3.6 Dimension reduction techniques
As we have seen in Section 3.2, the selection of the best linear model from a set of \(p\) predictors is a challenging task that increases with the dimension of the problem, that is, with \(p.\) In addition to the growth of the set of possible models as \(p\) grows, the model space becomes more complicated to explore due to the potential multicollinearity among the predictors. We will see in this section two methods to deal with these two problems simultaneously.
3.6.1 Review on principal component analysis
Principal Component Analysis (PCA) is a multivariate technique designed to summarize the most important features and relations of \(p\) numerical random variables \(X_1,\ldots,X_p.\) PCA computes a new set of variables, the principal components \(\Gamma_1,\ldots, \Gamma_p,\) that contain the same information as \(X_1,\ldots,X_p\) but expressed in a more convenient way. The goal of PCA is to retain only a limited number \(\ell,\) \(1\leq \ell\leq p,\) of principal components that explain most of the information, therefore performing dimension reduction.
If \(X_1,\ldots,X_p\) are centered,81 then the principal components are orthonormal linear combinations of \(X_1,\ldots,X_p\):
\[\begin{align} \Gamma_j:=a_{1j}X_1+a_{2j}X_2+\cdots+a_{pj}X_p=\mathbf{a}_j'\mathbf{X},\quad j=1,\ldots,p,\tag{3.9} \end{align}\]
where \(\mathbf{a}_j:=(a_{1j},\ldots,a_{pj})',\) \(\mathbf{X}:=(X_1,\ldots,X_p)',\) and the orthonormality condition is
\[\begin{align*} \mathbf{a}_i'\mathbf{a}_j=\begin{cases}1,&\text{if } i=j,\\0,&\text{if } i\neq j.\end{cases} \end{align*}\]
Remarkably, PCA computes the principal components in an ordered way: \(\Gamma_1\) is the principal component that explains the most of the information (quantified as the variance) of \(X_1,\ldots,X_p,\) and then the explained information decreases monotonically down to \(\Gamma_p,\) the principal component that explains the least information. Precisely:
\[\begin{align} \mathbb{V}\mathrm{ar}[\Gamma_1]\geq \mathbb{V}\mathrm{ar}[\Gamma_2]\geq\ldots\geq\mathbb{V}\mathrm{ar}[\Gamma_p].\tag{3.10} \end{align}\]
Mathematically, PCA computes the spectral decomposition82 of the covariance matrix \(\boldsymbol\Sigma:=\mathbb{V}\mathrm{ar}[\mathbf{X}]\):
\[\begin{align*} \boldsymbol\Sigma=\mathbf{A}\boldsymbol\Lambda\mathbf{A}', \end{align*}\]
where \(\boldsymbol\Lambda=\mathrm{diag}(\lambda_1,\ldots,\lambda_p)\) contains the eigenvalues of \(\boldsymbol\Sigma\) and \(\mathbf{A}\) is the orthogonal matrix83 whose columns are the unit-norm eigenvectors of \(\boldsymbol\Sigma.\) The matrix \(\mathbf{A}\) gives, thus, the coefficients of the orthonormal linear combinations:
\[\begin{align*} \mathbf{A}=\begin{pmatrix} \mathbf{a}_1 & \mathbf{a}_2 & \cdots & \mathbf{a}_p \end{pmatrix}=\begin{pmatrix} a_{11} & a_{12} & \cdots & a_{1p} \\ a_{21} & a_{22} & \cdots & a_{2p} \\ \vdots & \vdots & \ddots & \vdots \\ a_{p1} & a_{p2} & \cdots & a_{pp} \end{pmatrix}. \end{align*}\]
If the variables are not centered, the computation of the principal components is done by first subtracting \(\boldsymbol\mu=\mathbb{E}[\mathbf{X}]\) and then premultiplying with \(\mathbf{A}'\):
\[\begin{align} \boldsymbol{\Gamma}=\mathbf{A}'(\mathbf{X}-\boldsymbol{\mu}),\tag{3.11} \end{align}\]
where \(\boldsymbol{\Gamma}=(\Gamma_1,\ldots,\Gamma_p)',\) \(\mathbf{X}=(X_1,\ldots,X_p)',\) and \(\boldsymbol{\mu}=(\mu_1,\ldots,\mu_p)'.\) Note that, because of (1.3) and (3.11),
\[\begin{align} \mathbb{V}\mathrm{ar}[\boldsymbol{\Gamma}]=\mathbf{A}'\boldsymbol{\Sigma}\mathbf{A}=\mathbf{A}'\mathbf{A}\boldsymbol{\Lambda}\mathbf{A}'\mathbf{A}=\boldsymbol{\Lambda}. \end{align}\]
Therefore, \(\mathbb{V}\mathrm{ar}[\Gamma_j]=\lambda_j,\) \(j=1,\ldots,p,\) and as a consequence (3.10) indeed holds.
Also, from (3.11), it is evident that we can express the random vector \(\mathbf{X}\) in terms of \(\boldsymbol{\Gamma}\):
\[\begin{align} \mathbf{X}=\boldsymbol{\mu}+\mathbf{A}\boldsymbol{\Gamma},\tag{3.12} \end{align}\]
which admits an insightful interpretation: \(\boldsymbol{\Gamma}\) is an uncorrelated84 vector that, once rotated by \(\mathbf{A}\) and translated to the location \(\boldsymbol{\mu},\) produces exactly \(\mathbf{X}.\) Therefore, \(\boldsymbol{\Gamma}\) contains the same information as \(\mathbf{X}\) but rearranged in a more convenient way, because the principal components are centered and uncorrelated between them:
\[\begin{align*} \mathbb{E}[\Gamma_i]=0\text{ and }\mathrm{Cov}[\Gamma_i,\Gamma_j]=\begin{cases}\lambda_i,&\text{if } i=j,\\0,&\text{if } i\neq j.\end{cases} \end{align*}\]
Due to the uncorrelation of \(\boldsymbol{\Gamma},\) we can measure the total variance of \(\boldsymbol{\Gamma}\) as \(\sum_{j=1}^p\mathbb{V}\mathrm{ar}[\Gamma_j]=\sum_{j=1}^p\lambda_j.\) Consequently, we can define the proportion of variance explained by the first \(\ell\) principal components, \(1\leq \ell\leq p,\) as
\[\begin{align*} \frac{\sum_{j=1}^\ell\lambda_j}{\sum_{j=1}^p\lambda_j}. \end{align*}\]
In the sample case85 where a sample \(\mathbf{X}_1,\ldots,\mathbf{X}_n\) is given and \(\boldsymbol\mu\) and \(\boldsymbol\Sigma\) are unknown, \(\boldsymbol\mu\) is replaced by the sample mean \(\bar{\mathbf{X}}\) and \(\boldsymbol\Sigma\) by the sample variance-covariance matrix \(\mathbf{S}=\frac{1}{n}\sum_{i=1}^n(\mathbf{X}_i-\bar{\mathbf{X}})(\mathbf{X}_i-\bar{\mathbf{X}})'.\) Then, the spectral decomposition of \(\mathbf{S}\) is computed86. This gives \(\hat{\mathbf{A}},\) the matrix of loadings, and produces the scores of the data:
\[\begin{align*} \hat{\boldsymbol{\Gamma}}_1:=\hat{\mathbf{A}}'(\mathbf{X}_1-\bar{\mathbf{X}}),\ldots,\hat{\boldsymbol{\Gamma}}_n:=\hat{\mathbf{A}}'(\mathbf{X}_n-\bar{\mathbf{X}}). \end{align*}\]
The scores are centered, uncorrelated, and have sample variances in each vector’s entry that are sorted in a decreasing way. The scores are the data coordinates with respect to the principal component basis.
The maximum number of principal components that can be determined from a sample \(\mathbf{X}_1,\ldots,\mathbf{X}_n\) is \(\min(n-1,p),\) assuming that the matrix formed by \(\left(\mathbf{X}_1 \cdots \mathbf{X}_n\right)\) is of full rank (i.e., if the rank is \(\min(n,p)\)). If \(n\geq p\) and the variables \(X_1,\ldots,X_p\) are such that only \(r\) of them are linearly independent, then the maximum is \(\min(n-1,r).\)
Let’s see an example of these concepts in La Liga 2015/2016 dataset. It contains the standings and team statistics for La Liga 2015/2016.
