library(nimble)
library(GDINA)
library(MCMCvis)
library(nimble, warn.conflicts = FALSE)
DINA.mcmc <- nimbleCode({
for (n in 1:N) {
for (j in 1:J) {
prob[n, j] <- g[j] + (1 - s[j] - g[j]) * 1 * (sum(alpha[n,
1:K] * Q[j, 1:K]) >= sum(Q[j, 1:K]))
score[n, j] ~ dbern(prob[n, j])
}
latent.group.index[n] ~ dcat(pi[1:C])
alpha[n, 1:K] <- all.patterns[latent.group.index[n], 1:K]
}
pi[1:C] ~ ddirch(delta[1:C])
for (j in 1:J) {
s[j] ~ dbeta(1, 1)
g[j] ~ T(dbeta(1, 1), , 1 - s[j])
}
})
score <- sim10GDINA$simdat
Q <- sim10GDINA$simQ
J <- ncol(score)
K <- ncol(Q)
N <- nrow(score)
all.patterns <- GDINA::attributepattern(K)
C <- nrow(all.patterns)
DINAconstants <- list(Q = Q, N = N, J = J, K = K, C = C, delta = rep(1,
C))
DINAdata <- list(score = score, all.patterns = as.matrix(all.patterns))
initials <- list(list(s = rep(0.2, J), g = rep(0.2, J), pi = rep(1/C, C),
latent.group.index = sample(1:C, N, replace = TRUE)), list(s = rep(0.1,
J), g = rep(0.1, J), pi = rep(1/C, C), latent.group.index = sample(1:C,
N, replace = TRUE)))
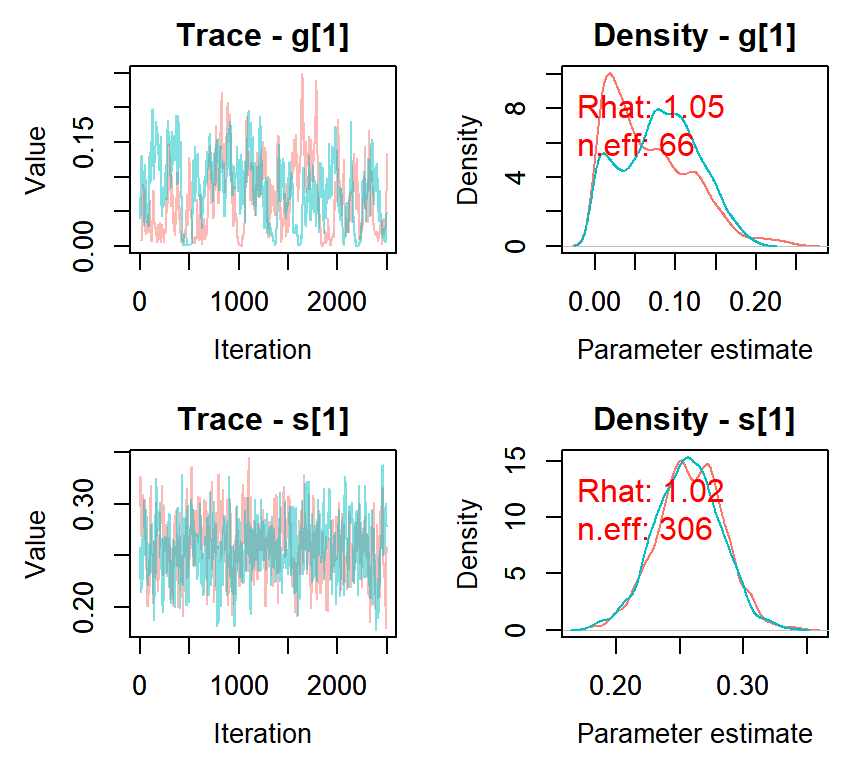
nimbleMCMC_samples <- nimbleMCMC(code = DINA.mcmc, constants = DINAconstants,
data = DINAdata, inits = initials, monitors = c("s", "g"), nburnin = 2500,
niter = 5000, nchains = 2, setSeed = c(123, 456))