第 64 章 贝叶斯推断
library(tidyverse)
library(tidybayes)
library(rstan)
library(brms)
rstan_options(auto_write = TRUE)
options(mc.cores = parallel::detectCores())之前我们讲了线性模型和混合线性模型,今天我们往前一步,应该说是一大步。因为这一步迈向了贝叶斯分析,与频率学派的分析有本质的区别,这种区别类似经典物理和量子物理的区别。
- 频率学派,是从数据出发
- 贝叶斯。先假定参数有一个分布,看到数据后,再重新分配可能性。
Statistical inference is the process of using observed data to infer properties of the statistical distributions that generated that data.
简单点说
\[ \Pr(\text{parameters} | \text{data}). \]
这个量实际上贝叶斯定理中的后验概率分布(posterior distribution)
\[ \underbrace{\Pr(\text{parameters} | \text{data})}_{\text{posterior}} = \frac{\overbrace{\Pr(\text{data} | \text{parameters})}^{\text{likelihood}} \overbrace{\Pr(\text{parameters})}^{\text{prior}}}{\underbrace{\Pr(\text{data})}_{evidence}} . \]
下面,通过具体的案例演示简单的贝叶斯推断(Bayesian inference)
64.1 学生身高的分布?
假定这是收集的200位学生身高和体重数据
## sex height weight
## 1 boy 173.7173 59.93405
## 2 boy 170.8879 60.03269
## 3 boy 182.1087 62.76687
## 4 boy 176.2107 55.53530
## 5 boy 167.0802 56.64638
## 6 boy 183.1166 60.60861用dplyr函数很容易得到样本的统计量
d %>%
summarise(
across(height, list(mean = mean, median = median, max = max, min = min, sd = sd))