laliga <- readxl::read_excel("la-liga-2015-2016.xlsx", sheet = 1, col_names = TRUE)
laliga <- as.data.frame(laliga) # Avoid tibble since it drops row.namesA quick preprocessing gives:
rownames(laliga) <- laliga$Team # Set teams as case names to avoid factors
laliga$Team <- NULL
laliga <- laliga[, -c(2, 8)] # Do not add irrelevant information
summary(laliga)
## Points Wins Draws Loses Goals.scored Goals.conceded
## Min. :32.00 Min. : 8.00 Min. : 4.00 Min. : 4.00 Min. : 34.00 Min. :18.00
## 1st Qu.:41.25 1st Qu.:10.00 1st Qu.: 8.00 1st Qu.:12.00 1st Qu.: 40.00 1st Qu.:42.50
## Median :44.50 Median :12.00 Median : 9.00 Median :15.50 Median : 45.50 Median :52.50
## Mean :52.40 Mean :14.40 Mean : 9.20 Mean :14.40 Mean : 52.15 Mean :52.15
## 3rd Qu.:60.50 3rd Qu.:17.25 3rd Qu.:10.25 3rd Qu.:18.25 3rd Qu.: 51.25 3rd Qu.:63.25
## Max. :91.00 Max. :29.00 Max. :18.00 Max. :22.00 Max. :112.00 Max. :74.00
## Percentage.scored.goals Percentage.conceded.goals Shots Shots.on.goal Penalties.scored Assistances
## Min. :0.890 Min. :0.470 Min. :346.0 Min. :129.0 Min. : 1.00 Min. :23.00
## 1st Qu.:1.050 1st Qu.:1.115 1st Qu.:413.8 1st Qu.:151.2 1st Qu.: 1.00 1st Qu.:28.50
## Median :1.195 Median :1.380 Median :438.0 Median :165.0 Median : 3.00 Median :32.50
## Mean :1.371 Mean :1.371 Mean :452.4 Mean :173.1 Mean : 3.45 Mean :37.85
## 3rd Qu.:1.347 3rd Qu.:1.663 3rd Qu.:455.5 3rd Qu.:180.0 3rd Qu.: 4.50 3rd Qu.:36.75
## Max. :2.950 Max. :1.950 Max. :712.0 Max. :299.0 Max. :11.00 Max. :90.00
## Fouls.made Matches.without.conceding Yellow.cards Red.cards Offsides
## Min. :385.0 Min. : 4.0 Min. : 66.0 Min. :1.00 Min. : 72.00
## 1st Qu.:483.8 1st Qu.: 7.0 1st Qu.: 97.0 1st Qu.:4.00 1st Qu.: 83.25
## Median :530.5 Median :10.5 Median :108.5 Median :5.00 Median : 88.00
## Mean :517.6 Mean :10.7 Mean :106.2 Mean :5.05 Mean : 92.60
## 3rd Qu.:552.8 3rd Qu.:13.0 3rd Qu.:115.2 3rd Qu.:6.00 3rd Qu.:103.75
## Max. :654.0 Max. :24.0 Max. :141.0 Max. :9.00 Max. :123.00Let’s check that R’s function for PCA, princomp, returns the same principal components we outlined in the theory.
# PCA
pcaLaliga <- princomp(laliga, fix_sign = TRUE)
summary(pcaLaliga)
## Importance of components:
## Comp.1 Comp.2 Comp.3 Comp.4 Comp.5 Comp.6 Comp.7 Comp.8
## Standard deviation 104.7782561 48.5461449 22.13337511 12.66692413 8.234215354 7.83426116 6.068864168 4.137079559
## Proportion of Variance 0.7743008 0.1662175 0.03455116 0.01131644 0.004782025 0.00432876 0.002597659 0.001207133
## Cumulative Proportion 0.7743008 0.9405183 0.97506949 0.98638593 0.991167955 0.99549671 0.998094374 0.999301507
## Comp.9 Comp.10 Comp.11 Comp.12 Comp.13 Comp.14 Comp.15
## Standard deviation 2.0112480391 1.8580509157 1.126111e+00 9.568824e-01 4.716064e-01 1.707105e-03 8.365534e-04
## Proportion of Variance 0.0002852979 0.0002434908 8.943961e-05 6.457799e-05 1.568652e-05 2.055361e-10 4.935768e-11
## Cumulative Proportion 0.9995868048 0.9998302956 9.999197e-01 9.999843e-01 1.000000e+00 1.000000e+00 1.000000e+00
## Comp.16 Comp.17
## Standard deviation 0 0
## Proportion of Variance 0 0
## Cumulative Proportion 1 1
# The standard deviations are the square roots of the eigenvalues
# The cumulative proportion of variance explained accumulates the
# variance explained starting at the first component
# We use fix_sign = TRUE so that the signs of the loadings are
# determined by the first element of each loading, set to be
# nonnegative. Otherwise, the signs could change for different OS /
# R versions yielding to opposite interpretations of the PCs
# Plot of variances of each component (screeplot)
plot(pcaLaliga, type = "l")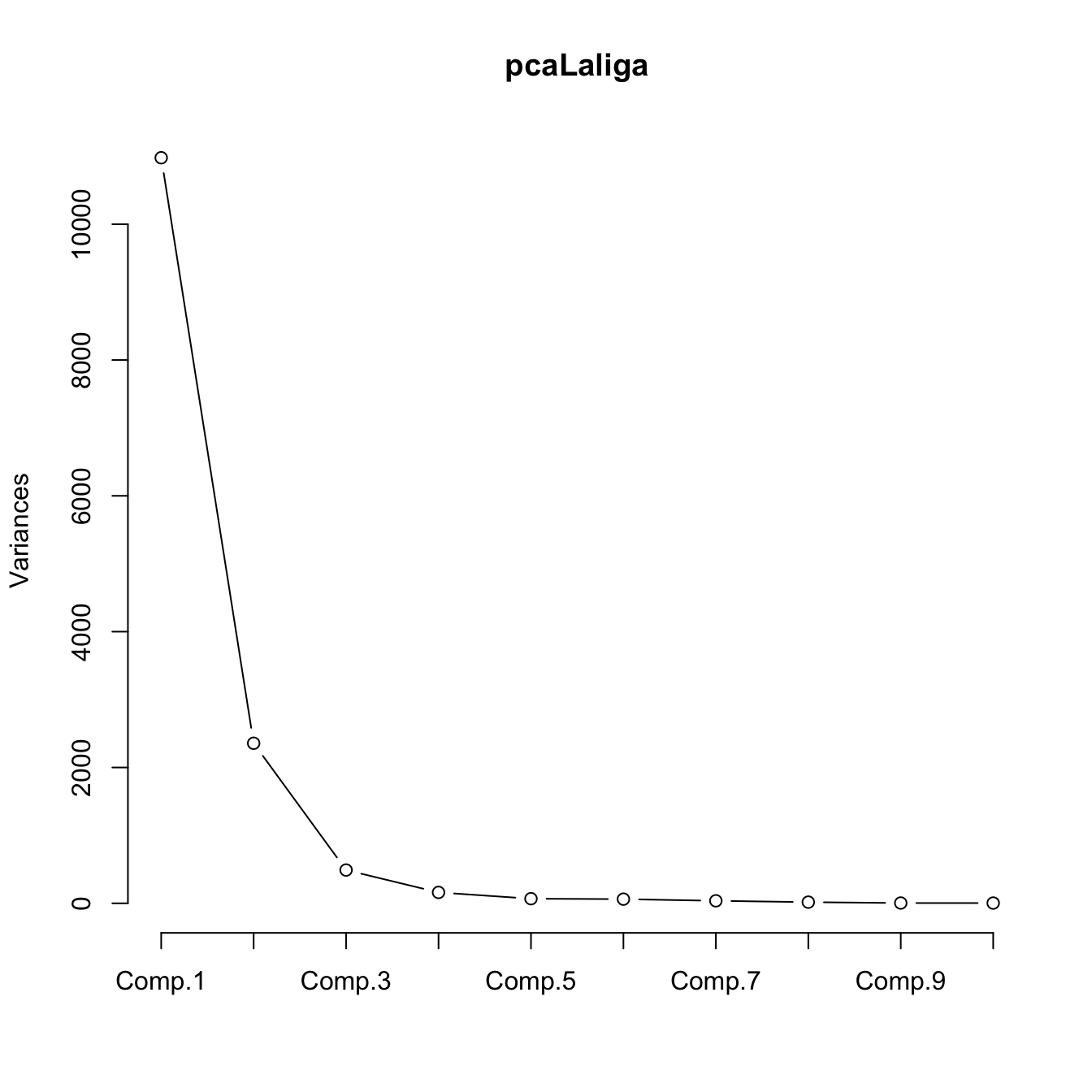
