第 95 章 懒人系列
R社区上很多大神,贡献了很多非常优秀的工具,节省了我们的时间,也给我们的生活增添了无限乐趣。我平时逛github的时候时整理一些,现在分享出来供像我一样的懒人用,因此本文档叫“懒人系列”。欢迎大家补充。
95.1 列名太乱了
library(tidyverse)
library(janitor)
## install.packages("janitor")
## https://github.com/sfirke/janitor
fake_raw <- tibble::tribble(
~id, ~`count/num`, ~W.t, ~Case, ~`time--d`, ~`%percent`,
1L, "china", 3L, "w", 5L, 25L,
2L, "us", 4L, "f", 6L, 34L,
3L, "india", 5L, "q", 8L, 78L
)
fake_raw## # A tibble: 3 × 6
## id `count/num` W.t Case `time--d` `%percent`
## <int> <chr> <int> <chr> <int> <int>
## 1 1 china 3 w 5 25
## 2 2 us 4 f 6 34
## 3 3 india 5 q 8 78
fake_raw %>% janitor::clean_names()## # A tibble: 3 × 6
## id count_num w_t case time_d percent_percent
## <int> <chr> <int> <chr> <int> <int>
## 1 1 china 3 w 5 25
## 2 2 us 4 f 6 34
## 3 3 india 5 q 8 7895.3 比distinct()更知我心
df <- tribble(
~id, ~date, ~store_id, ~sales,
1, "2020-03-01", 1, 100,
2, "2020-03-01", 2, 100,
3, "2020-03-01", 3, 150,
4, "2020-03-02", 1, 110,
5, "2020-03-02", 3, 101
)
df %>%
janitor::get_dupes(store_id)## # A tibble: 4 × 5
## store_id dupe_count id date sales
## <dbl> <int> <dbl> <chr> <dbl>
## 1 1 2 1 2020-03-01 100
## 2 1 2 4 2020-03-02 110
## 3 3 2 3 2020-03-01 150
## 4 3 2 5 2020-03-02 101## # A tibble: 5 × 5
## date dupe_count id store_id sales
## <chr> <int> <dbl> <dbl> <dbl>
## 1 2020-03-01 3 1 1 100
## 2 2020-03-01 3 2 2 100
## 3 2020-03-01 3 3 3 150
## 4 2020-03-02 2 4 1 110
## 5 2020-03-02 2 5 3 10195.4 代码太乱了,谁帮我整理下
## install.packages("styler")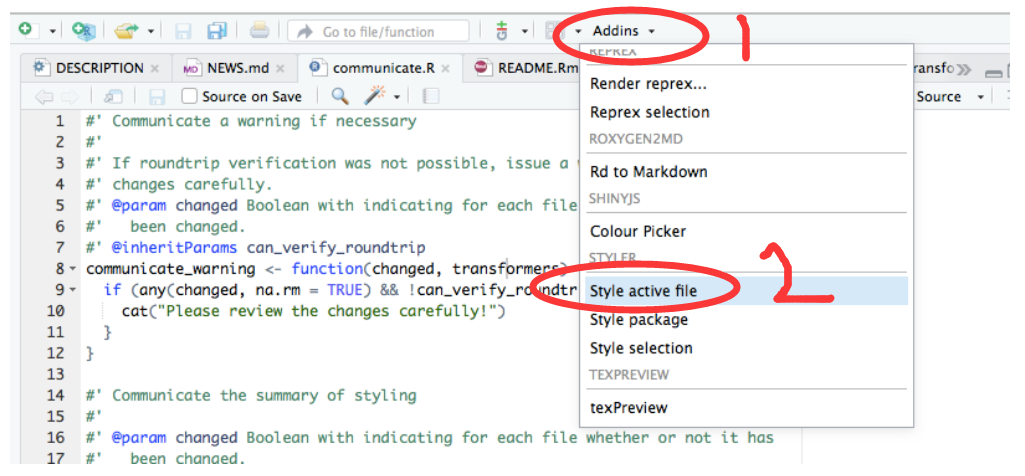
安装后,然后这两个地方点两下,就发现你的代码整齐很多了。或者直接输入
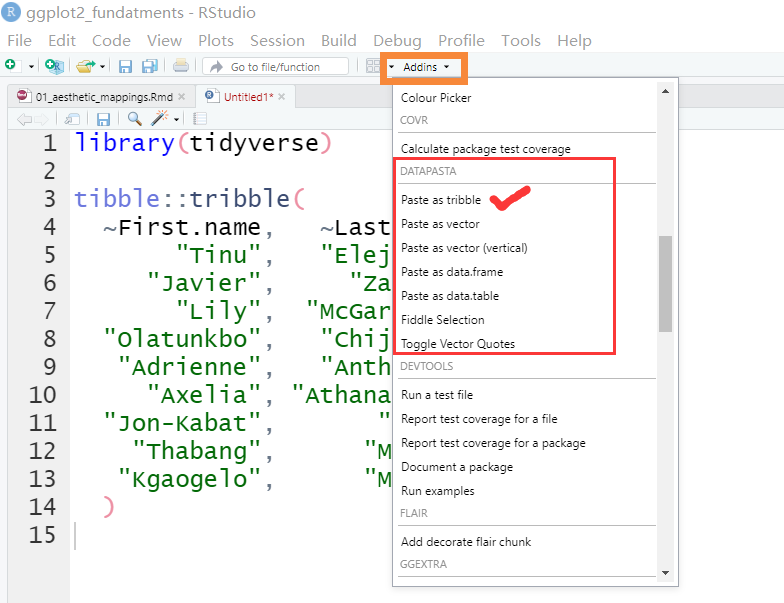
styler:::style_active_file()95.5 用datapasta粘贴小表格
有时候想把excel或者网页上的小表格,放到R里测试下,如果用readr读取excel小数据,可能觉得麻烦,或者大材小用。比如网页https://en.wikipedia.org/wiki/Table_(information)有个表格,有偷懒的办法弄到R?