)## height_mean height_median height_max height_min height_sd
## 1 164.8933 164.1618 185.2429 142.3032 7.305997
d %>%
ggplot(aes(x = height)) +
geom_density()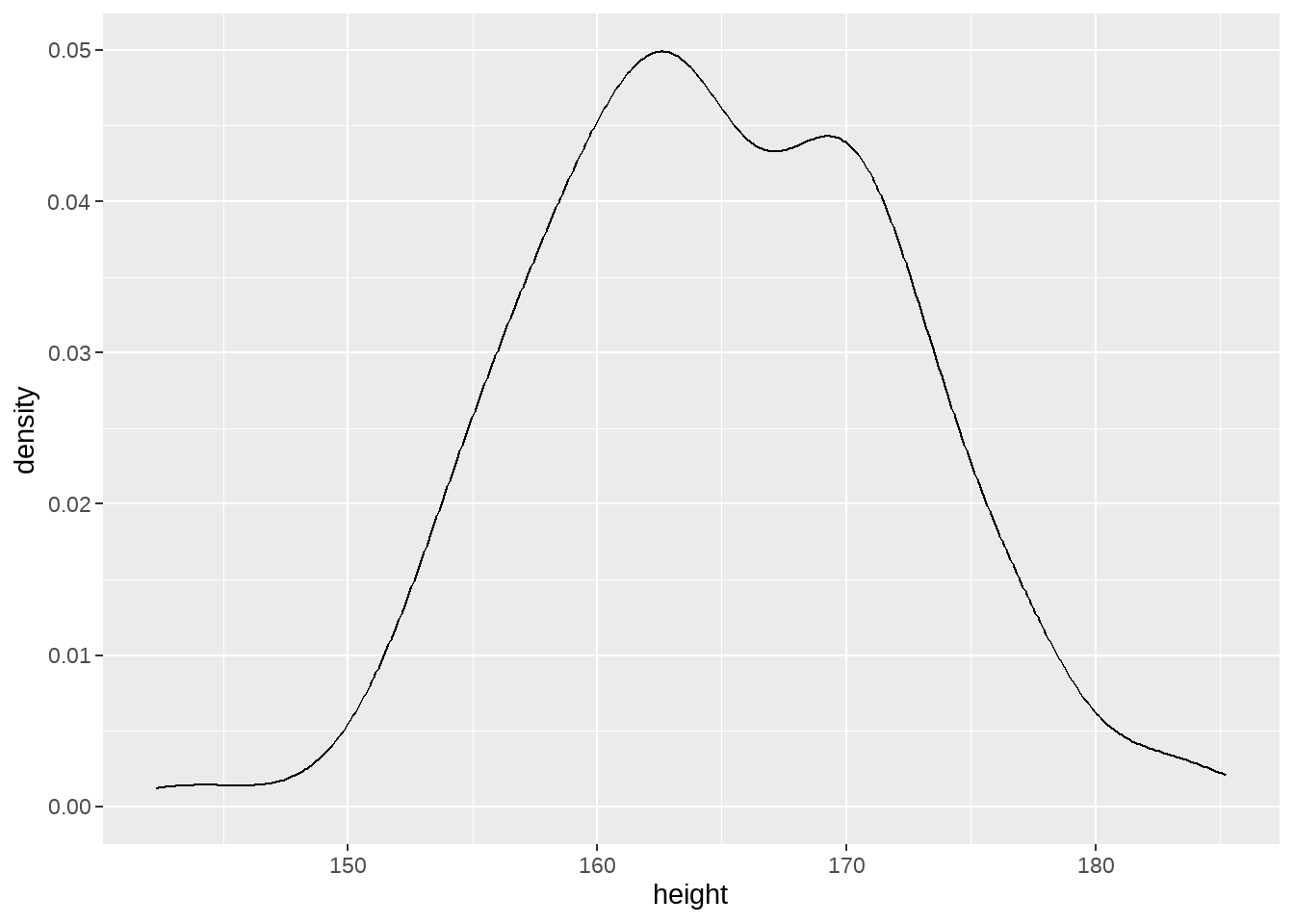
64.2 推断
注意到,我们的数据只是样本,不代表全体分布。我们只有通过样本去推断全体分布情况。
通过前面的身高的统计量,我们可以合理的猜测:
均值可能是160,162,170,172,…, 或者说这个均值在一个范围之内,在这个范围内,有些值的可能性大,有些值可能性较低。比如,认为这值游离在(150,180)范围,其中168左右的可能最大,两端的可能性最低。如果寻求用数学语言来描述,它符合正态分布的特征
方差也可以假设在(0, 50)范围内都有可能,而且每个位置上的概率都相等
把我们的猜测画出来就是这样的,
library(patchwork)
p1 <-
ggplot(data = tibble(x = seq(from = 100, to = 230, by = .1)),
aes(x = x, y = dnorm(x, mean = 168, sd = 20))) +
geom_line() +
xlab("height_mean") +
ylab("density")
p2 <-
ggplot(data = tibble(x = seq(from = -10, to = 55, by = .1)),
aes(x = x, y = dunif(x, min = 0, max = 50))) +
geom_line() +
xlab("height_sd") +
ylab("density")
p1 + p2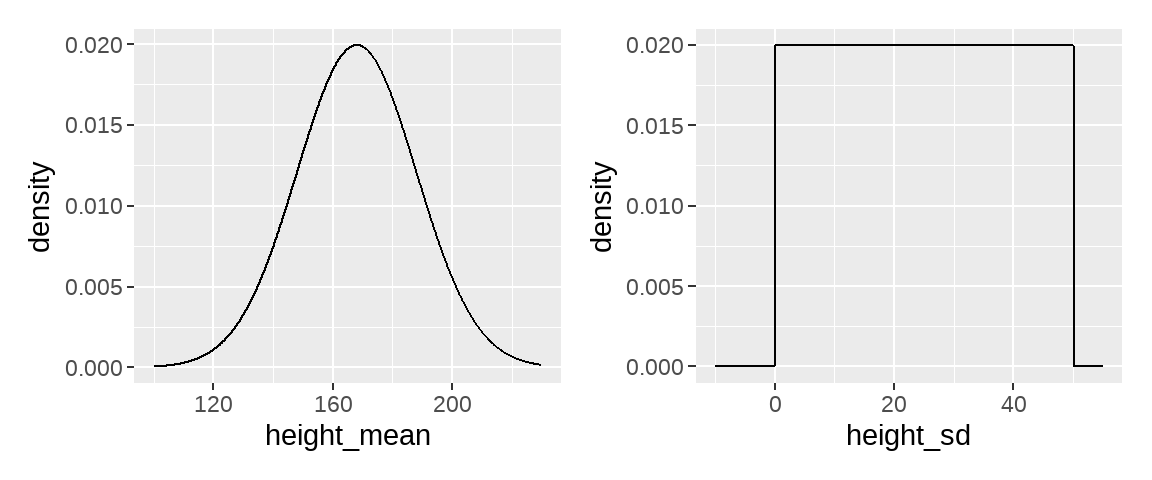
64.2.1 参数空间
我们这里构建 1000*1000个 (mu, sigma) 参数空间
d_grid <- crossing(
mu = seq(from = 150, to = 190, length.out = 1000),
sigma = seq(from = 4, to = 9, length.out = 1000)
)
d_grid## # A tibble: 1,000,000 × 2