# Useful for detecting an "elbow" in the graph whose location gives the
# "right" number of components to retain. Ideally, this elbow appears
# when the next variances are almost similar and notably smaller when
# compared with the previous
# Alternatively: plot of the cumulated percentage of variance
# barplot(cumsum(pcaLaliga$sdev^2) / sum(pcaLaliga$sdev^2))
# Computation of PCA from the spectral decomposition
n <- nrow(laliga)
eig <- eigen(cov(laliga) * (n - 1) / n) # By default, cov() computes the
# quasi-variance-covariance matrix that divides by n - 1, but PCA and princomp
# consider the sample variance-covariance matrix that divides by n
A <- eig$vectors
# Same eigenvalues
pcaLaliga$sdev^2 - eig$values
## Comp.1 Comp.2 Comp.3 Comp.4 Comp.5 Comp.6 Comp.7 Comp.8
## -5.456968e-12 -1.364242e-12 0.000000e+00 -4.263256e-13 -2.415845e-13 -1.278977e-13 -3.552714e-14 -4.263256e-14
## Comp.9 Comp.10 Comp.11 Comp.12 Comp.13 Comp.14 Comp.15 Comp.16
## -2.025047e-13 -7.549517e-14 -1.088019e-14 -3.108624e-15 -1.626477e-14 -3.685726e-15 1.036604e-15 -2.025796e-13
## Comp.17
## 1.435137e-12
# The eigenvectors (the a_j vectors) are the column vectors in $loadings
pcaLaliga$loadings
##
## Loadings:
## Comp.1 Comp.2 Comp.3 Comp.4 Comp.5 Comp.6 Comp.7 Comp.8 Comp.9 Comp.10 Comp.11 Comp.12
## Points 0.125 0.497 0.195 0.139 0.340 0.425 0.379 0.129 0.166
## Wins 0.184 0.175 0.134 -0.198 0.139
## Draws 0.101 -0.186 0.608 0.175 0.185 -0.251
## Loses -0.129 -0.114 -0.157 -0.410 -0.243 -0.166 0.112
## Goals.scored 0.181 0.251 -0.186 -0.169 0.399 0.335 -0.603 -0.155 0.129 0.289 -0.230
## Goals.conceded -0.471 -0.493 -0.277 0.257 0.280 0.441 -0.118 0.297
## Percentage.scored.goals
## Percentage.conceded.goals
## Shots 0.718 0.442 -0.342 0.255 0.241 0.188
## Shots.on.goal 0.386 0.213 0.182 -0.287 -0.532 -0.163 -0.599
## Penalties.scored -0.350 0.258 0.378 -0.661 0.456
## Assistances 0.148 0.198 -0.173 0.362 0.216 0.215 0.356 -0.685 -0.265
## Fouls.made -0.480 0.844 0.166 -0.110
## Matches.without.conceding 0.151 0.129 -0.182 0.176 -0.369 -0.376 -0.411
## Yellow.cards -0.141 0.144 -0.363 0.113 0.225 0.637 -0.550 -0.126 0.156
## Red.cards -0.123 -0.157 0.405 0.666
## Offsides 0.108 0.202 -0.696 0.647 -0.106
## Comp.13 Comp.14 Comp.15 Comp.16 Comp.17
## Points 0.138 0.240 0.345
## Wins 0.147 -0.904
## Draws -0.304 0.554 -0.215
## Loses 0.156 0.794 0.130
## Goals.scored -0.153
## Goals.conceded
## Percentage.scored.goals -0.760 -0.650
## Percentage.conceded.goals 0.650 -0.760
## Shots
## Shots.on.goal
## Penalties.scored -0.114
## Assistances -0.102
## Fouls.made
## Matches.without.conceding -0.664
## Yellow.cards
## Red.cards -0.587
## Offsides
##
## Comp.1 Comp.2 Comp.3 Comp.4 Comp.5 Comp.6 Comp.7 Comp.8 Comp.9 Comp.10 Comp.11 Comp.12 Comp.13 Comp.14
## SS loadings 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000
## Proportion Var 0.059 0.059 0.059 0.059 0.059 0.059 0.059 0.059 0.059 0.059 0.059 0.059 0.059 0.059
## Cumulative Var 0.059 0.118 0.176 0.235 0.294 0.353 0.412 0.471 0.529 0.588 0.647 0.706 0.765 0.824
## Comp.15 Comp.16 Comp.17
## SS loadings 1.000 1.000 1.000
## Proportion Var 0.059 0.059 0.059
## Cumulative Var 0.882 0.941 1.000
# The scores is the representation of the data in the principal components -
# it has the same information as laliga
head(round(pcaLaliga$scores, 4))