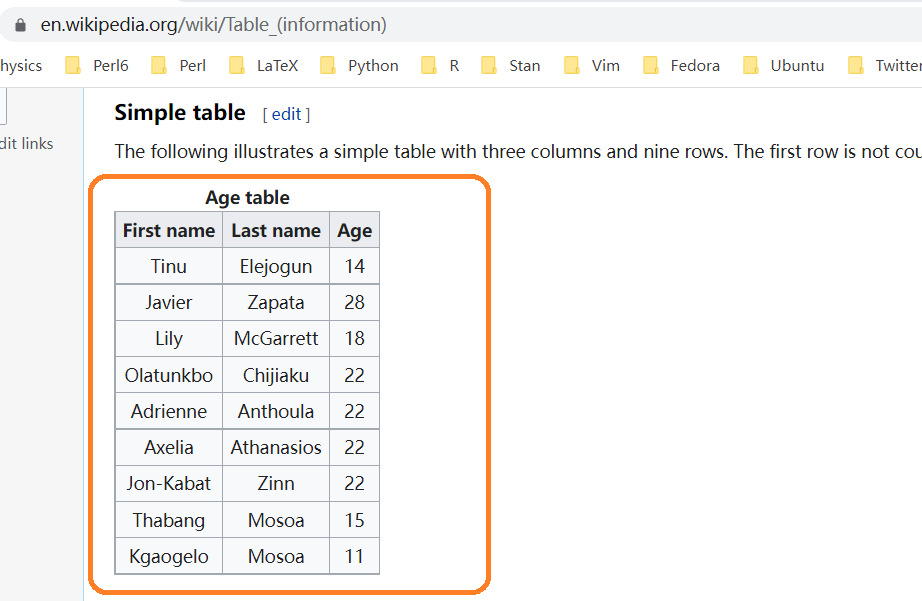
推荐一个方法
- 安装
install.packages("datapasta") - 鼠标选中并复制网页中的表格
- 在 Rstudio 中的
Addins找到datapasta,并点击paste as tribble
95.6 谁帮我敲模型的公式
library(equatiomatic)
## https://github.com/datalorax/equatiomatic
mod1 <- lm(mpg ~ cyl + disp, mtcars)
extract_eq(mod1)\[ \operatorname{mpg} = \alpha + \beta_{1}(\operatorname{cyl}) + \beta_{2}(\operatorname{disp}) + \epsilon \]
extract_eq(mod1, use_coefs = TRUE)\[ \operatorname{\widehat{mpg}} = 34.66 - 1.59(\operatorname{cyl}) - 0.02(\operatorname{disp}) \]
95.7 模型有了,不知道怎么写论文?
We fitted a linear model (estimated using OLS) to predict Sepal.Length with Species (formula: Sepal.Length ~ Species). The model explains a statistically significant and substantial proportion of variance (R2 = 0.62, F(2, 147) = 119.26, p < .001, adj. R2 = 0.61). The model’s intercept, corresponding to Species = setosa, is at 5.01 (95% CI [4.86, 5.15], t(147) = 68.76, p < .001). Within this model:
- The effect of Species [versicolor] is statistically significant and positive (beta = 0.93, 95% CI [0.73, 1.13], t(147) = 9.03, p < .001; Std. beta = 1.12, 95% CI [0.88, 1.37])
- The effect of Species [virginica] is statistically significant and positive (beta = 1.58, 95% CI [1.38, 1.79], t(147) = 15.37, p < .001; Std. beta = 1.91, 95% CI [1.66, 2.16])
Standardized parameters were obtained by fitting the model on a standardized version of the dataset. 95% Confidence Intervals (CIs) and p-values were computed using a Wald t-distribution approximation.
