,
family = binomial(link = "logit")
)
summary(brms_mod2)84.2.10.3 model_03
penguins %>%
ggplot(aes(x = flipper_length_mm, y = bill_length_mm, color = species)) +
geom_point()
brms_mod3 <- brm(bill_length_mm ~ flipper_length_mm + (1|species),
data = penguins
)
penguins %>%
group_by(species) %>%
modelr::data_grid(flipper_length_mm) %>%
tidybayes::add_fitted_draws(brms_mod3, n = 100) %>%
ggplot() +
geom_point(
data = penguins,
aes(flipper_length_mm, bill_length_mm, color = species, shape = species)
) +
geom_line(aes(flipper_length_mm, .value, group = interaction(.draw, species), color = species), alpha = 0.1)