## Comp.1 Comp.2 Comp.3 Comp.4 Comp.5 Comp.6 Comp.7 Comp.8 Comp.9 Comp.10 Comp.11 Comp.12
## Barcelona 242.2392 -21.5810 25.3801 -17.4375 -7.1797 9.0814 -5.5920 -7.3615 -0.3716 1.7161 0.0265 -1.0948
## Real Madrid 313.6026 63.2024 -8.7570 8.5558 0.7119 0.2221 6.7035 2.4456 1.8388 -2.9661 -0.1345 0.3410
## Atlético Madrid 45.9939 -0.6466 38.5410 31.3888 3.9163 3.2904 0.2432 5.0913 -3.0444 2.0975 0.6771 -0.3986
## Villarreal -96.2201 -42.9329 50.0036 -11.2420 10.4733 2.4294 -3.0183 0.1958 1.2106 -1.7453 0.1351 -0.5735
## Athletic 14.5173 -16.1897 18.8840 -0.4122 -5.6491 -6.9330 8.0653 2.4783 -2.6921 0.8950 0.1542 1.4715
## Celta -13.0748 6.7925 5.2271 -9.0945 6.1265 11.8795 -2.6148 6.9707 3.0826 0.3130 0.0860 1.9159
## Comp.13 Comp.14 Comp.15 Comp.16 Comp.17
## Barcelona 0.1516 -0.0010 -2e-04 0 0
## Real Madrid -0.0332 0.0015 4e-04 0 0
## Atlético Madrid -0.1809 0.0005 -1e-04 0 0
## Villarreal -0.5894 -0.0001 -2e-04 0 0
## Athletic 0.1109 -0.0031 -3e-04 0 0
## Celta 0.3722 -0.0018 3e-04 0 0
# Uncorrelated
corrplot::corrplot(cor(pcaLaliga$scores), addCoef.col = "gray")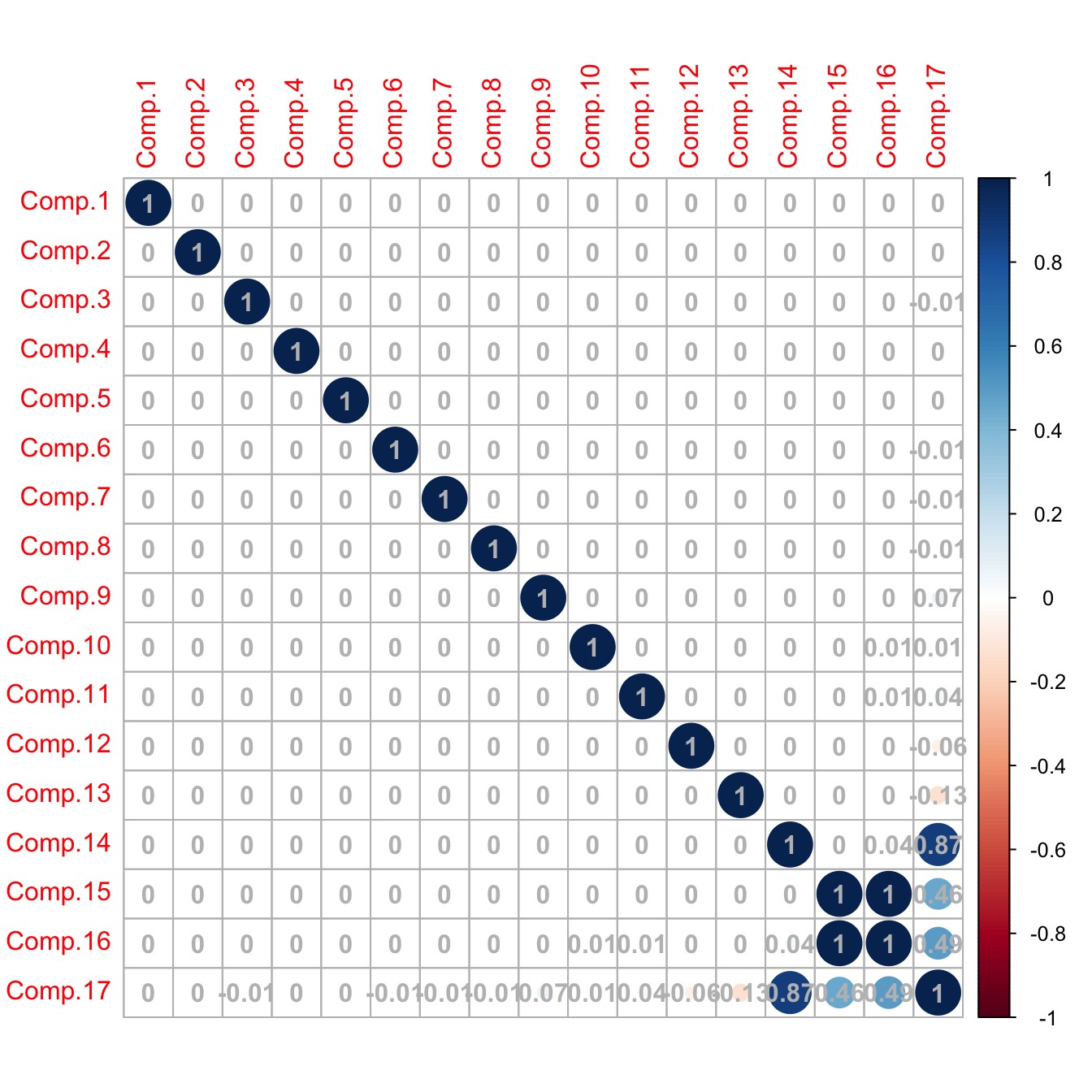
# Caution! What happened in the last columns? What happened is that the
# variance for the last principal components is close to zero (because there
# are linear dependencies on the variables; e.g., Points, Wins, Loses, Draws),
# so the computation of the correlation matrix becomes unstable for those
# variables (a 0/0 division takes place)
# Better to inspect the covariance matrix
corrplot::corrplot(cov(pcaLaliga$scores), addCoef.col = "gray", is.corr = FALSE)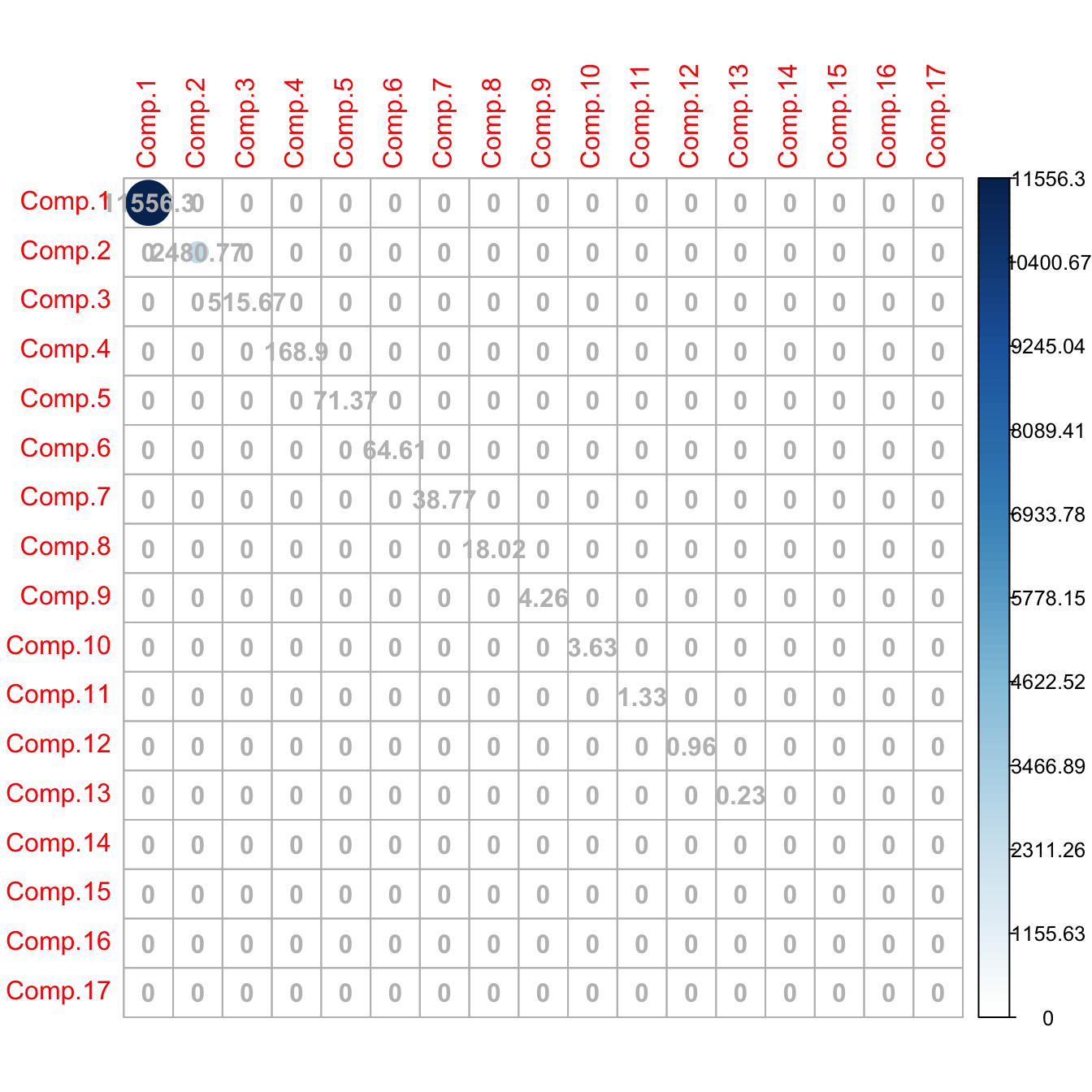
# The scores are A' * (X_i - mu). We center the data with scale()
# and then multiply each row by A'
scores <- scale(laliga, center = TRUE, scale = FALSE) %*% A
# Same as (but this is much slower)
# scores <- t(apply(scale(laliga, center = TRUE, scale = FALSE), 1,
# function(x) t(A) %*% x))
# Same scores (up to possible changes in signs)
max(abs(abs(pcaLaliga$scores) - abs(scores)))
## [1] 2.278507e-11
# Reconstruct the data from all the principal components
head(
sweep(pcaLaliga$scores %*% t(pcaLaliga$loadings), 2, pcaLaliga$center, "+")
)
## Points Wins Draws Loses Goals.scored Goals.conceded Percentage.scored.goals Percentage.conceded.goals
## Barcelona 91 29 4 5 112 29 2.95 0.76
## Real Madrid 90 28 6 4 110 34 2.89 0.89
## Atlético Madrid 88 28 4 6 63 18 1.66 0.47
## Villarreal 64 18 10 10 44 35 1.16 0.92
## Athletic 62 18 8 12 58 45 1.53 1.18
## Celta 60 17 9 12 51 59 1.34 1.55
## Shots Shots.on.goal Penalties.scored Assistances Fouls.made Matches.without.conceding Yellow.cards
## Barcelona 600 277 11 79 385 18 66
## Real Madrid 712 299 6 90 420 14 72
## Atlético Madrid 481 186 1 49 503 24 91
## Villarreal 346 135 3 32 534 17 100
## Athletic 450 178 3 42 502 13 84
## Celta 442 170 4 43 528 10 116
## Red.cards Offsides
## Barcelona 1 120
## Real Madrid 5 114
## Atlético Madrid 3 84
## Villarreal 4 106
## Athletic 5 92
## Celta 6 103An important issue when doing PCA is the scale of the variables, since the variance depends on the units in which the variable is measured.87 Therefore, when variables with different ranges are mixed, the variability of one may dominate the other merely due to a scale artifact. To prevent this, we standardize the dataset prior to do a PCA.
# Use cor = TRUE to standardize variables (all have unit variance)
# and avoid scale distortions
pcaLaligaStd <- princomp(x = laliga, cor = TRUE, fix_sign = TRUE)
summary(pcaLaligaStd)
## Importance of components:
## Comp.1 Comp.2 Comp.3 Comp.4 Comp.5 Comp.6 Comp.7 Comp.8
## Standard deviation 3.2918365 1.5511043 1.13992451 0.91454883 0.85765282 0.59351138 0.45780827 0.370649324
## Proportion of Variance 0.6374228 0.1415250 0.07643694 0.04919997 0.04326873 0.02072093 0.01232873 0.008081231
## Cumulative Proportion 0.6374228 0.7789478 0.85538472 0.90458469 0.94785342 0.96857434 0.98090308 0.988984306
## Comp.9 Comp.10 Comp.11 Comp.12 Comp.13 Comp.14 Comp.15
## Standard deviation 0.327182806 0.217470830 0.128381750 0.0976778705 0.083027923 2.582795e-03 1.226924e-03
## Proportion of Variance 0.006296976 0.002781974 0.000969522 0.0005612333 0.000405508 3.924019e-07 8.854961e-08
## Cumulative Proportion 0.995281282 0.998063256 0.999032778 0.9995940111 0.999999519 9.999999e-01 1.000000e+00
## Comp.16 Comp.17
## Standard deviation 9.256461e-09 0
## Proportion of Variance 5.040122e-18 0
## Cumulative Proportion 1.000000e+00 1
# The effects of the distortion can be clearly seen with the biplot
# Variability absorbed by Shots, Shots.on.goal, Fouls.made
biplot(pcaLaliga, cex = 0.75)