95.8 模型评估一步到位
library(performance)
model <- lm(mpg ~ wt * cyl + gear, data = mtcars)
performance::check_model(model)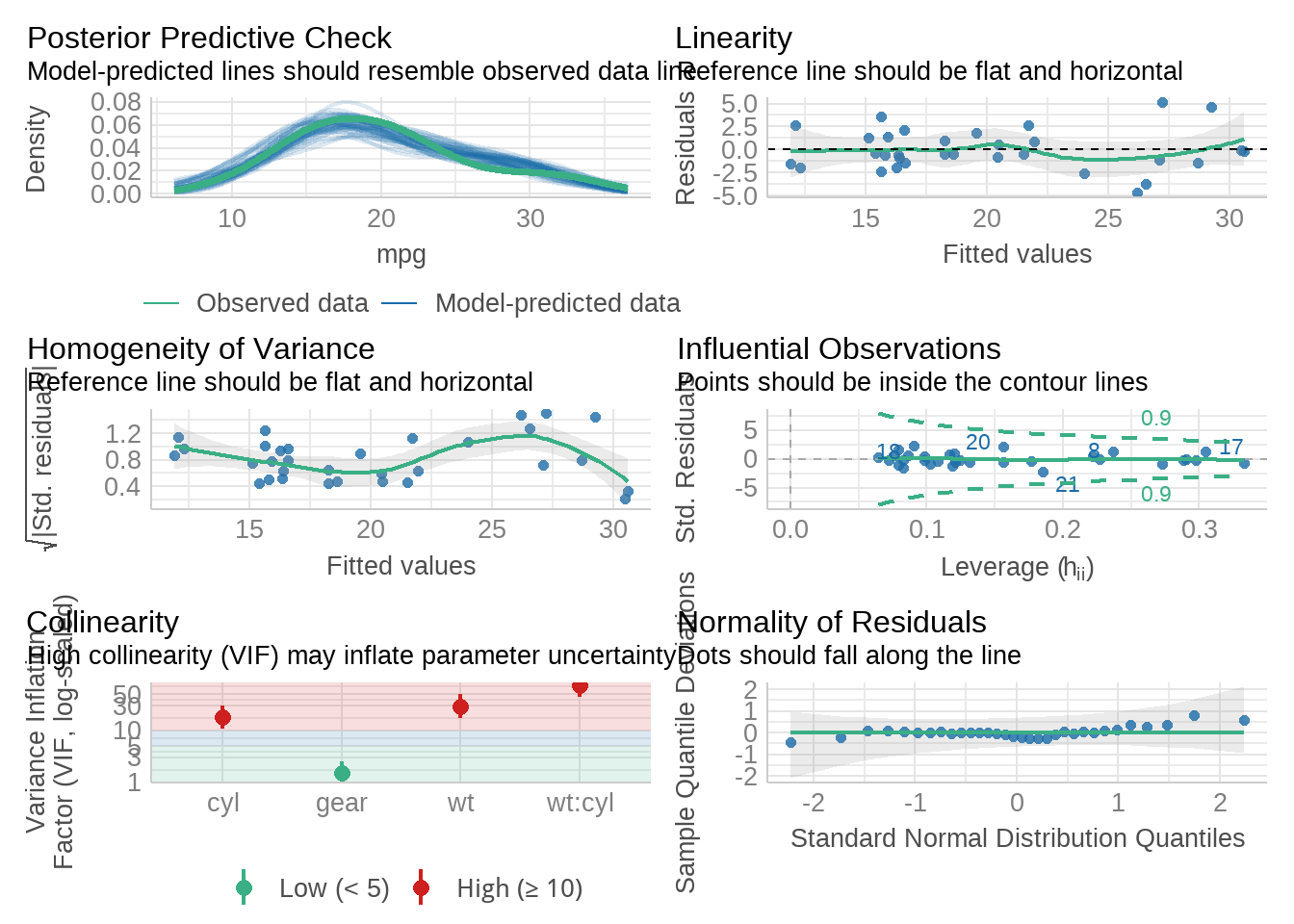
95.9 统计表格不用愁
这个确实省力不少
library(palmerpenguins)
library(gtsummary)
## https://github.com/ddsjoberg/gtsummary
penguins %>%
drop_na() %>%
select(starts_with("bill_")) %>%
tbl_summary(
type = list(everything() ~ "continuous"),
statistic = list(all_continuous() ~ "{mean} ({sd})")
)| Characteristic | N = 3331 |
|---|---|
| bill_length_mm | 44.0 (5.5) |
| bill_depth_mm | 17.16 (1.97) |
| 1 Mean (SD) | |
gtsummary::trial %>%
dplyr::select(trt, age, grade, response) %>%
gtsummary::tbl_summary(
by = trt,
missing = "no"
) %>%
gtsummary::add_p() %>%
gtsummary::add_overall() %>%
gtsummary::add_n() %>%
gtsummary::bold_labels()| Characteristic | N |
Overall N = 2001 |
Drug A N = 981 |
Drug B N = 1021 |
p-value2 |
|---|---|---|---|---|---|
| Age | 189 | 47 (38, 57) | 46 (37, 60) | 48 (39, 56) | 0.7 |
| Grade | 200 | 0.9 | |||
| I | 68 (34%) | 35 (36%) | 33 (32%) | ||
| II | 68 (34%) | 32 (33%) | 36 (35%) | ||
| III | 64 (32%) | 31 (32%) | 33 (32%) | ||
| Tumor Response | 193 | 61 (32%) | 28 (29%) | 33 (34%) | 0.5 |
| 1 Median (Q1, Q3); n (%) | |||||
| 2 Wilcoxon rank sum test; Pearson’s Chi-squared test | |||||
直接复制到论文即可
t1 <-
glm(response ~ trt + age + grade, trial, family = binomial) %>%
gtsummary::tbl_regression(exponentiate = TRUE)
t2 <-