## mu sigma
## <dbl> <dbl>
## 1 150 4
## 2 150 4.01
## 3 150 4.01
## 4 150 4.02
## 5 150 4.02
## 6 150 4.03
## 7 150 4.03
## 8 150 4.04
## 9 150 4.04
## 10 150 4.05
## # ℹ 999,990 more rows64.2.2 likelihood
参数空间里,计算在每个(mu, sigma)组合下,身高值(d$height)出现的概率密度dnorm(d2$height, mean = mu, sd = sigma),然后加起来。
很显然,不同的(mu, sigma),概率密度之和是不一样的,我们这里有10001000 个(mu, sigma)组合,
所以会产生 10001000 个值
64.2.4 posterior
d_grid <-
d_grid %>%
mutate(log_likelihood = map2_dbl(mu, sigma, grid_function)) %>%
mutate(prior_mu = dnorm(mu, mean = 168, sd = 20, log = T),
prior_sigma = dunif(sigma, min = 0, max = 50, log = T)) %>%
mutate(product = log_likelihood + prior_mu + prior_sigma) %>%
mutate(probability = exp(product - max(product)))
head(d_grid)## # A tibble: 6 × 7
## mu sigma log_likelihood prior_mu prior_sigma product probability
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 150 4 -2179. -4.32 -3.91 -2188. 0
## 2 150 4.01 -2175. -4.32 -3.91 -2183. 0
## 3 150 4.01 -2171. -4.32 -3.91 -2179. 0
## 4 150 4.02 -2167. -4.32 -3.91 -2175. 0
## 5 150 4.02 -2163. -4.32 -3.91 -2171. 0
## 6 150 4.03 -2159. -4.32 -3.91 -2167. 0
d_grid %>%
ggplot(aes(x = mu, y = sigma, z = probability)) +
geom_contour() +
labs(
x = expression(mu),
y = expression(sigma)
) +
coord_cartesian(
xlim = range(d_grid$mu),