# The effects of the variables are more balanced
biplot(pcaLaligaStd, cex = 0.75)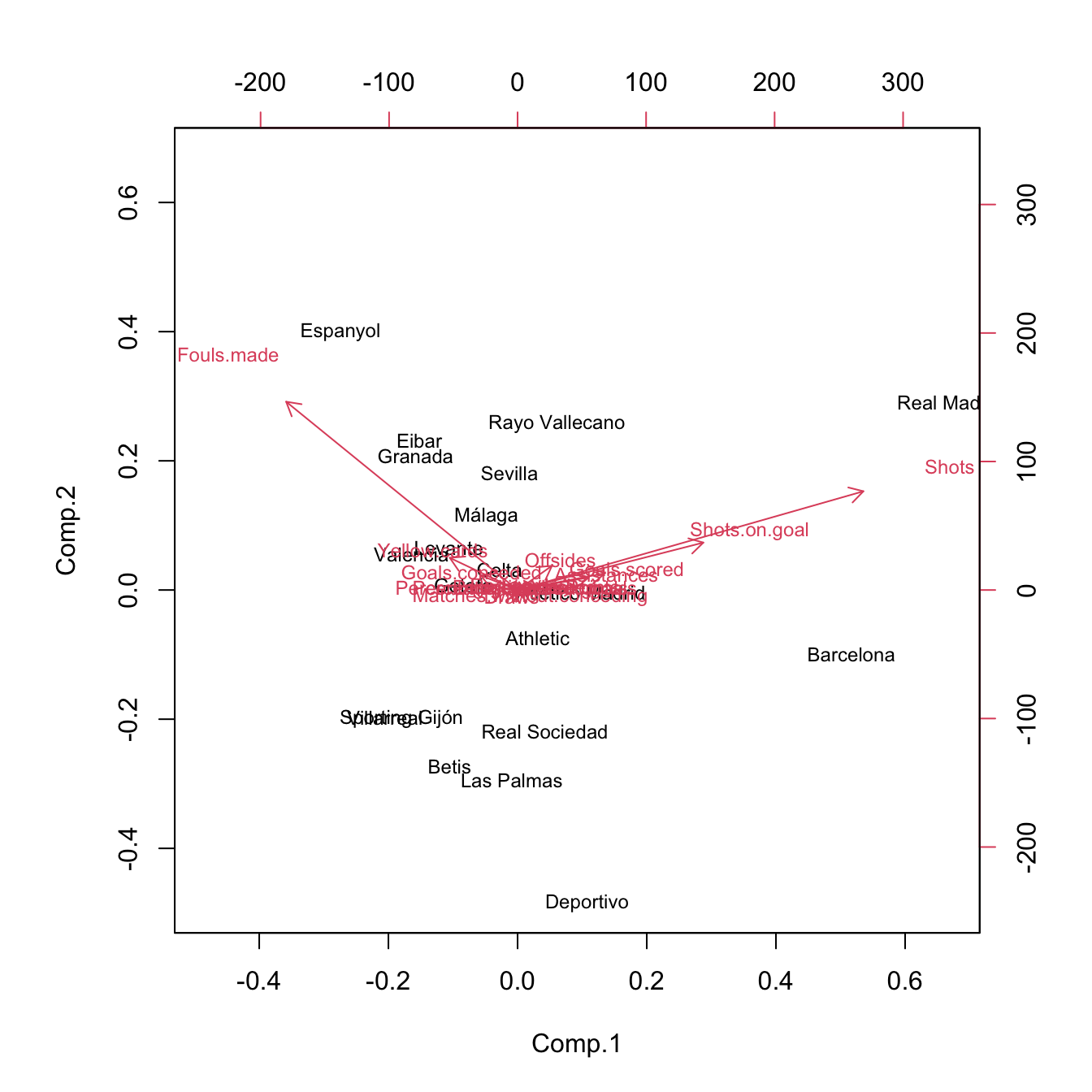
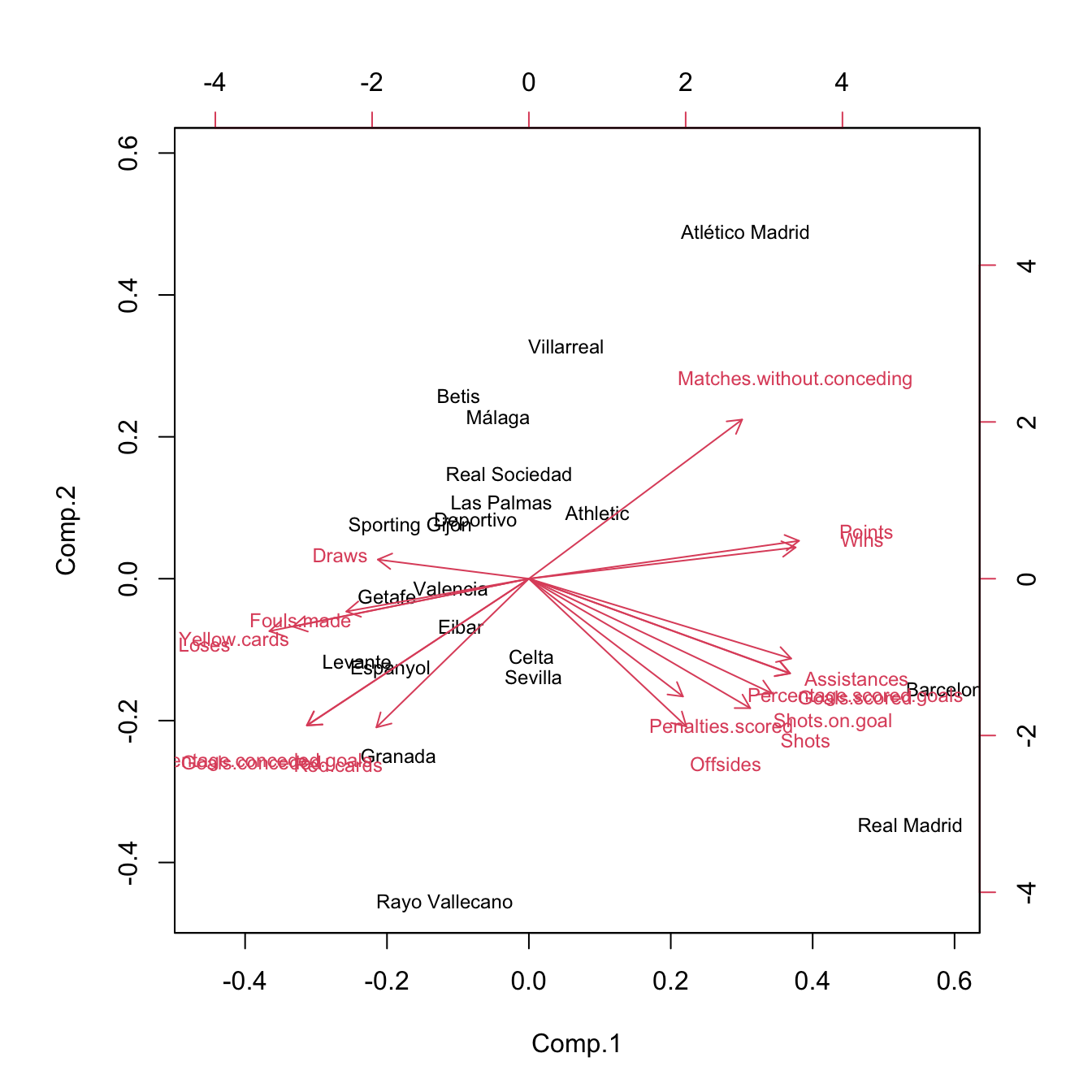
Figure 3.32: Biplots for laliga dataset, with unstandardized and standardized data, respectively.
The biplot88 provides a powerful and succinct way of displaying the relevant information contained in the first two principal components. It shows:
- The scores \(\{(\hat{\Gamma}_{i1},\hat{\Gamma}_{i2})\}_{i=1}^n\) by points (with optional text labels, depending if there are case names). These are the representations of the data in the first two principal components.
- The loadings \(\{(\hat{a}_{j1},\hat{a}_{j2})\}_{j=1}^p\) by the arrows, centered at \((0,0).\) These are the representations of the variables in the first two principal components.
Let’s examine the population89 arrow associated to the variable \(X_j.\) \(X_j\) is expressed in terms of \(\Gamma_1\) and \(\Gamma_2\) by the loadings \(a_{j1}\) and \(a_{j2}\;\)90:
\[\begin{align*} X_j=a_{j1}\Gamma_{1} + a_{j2}\Gamma_{2} + \cdots + a_{jp}\Gamma_{p}\approx a_{j1}\Gamma_{1} + a_{j2}\Gamma_{2}. \end{align*}\]
The weights \(a_{j1}\) and \(a_{j2}\) have the same sign as \(\mathrm{Cor}(X_j,\Gamma_{1})\) and \(\mathrm{Cor}(X_j,\Gamma_{2}),\) respectively. The arrow associated to \(X_j\) is given by the segment joining \((0,0)\) and \((a_{j1},a_{j2}).\) Therefore:
- If the arrow points right (\(a_{j1}>0\)), there is positive correlation between \(X_j\) and \(\Gamma_1.\) Analogous if the arrow points left.
- If the arrow is approximately vertical (\(a_{j1}\approx0\)), there is uncorrelation between \(X_j\) and \(\Gamma_1.\)
Analogously:
- If the arrow points up (\(a_{j2}>0\)), there is positive correlation between \(X_j\) and \(\Gamma_2.\) Analogous if the arrow points down.
- If the arrow is approximately horizontal (\(a_{j2}\approx0\)), there is uncorrelation between \(X_j\) and \(\Gamma_2.\)
In addition, the magnitude of the arrow informs us about the strength of the correlation of \(X_j\) with \((\Gamma_{1}, \Gamma_{2}).\)
The biplot also informs about the direct relation between variables, at sight of their expressions in \(\Gamma_1\) and \(\Gamma_2.\) The angle of the arrows of variable \(X_j\) and \(X_k\) gives an approximation to the correlation between them, \(\mathrm{Cor}(X_j,X_k)\):
- If \(\text{angle}\approx 0^\circ,\) the two variables are highly positively correlated.
- If \(\text{angle}\approx 90^\circ,\) they are approximately uncorrelated.
- If \(\text{angle}\approx 180^\circ,\) the two variables are highly negatively correlated.
The insights obtained from the approximate correlations between variables are as precise as the percentage of variance explained by the first two principal components.
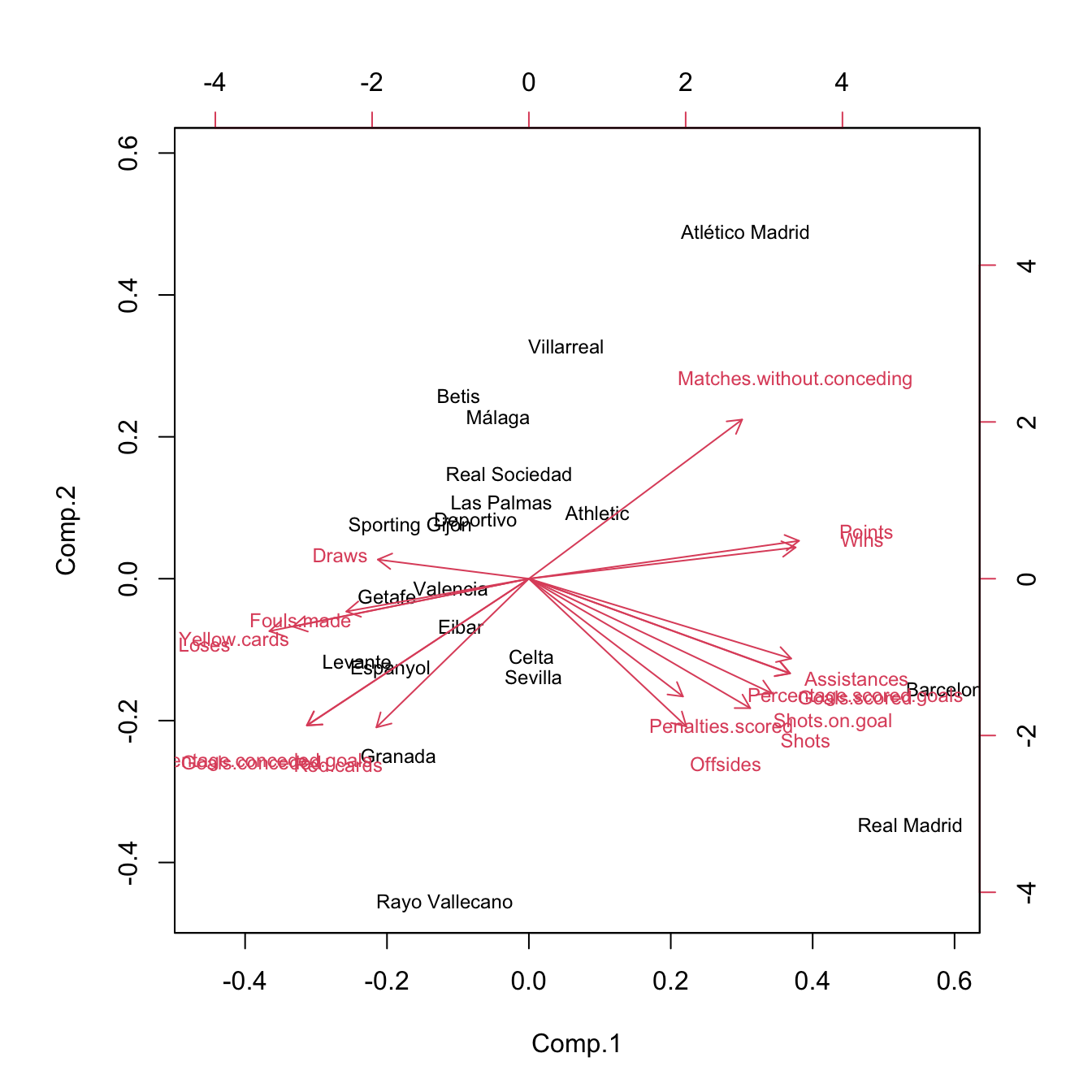
Figure 3.33: Biplot for laliga dataset. The interpretations of the first two principal components are driven by the signs of the variables inside them (directions of the arrows) and by the strength of their correlations with the variables (length of the arrows). The scores of the data serve also to cluster similar observations according to their proximity in the biplot.