survival::coxph(survival::Surv(ttdeath, death) ~ trt + grade + age, trial) %>%
gtsummary::tbl_regression(exponentiate = TRUE)
gtsummary::tbl_merge(
tbls = list(t1, t2),
tab_spanner = c("**Tumor Response**", "**Time to Death**")
)| Characteristic |
Tumor Response
|
Time to Death
|
||||
|---|---|---|---|---|---|---|
| OR1 | 95% CI1 | p-value | HR1 | 95% CI1 | p-value | |
| Chemotherapy Treatment | ||||||
| Drug A | — | — | — | — | ||
| Drug B | 1.13 | 0.60, 2.13 | 0.7 | 1.30 | 0.88, 1.92 | 0.2 |
| Age | 1.02 | 1.00, 1.04 | 0.10 | 1.01 | 0.99, 1.02 | 0.3 |
| Grade | ||||||
| I | — | — | — | — | ||
| II | 0.85 | 0.39, 1.85 | 0.7 | 1.21 | 0.73, 1.99 | 0.5 |
| III | 1.01 | 0.47, 2.15 | >0.9 | 1.79 | 1.12, 2.86 | 0.014 |
| 1 OR = Odds Ratio, CI = Confidence Interval, HR = Hazard Ratio | ||||||
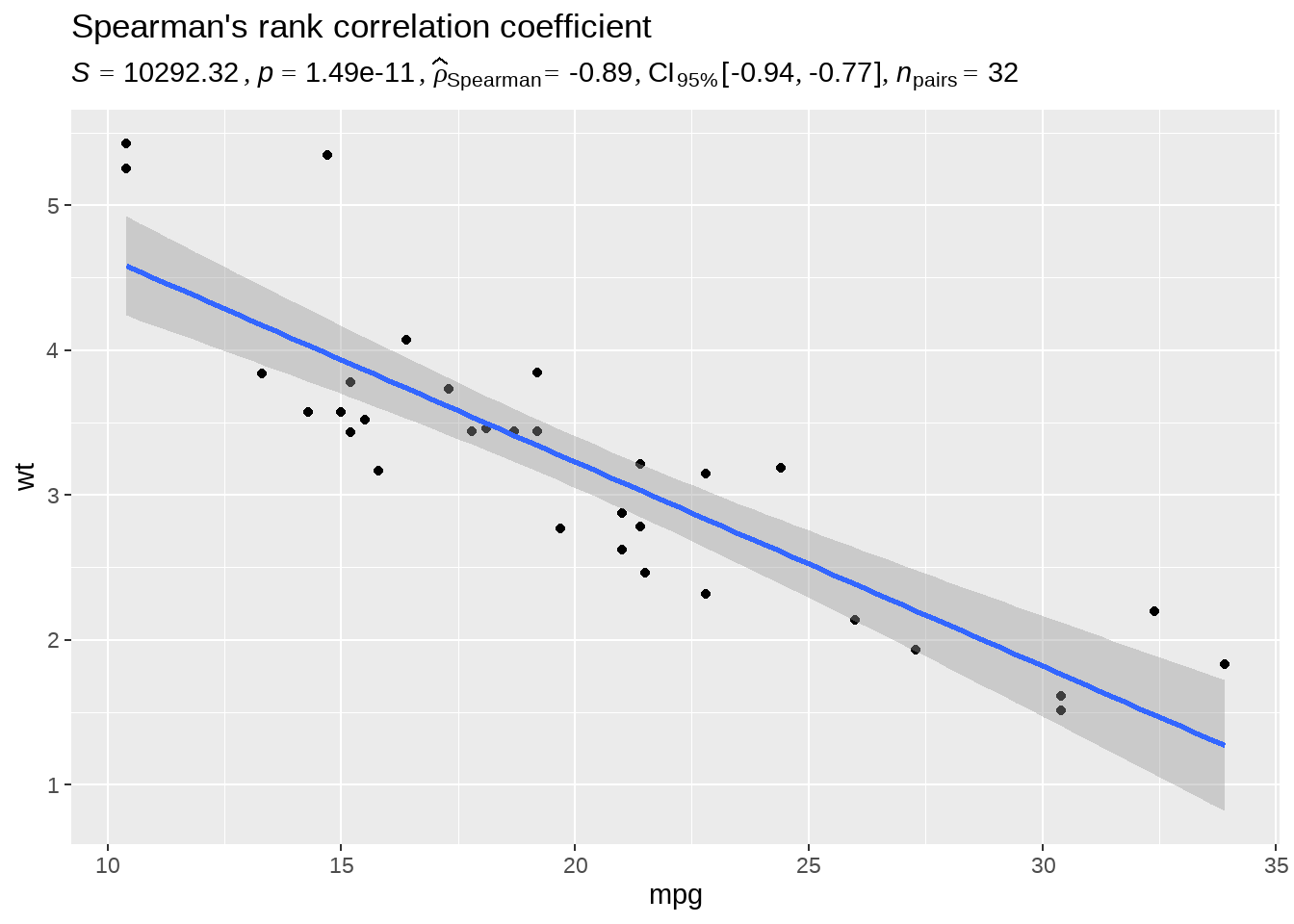
95.10 统计结果写图上
library(ggplot2)
library(statsExpressions)
# https://github.com/IndrajeetPatil/statsExpressions
# dataframe with results
results_data <- corr_test(mtcars, mpg, wt, type = "nonparametric")
# create a scatter plot
ggplot(mtcars, aes(mpg, wt)) +
geom_point() +
geom_smooth(method = "lm", formula = y ~ x) +
labs(
title = "Spearman's rank correlation coefficient",
subtitle = parse(text = results_data$expression)
)
95.11 正则表达式太南了
library(inferregex)
## remotes::install_github("daranzolin/inferregex")
s <- "abcd-9999-ab9"
infer_regex(s)$regex## [1] "^[a-z]{4}-\\d{4}-[a-z]{2}\\d$"有了它,妈妈再也不担心我的正则表达式了
95.12 颜控怎么配色?