ylim = range(d_grid$sigma)
) +
theme(panel.grid = element_blank())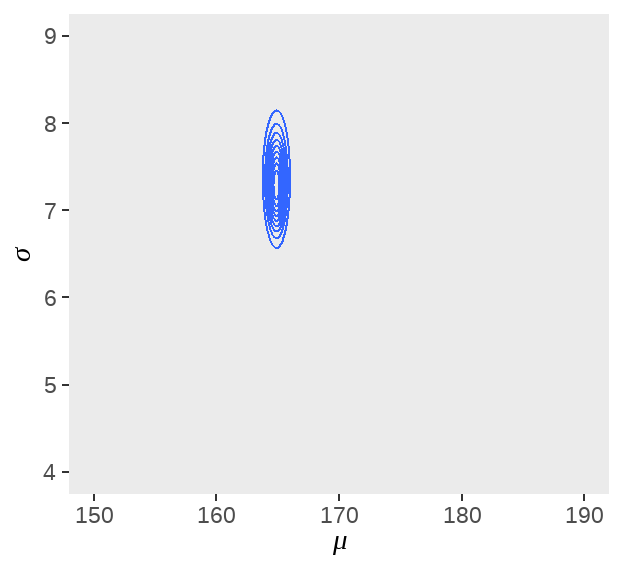
d_grid %>%
ggplot(aes(x = mu, y = sigma)) +
geom_raster(
aes(fill = probability),
interpolate = T
) +
scale_fill_viridis_c(option = "A") +
labs(
x = expression(mu),
y = expression(sigma)
) +
theme(panel.grid = element_blank())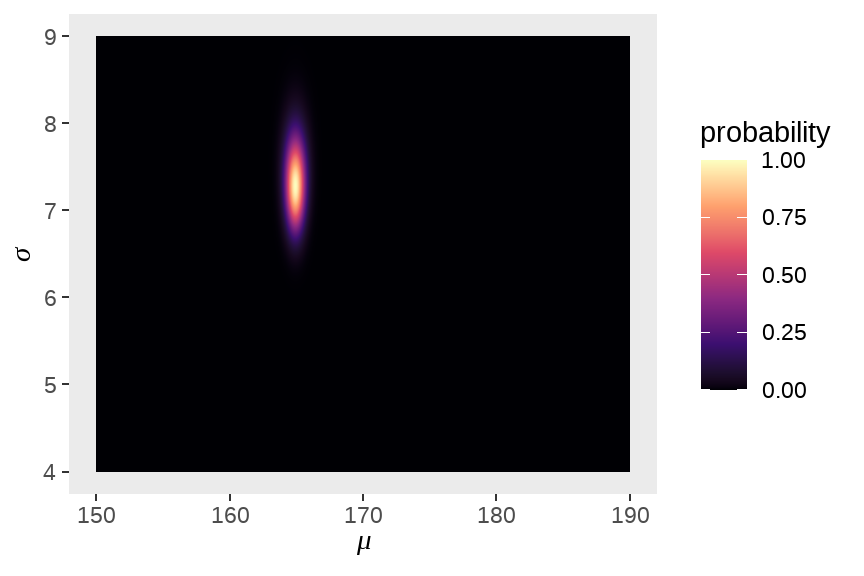
64.2.5 sampling from posterior
后验分布按照probability值的大小来抽样。
d_grid_samples %>%
ggplot(aes(x = mu, y = sigma)) +
geom_point(size = .9, alpha = 1/15) +
scale_fill_viridis_c() +
labs(x = expression(mu[samples]),
y = expression(sigma[samples])) +
theme(panel.grid = element_blank())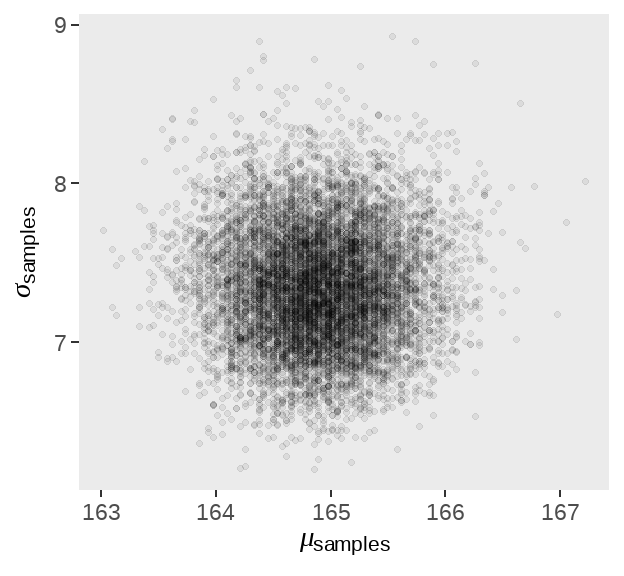
d_grid_samples %>%
select(mu, sigma) %>%
pivot_longer(
cols = everything(),
names_to = "key",
values_to = "value"
) %>%
ggplot(aes(x = value)) +
geom_density(fill = "grey33", size = 0) +
scale_y_continuous(NULL, breaks = NULL) +
xlab(NULL) +
theme(panel.grid = element_blank()) +
facet_wrap(~key, scales = "free")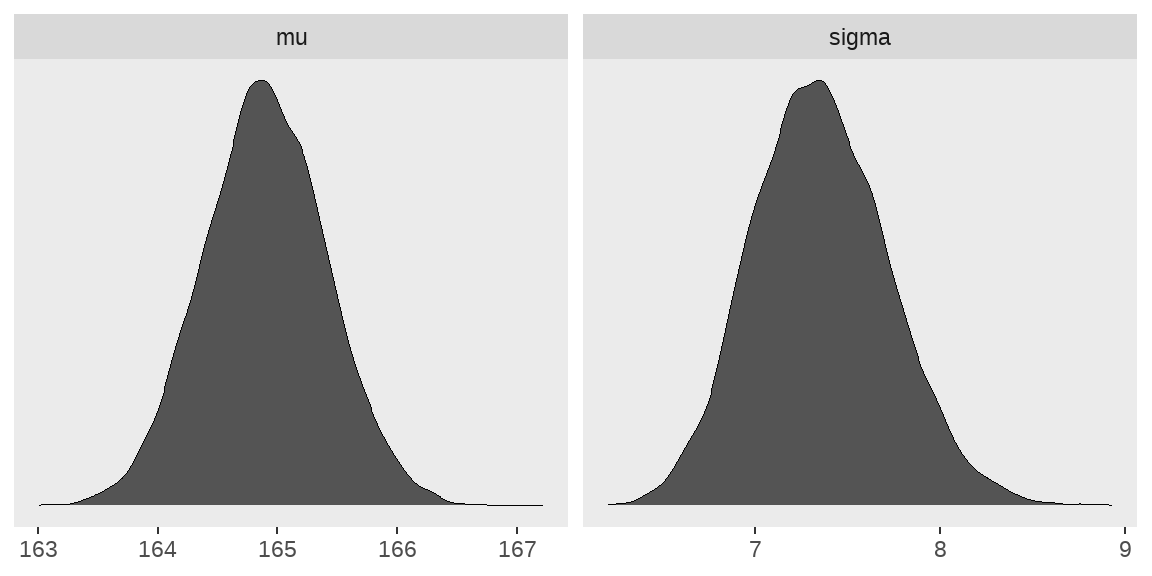
64.2.6 最高密度区间
也可以用tidybayes::mode_hdi()得到后验概率的最高密度区间
library(tidybayes)
d_grid_samples %>%
select(mu, sigma) %>%
pivot_longer(