Some interesting insights in La Liga 2015/2016 biplot, obtained from the previous remarks, are:91
- The first component can be regarded as the performance of a team during the season. It is positively correlated with
Wins,Points, etc. and negatively correlated withDraws,Loses,Yellow.cards, etc. The best performing teams are not surprising: Barcelona, Real Madrid, and Atlético Madrid. On the other hand, among the worst-performing teams are Levante, Getafe, and Granada. - The second component can be seen as the efficiency of a team (obtaining points with little participation in the game). Using this interpretation we can see that Atlético Madrid and Villareal were the most efficient teams and that Rayo Vallecano and Real Madrid were the most inefficient.
Offsidesis approximately uncorrelated withRed.cardsandMatches.without.conceding.
A 3D representation of the biplot can be computed through:
pca3d::pca3d(pcaLaligaStd, show.labels = TRUE, biplot = TRUE)
## [1] 0.1747106 0.0678678 0.0799681
rgl::rglwidget()
Finally, the biplot function allows to construct the biplot using two arbitrary principal components using the choices argument. Keep in mind than these pseudo-biplots will explain a lower proportion of variance than the default choices = 1:2:
biplot(pcaLaligaStd, choices = c(1, 3)) # 0.7138 proportion of variance
biplot(pcaLaligaStd, choices = c(2, 3)) # 0.2180 proportion of variance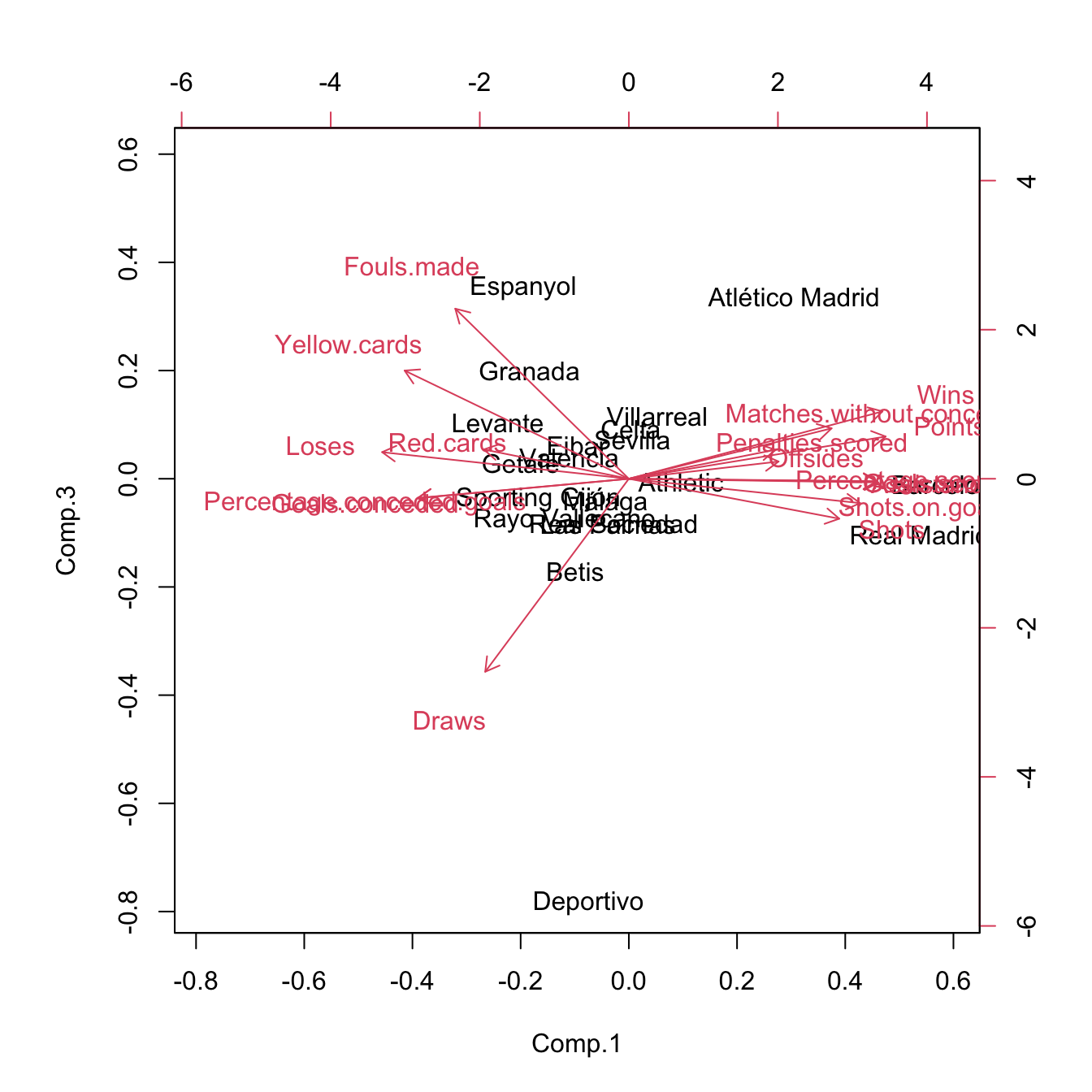
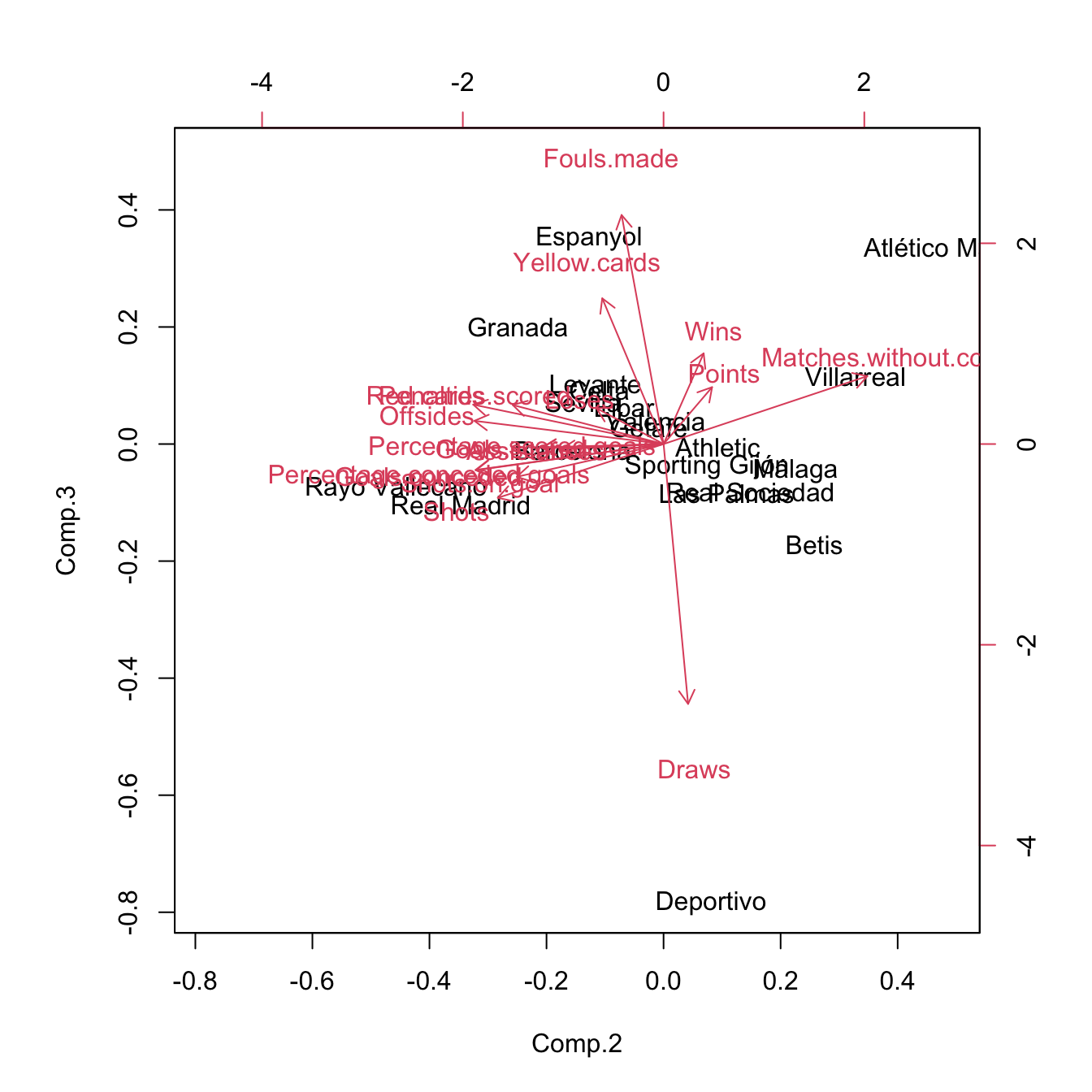
Figure 3.34: Pseudo-biplots for laliga dataset based on principal components 1 and 3, and principal components 2 and 3, respectively.