mtcars %>%
mutate(cyl = factor(cyl)) %>%
ggplot(aes(x = mpg, fill = cyl, colour = cyl)) +
geom_density(alpha = 0.75) +
labs(fill = "Cylinders", colour = "Cylinders", x = "MPG", y = "Density") +
legend_top()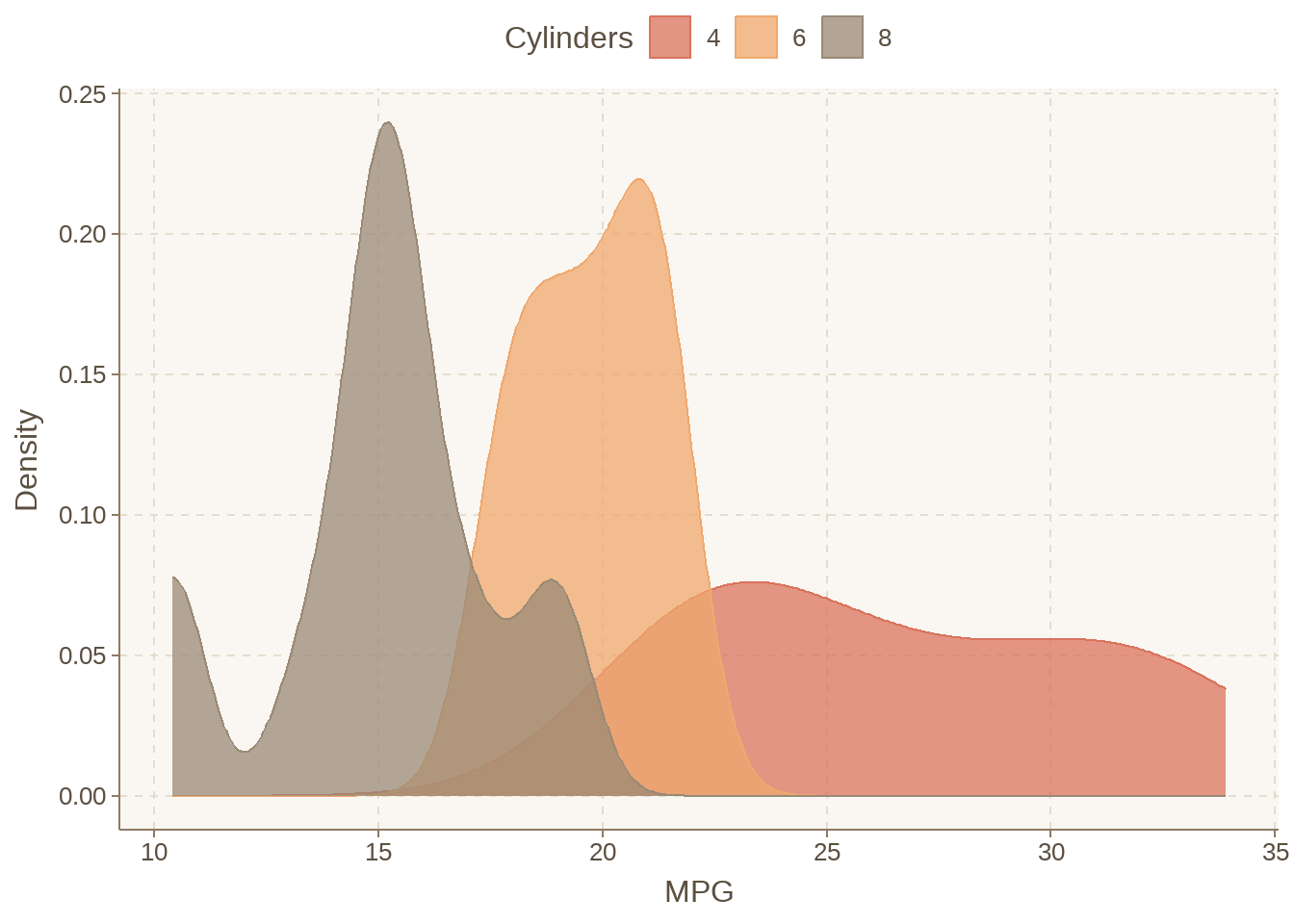
用完别忘了
95.13 画图颜色好看不
scales也是大神的作品,功能多多
## https://github.com/r-lib/scales
library(scales)
show_col(viridis_pal()(10))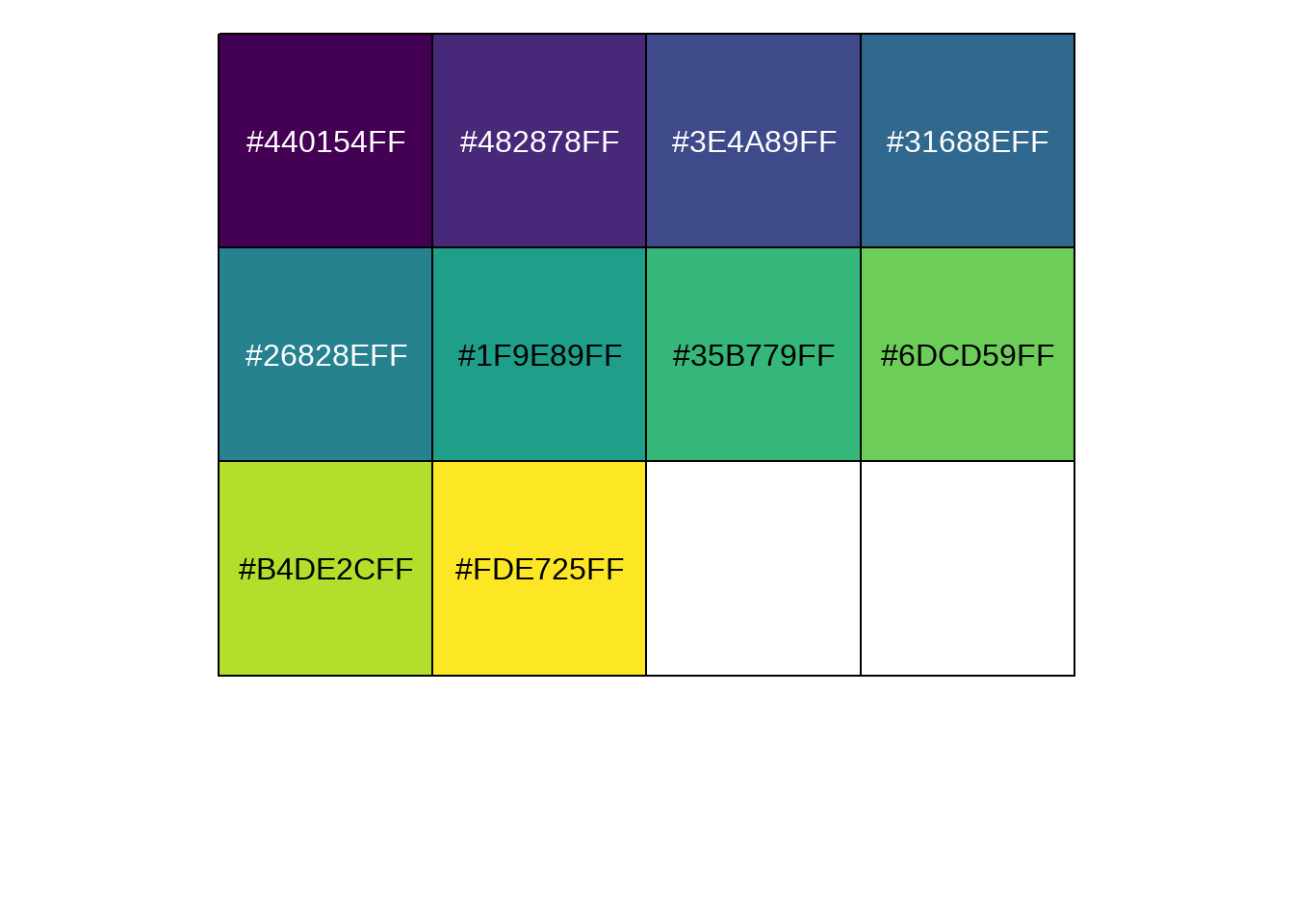
不推荐个人配色,因为我们不专业。直接用专业的配色网站 colorbrewer
先看看颜色,再选择
95.15 犹抱琵琶半遮面
## https://github.com/EmilHvitfeldt/gganonymize
library(ggplot2)