cols = everything(),
names_to = "key",
values_to = "value"
) %>%
group_by(key) %>%
mode_hdi(value)## # A tibble: 2 × 7
## key value .lower .upper .width .point .interval
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
## 1 mu 165. 164. 166. 0.95 mode hdi
## 2 sigma 7.33 6.62 8.05 0.95 mode hdi以上是通过网格近似的方法得到height分布的后验概率,但这种方法需要构建参数网格,对于较复杂的模型,计算量会陡增,内存占用大、比较费时,因此在实际的数据中,一般不采用这种方法,但网格近似的方法可以帮助我们很好地理解贝叶斯数据分析。
64.3 参考资料
- https://mc-stan.org/
- https://github.com/jgabry/bayes-workflow-book
- https://github.com/XiangyunHuang/masr/
- https://github.com/ASKurz/Statistical_Rethinking_with_brms_ggplot2_and_the_tidyverse_2_ed/
- 《Regression and Other Stories》, Andrew Gelman, Cambridge University Press. 2020
- 《A Student’s Guide to Bayesian Statistics》, Ben Lambert, 2018
- 《Statistical Rethinking: A Bayesian Course with Examples in R and STAN》 ( 2nd Edition), by Richard McElreath, 2020
- 《Bayesian Data Analysis》, Third Edition, 2013
- 《Doing Bayesian Data Analysis: A Tutorial with R, JAGS, and Stan》 (2nd Edition) John Kruschke, 2014
- 《Bayesian Models for Astrophysical Data: Using R, JAGS, Python, and Stan》, Joseph M. Hilbe, Cambridge University Press, 2017