3.6.2 Principal components regression
The key idea behind Principal Components Regression (PCR) is to regress the response \(Y\) in a set of principal components \(\Gamma_1,\ldots,\Gamma_\ell\) obtained from the predictors \(X_1,\ldots,X_p,\) where \(\ell<p.\) The motivation is that often a small number of principal components is enough to explain most of the variability of the predictors and consequently their relationship with \(Y.\)92 Therefore, we look for fitting the linear model93
\[\begin{align} Y=\alpha_0+\alpha_1\Gamma_1+\cdots+\alpha_\ell\Gamma_\ell+\varepsilon.\tag{3.13} \end{align}\]
The main advantages of PCR are two:
- Multicollinearity is avoided by design: \(\Gamma_1,\ldots,\Gamma_\ell\) are uncorrelated between them.
- There are less coefficients to estimate (\(\ell\) instead of \(p\)), hence the accuracy of the estimation increases.
However, keep in mind that PCR affects the linear model in two fundamental ways:
- Interpretation of the coefficients is not directly related with the predictors, but with the principal components. Hence, the interpretability of a given coefficient in the regression model is tied to the interpretability of the associated principal component.
- Prediction needs an extra step, since it is required to obtain the scores of the new observations of the predictors.
The first point is elaborated next. The PCR model (3.13) can be seen as a linear model expressed in terms of the original predictors. To make this point clearer, let’s re-express (3.13) as
\[\begin{align} Y=\alpha_0+\boldsymbol{\Gamma}_{1:\ell}'\boldsymbol{\alpha}_{1:\ell}+\varepsilon,\tag{3.14} \end{align}\]
where the subindex \(1:\ell\) denotes the inclusion of the vector entries from \(1\) to \(\ell.\) Now, we can express the PCR problem (3.14) in terms of the original predictors:94
\[\begin{align*} Y&=\alpha_0+(\mathbf{A}_{1:\ell}'(\mathbf{X}-\boldsymbol{\mu}))'\boldsymbol{\alpha}_{1:\ell}+\varepsilon\\ &=(\alpha_0-\boldsymbol{\mu}'\mathbf{A}_{1:\ell}\boldsymbol{\alpha}_{1:\ell})+\mathbf{X}'\mathbf{A}_{1:\ell}\boldsymbol{\alpha}_{1:\ell}+\varepsilon\\ &=\gamma_0+\mathbf{X}'\boldsymbol{\gamma}_{1:p}+\varepsilon, \end{align*}\]
where \(\mathbf{A}_{1:\ell}\) represents the \(\mathbf{A}\) matrix with only its first \(\ell\) columns and
\[\begin{align} \gamma_0:=\alpha_0-\boldsymbol{\mu}'\mathbf{A}_{1:\ell}\boldsymbol{\alpha}_{1:\ell},\quad \boldsymbol{\gamma}_{1:p}:=\mathbf{A}_{1:\ell}\boldsymbol{\alpha}_{1:\ell}.\tag{3.15} \end{align}\]
In other words, the coefficients \(\boldsymbol{\alpha}_{1:\ell}\) of the PCR done with \(\ell\) principal components in (3.13) translate into the coefficients (3.15) of the linear model based on the \(p\) original predictors:
\[\begin{align*} Y=\gamma_0+\gamma_1X_1+\cdots+\gamma_pX_p+\varepsilon. \end{align*}\]
In the sample case, we have that
\[\begin{align} \hat{\gamma}_0=\hat\alpha_0-\bar{\mathbf{X}}'\hat{\mathbf{A}}_{1:\ell}\hat{\boldsymbol{\alpha}}_{1:\ell},\quad \hat{\boldsymbol{\gamma}}_{1:p}=\hat{\mathbf{A}}_{1:\ell}\hat{\boldsymbol{\alpha}}_{1:\ell}.\tag{3.16} \end{align}\]
Notice that \(\hat{\boldsymbol{\gamma}}\) is not the least squares estimator performed with the original predictors, that we use to denote by \(\hat{\boldsymbol{\beta}}.\) \(\hat{\boldsymbol{\gamma}}\) contains the coefficients of the PCR that are associated to the original predictors. Consequently, \(\hat{\boldsymbol{\gamma}}\) is useful for the interpretation of the linear model produced by PCR, as it can be interpreted in the same way \(\hat{\boldsymbol{\beta}}\) was.95
Finally, remember that the usefulness of PCR relies on how well we are able to reduce the dimensionality96 of the predictors and the veracity of the assumption that the \(\ell\) principal components are related with \(Y.\)
Keep in mind that PCR considers the PCA done in the set of
predictors, this is, we exclude the response for obvious
reasons (a perfect and useless fit). It is important to remove the
response from the call to princomp if we want to use the
output in lm.
We see now two approaches for performing PCR, which we illustrate with the laliga dataset. The common objective is to predict Points using the remaining variables (excluding those directly related: Wins, Draws, Loses, and Matches.without.conceding) in order to quantify, explain, and predict the final points of a team from its performance.
The first approach combines the use of the princomp and lm functions. Its strong points are that is both able to predict and explain, and is linked with techniques we have employed so far. The weak point is that it requires extra coding.
# A linear model is problematic
mod <- lm(Points ~ . - Wins - Draws - Loses - Matches.without.conceding,
data = laliga)
summary(mod) # Lots of non-significant predictors
##
## Call:
## lm(formula = Points ~ . - Wins - Draws - Loses - Matches.without.conceding,
## data = laliga)
##
## Residuals:
## Min 1Q Median 3Q Max
## -5.2117 -1.4766 0.0544 1.9515 4.1422
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 77.11798 26.12915 2.951 0.0214 *
## Goals.scored -28.21714 17.44577 -1.617 0.1498
## Goals.conceded -24.23628 15.45595 -1.568 0.1608
## Percentage.scored.goals 1066.98731 655.69726 1.627 0.1477
## Percentage.conceded.goals 896.94781 584.97833 1.533 0.1691
## Shots -0.10246 0.07754 -1.321 0.2279
## Shots.on.goal 0.02024 0.13656 0.148 0.8863
## Penalties.scored -0.81018 0.77600 -1.044 0.3312
## Assistances 1.41971 0.44103 3.219 0.0147 *
## Fouls.made -0.04438 0.04267 -1.040 0.3328
## Yellow.cards 0.27850 0.16814 1.656 0.1416
## Red.cards 0.68663 1.44229 0.476 0.6485
## Offsides -0.00486 0.14854 -0.033 0.9748
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 4.274 on 7 degrees of freedom
## Multiple R-squared: 0.9795, Adjusted R-squared: 0.9443
## F-statistic: 27.83 on 12 and 7 DF, p-value: 9.784e-05
# We try to clean the model
modBIC <- MASS::stepAIC(mod, k = log(nrow(laliga)), trace = 0)
summary(modBIC) # Better, but still unsatisfactory
##
## Call:
## lm(formula = Points ~ Goals.scored + Goals.conceded + Percentage.scored.goals +
## Percentage.conceded.goals + Shots + Assistances + Yellow.cards,
## data = laliga)
##
## Residuals:
## Min 1Q Median 3Q Max
## -5.4830 -1.4505 0.9008 1.1813 5.8662
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 62.91373 10.73528 5.860 7.71e-05 ***