library(gganonymize)
ggg <-
ggplot(mtcars, aes(as.factor(cyl))) +
geom_bar() +
labs(
title = "Test title",
subtitle = "Test subtitle, this one have a lot lot lot lot lot more text then the rest",
caption = "Test caption",
tag = 1
) +
facet_wrap(~vs)
gganonomize(ggg)
你可以看我的图,但就不想告诉你图什么意思,因为我加密了
95.16 整理Rmarkdown
# remotes::install_github("tjmahr/WrapRmd")
# remotes::install_github("fkeck/quickview")
# remotes::install_github("mwip/beautifyR")95.19 多张图摆放
library(patchwork)
p1 <- ggplot(mtcars) +
geom_point(aes(mpg, disp))
p2 <- ggplot(mtcars) +
geom_boxplot(aes(gear, disp, group = gear))
p3 <- ggplot(mtcars) +
geom_smooth(aes(disp, qsec))
# Side by side
p1 + p2 + p3
# On top of each other
p1 / p2 / p3
# Grid
p1 / (p2 + p3)
# plot_layout()
p1 + p2 + p3 +
plot_layout(
nrow = 2,
ncol = 2
)
# layout
layout <- "
AAAA
BCCC
"
p1 + p2 + p3 +
plot_layout(
design = layout
)
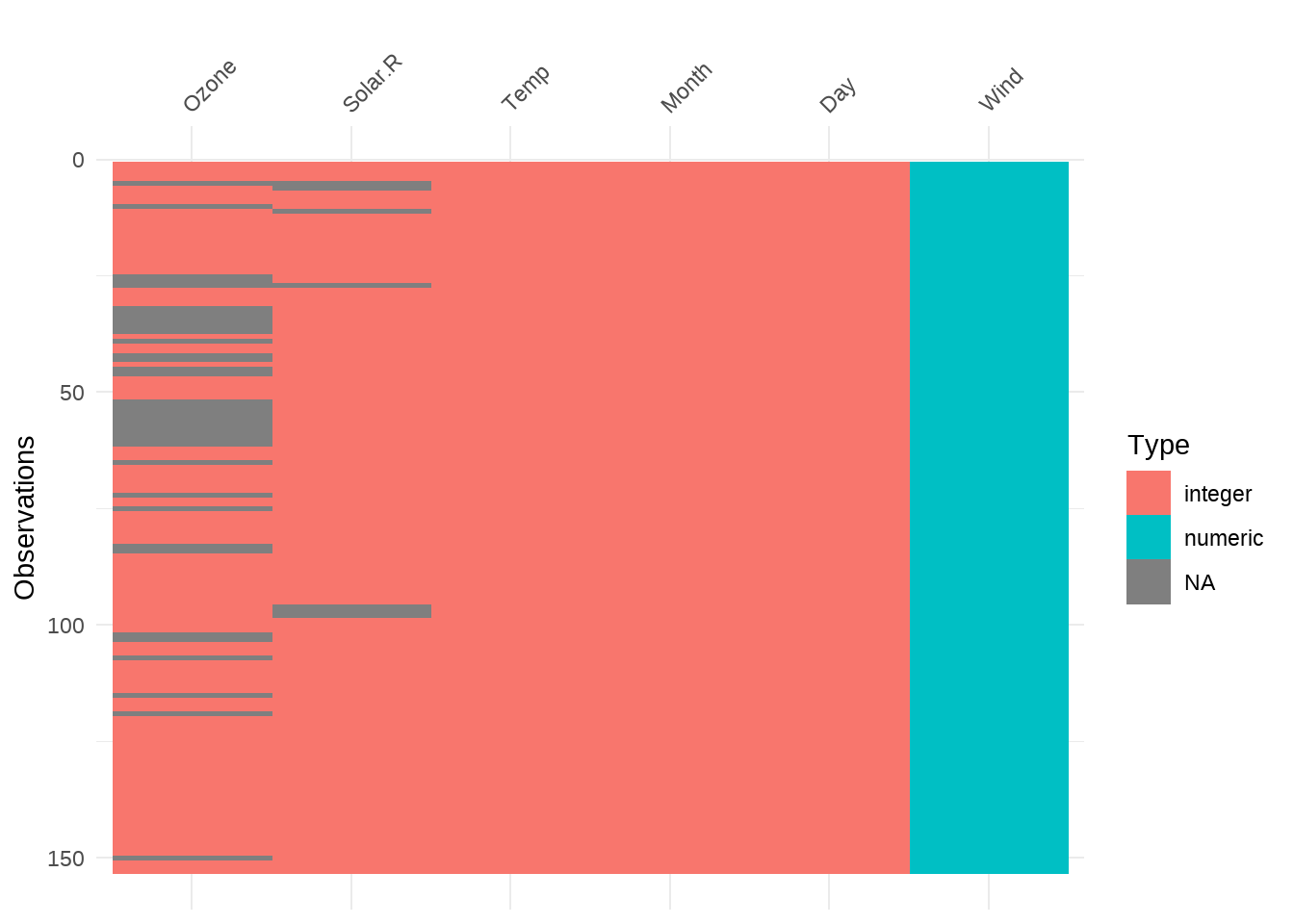
95.20 缺失值处理
library(naniar)
## https://github.com/njtierney/naniar
airquality %>%
group_by(Month) %>%
naniar::miss_var_summary()## # A tibble: 25 × 4
## # Groups: Month [5]
## Month variable n_miss pct_miss
## <int> <chr> <int> <num>
## 1 5 Ozone 5 16.1
## 2 5 Solar.R 4 12.9
## 3 5 Wind 0 0
## 4 5 Temp 0 0
## 5 5 Day 0 0
## 6 6 Ozone 21 70
## 7 6 Solar.R 0 0
## 8 6 Wind 0 0
## 9 6 Temp 0 0
## 10 6 Day 0 0
## # ℹ 15 more rows95.22 让妹纸激发你的热情
library(tidyverse)
library(cowplot)
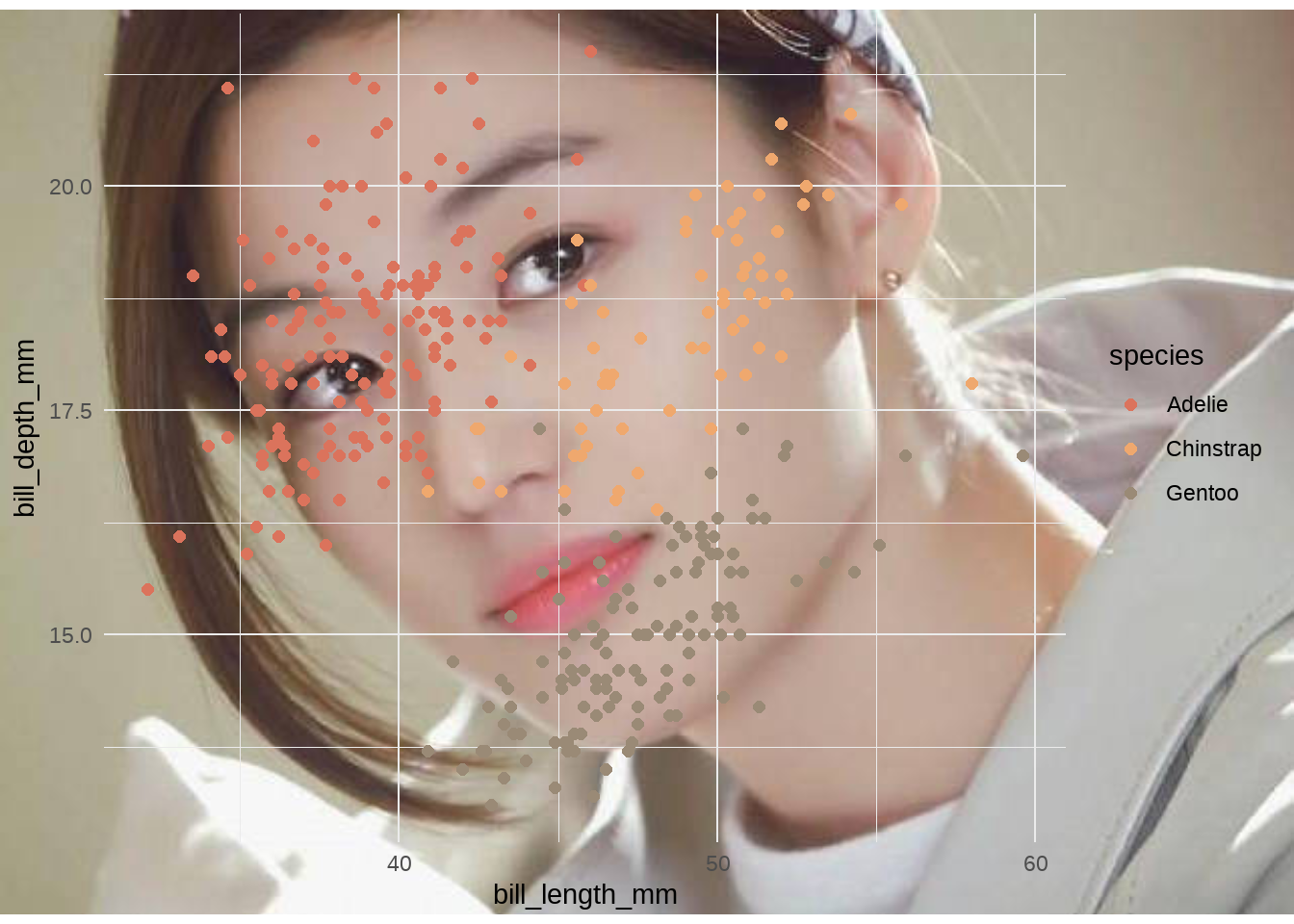
plot <- penguins %>%
ggplot(aes(x = bill_length_mm, y = bill_depth_mm)) +
geom_point(aes(colour = species), size = 2) +
theme_minimal()
ggdraw() +
draw_image("./images/mm.jpeg") +
draw_plot(plot)