## Goals.scored -23.90903 11.22573 -2.130 0.05457 .
## Goals.conceded -12.16610 8.11352 -1.499 0.15959
## Percentage.scored.goals 894.56861 421.45891 2.123 0.05528 .
## Percentage.conceded.goals 440.76333 307.38671 1.434 0.17714
## Shots -0.05752 0.02713 -2.120 0.05549 .
## Assistances 1.42267 0.28462 4.999 0.00031 ***
## Yellow.cards 0.11313 0.07868 1.438 0.17603
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 3.743 on 12 degrees of freedom
## Multiple R-squared: 0.973, Adjusted R-squared: 0.9572
## F-statistic: 61.77 on 7 and 12 DF, p-value: 1.823e-08
# Also, huge multicollinearity
car::vif(modBIC)
## Goals.scored Goals.conceded Percentage.scored.goals Percentage.conceded.goals
## 77998.044760 22299.952547 76320.612579 22322.307151
## Shots Assistances Yellow.cards
## 6.505748 32.505831 3.297224
# A quick way of removing columns without knowing its position
laligaRed <- subset(laliga, select = -c(Points, Wins, Draws, Loses,
Matches.without.conceding))
# PCA without Points, Wins, Draws, Loses, and Matches.without.conceding
pcaLaligaRed <- princomp(x = laligaRed, cor = TRUE, fix_sign = TRUE)
summary(pcaLaligaRed) # l = 3 gives 86% of variance explained
## Importance of components:
## Comp.1 Comp.2 Comp.3 Comp.4 Comp.5 Comp.6 Comp.7 Comp.8
## Standard deviation 2.7437329 1.4026745 0.91510249 0.8577839 0.65747209 0.5310954 0.332556029 0.263170555
## Proportion of Variance 0.6273392 0.1639580 0.06978438 0.0613161 0.03602246 0.0235052 0.009216126 0.005771562
## Cumulative Proportion 0.6273392 0.7912972 0.86108155 0.9223977 0.95842012 0.9819253 0.991141438 0.996913000
## Comp.9 Comp.10 Comp.11 Comp.12
## Standard deviation 0.146091551 0.125252621 3.130311e-03 1.801036e-03
## Proportion of Variance 0.001778562 0.001307352 8.165704e-07 2.703107e-07
## Cumulative Proportion 0.998691562 0.999998913 9.999997e-01 1.000000e+00
# Interpretation of PC1 and PC2
biplot(pcaLaligaRed)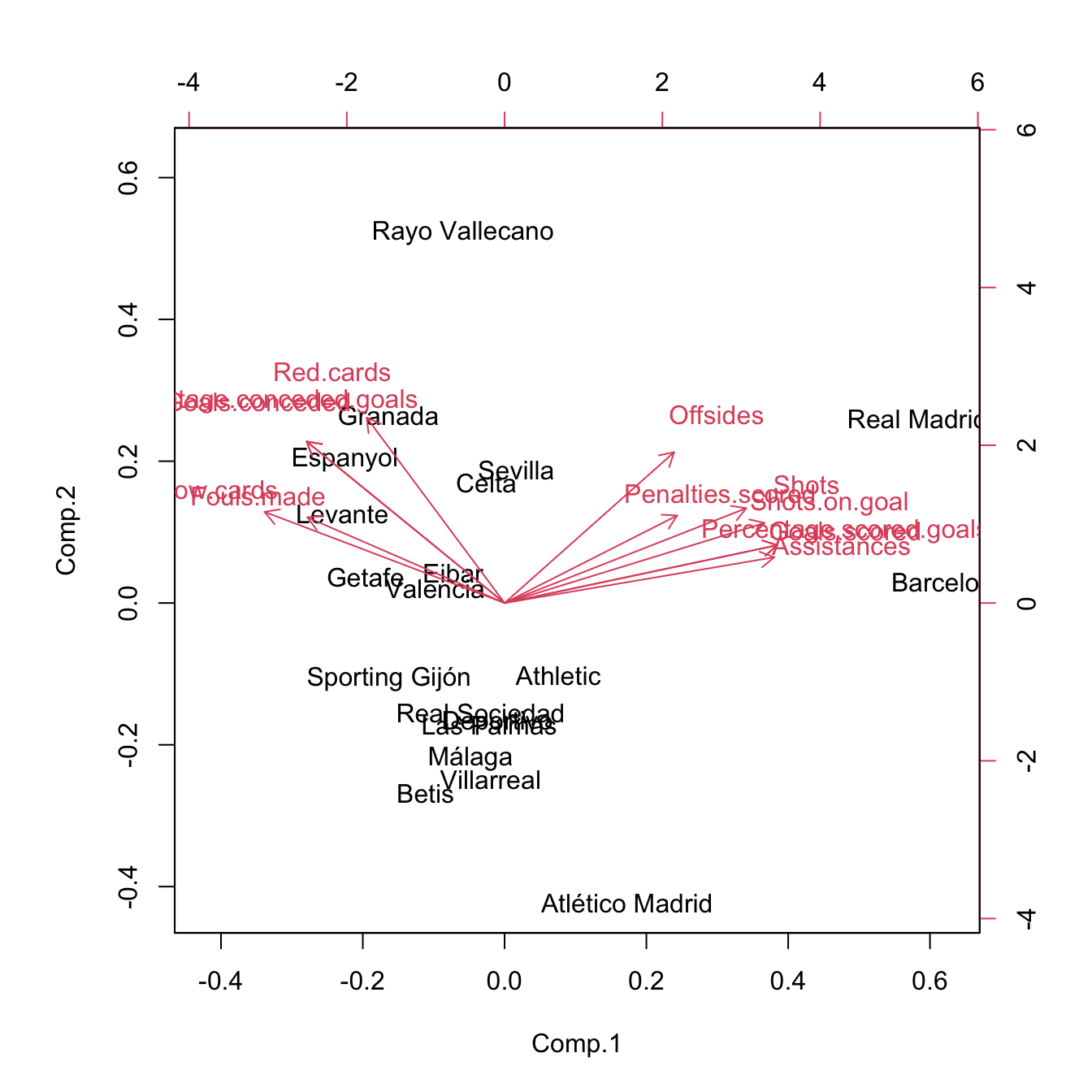
# PC1: attack performance of the team
# Create a new dataset with the response + principal components
laligaPCA <- data.frame("Points" = laliga$Points, pcaLaligaRed$scores)
# Regression on all the principal components
modPCA <- lm(Points ~ ., data = laligaPCA)
summary(modPCA) # Predictors clearly significative -- same R^2 as mod
##
## Call:
## lm(formula = Points ~ ., data = laligaPCA)
##
## Residuals:
## Min 1Q Median 3Q Max
## -5.2117 -1.4766 0.0544 1.9515 4.1422
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 52.4000 0.9557 54.831 1.76e-10 ***
## Comp.1 5.7690 0.3483 16.563 7.14e-07 ***
## Comp.2 -2.4376 0.6813 -3.578 0.0090 **
## Comp.3 3.4222 1.0443 3.277 0.0135 *
## Comp.4 -3.6079 1.1141 -3.238 0.0143 *
## Comp.5 1.9713 1.4535 1.356 0.2172
## Comp.6 5.7067 1.7994 3.171 0.0157 *
## Comp.7 -3.4169 2.8737 -1.189 0.2732
## Comp.8 9.0212 3.6313 2.484 0.0419 *
## Comp.9 -4.6455 6.5415 -0.710 0.5006
## Comp.10 -10.2087 7.6299 -1.338 0.2227
## Comp.11 222.0340 305.2920 0.727 0.4907
## Comp.12 -954.7650 530.6164 -1.799 0.1150
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 4.274 on 7 degrees of freedom
## Multiple R-squared: 0.9795, Adjusted R-squared: 0.9443
## F-statistic: 27.83 on 12 and 7 DF, p-value: 9.784e-05
car::vif(modPCA) # No problems at all
## Comp.1 Comp.2 Comp.3 Comp.4 Comp.5 Comp.6 Comp.7 Comp.8 Comp.9 Comp.10 Comp.11 Comp.12
## 1 1 1 1 1 1 1 1 1 1 1 1
# Using the first three components
modPCA3 <- lm(Points ~ Comp.1 + Comp.2 + Comp.3, data = laligaPCA)
summary(modPCA3)
##
## Call:
## lm(formula = Points ~ Comp.1 + Comp.2 + Comp.3, data = laligaPCA)
##
## Residuals:
## Min 1Q Median 3Q Max
## -8.178 -4.541 -1.401 3.501 16.093
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 52.4000 1.5672 33.435 3.11e-16 ***
## Comp.1 5.7690 0.5712 10.100 2.39e-08 ***
## Comp.2 -2.4376 1.1173 -2.182 0.0444 *
## Comp.3 3.4222 1.7126 1.998 0.0630 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 7.009 on 16 degrees of freedom
## Multiple R-squared: 0.8738, Adjusted R-squared: 0.8501
## F-statistic: 36.92 on 3 and 16 DF, p-value: 2.027e-07
# Coefficients associated to each original predictor (gamma)
alpha <- modPCA3$coefficients
gamma <- pcaLaligaRed$loadings[, 1:3] %*% alpha[-1] # Slopes
gamma <- c(alpha[1] - pcaLaligaRed$center %*% gamma, gamma) # Intercept
names(gamma) <- c("Intercept", rownames(pcaLaligaRed$loadings))
gamma
## Intercept Goals.scored Goals.conceded Percentage.scored.goals
## -44.2288551 1.7305124 -3.4048178 1.7416378
## Percentage.conceded.goals Shots Shots.on.goal Penalties.scored
## -3.3944235 0.2347716 0.8782162 2.6044699
## Assistances Fouls.made Yellow.cards Red.cards
## 1.4548813 -0.1171732 -1.7826488 -2.6423211
## Offsides
## 1.3755697
# We can overpenalize to have a simpler model -- also one single
# principal component does quite well
modPCABIC <- MASS::stepAIC(modPCA, k = 2 * log(nrow(laliga)), trace = 0)
summary(modPCABIC)
##
## Call:
## lm(formula = Points ~ Comp.1 + Comp.2 + Comp.3 + Comp.4 + Comp.6 +
## Comp.8, data = laligaPCA)
##
## Residuals:
## Min 1Q Median 3Q Max
## -6.6972 -2.6418 -0.3265 2.3535 8.4944
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 52.4000 1.0706 48.946 3.95e-16 ***
## Comp.1 5.7690 0.3902 14.785 1.65e-09 ***
## Comp.2 -2.4376 0.7632 -3.194 0.00705 **
## Comp.3 3.4222 1.1699 2.925 0.01182 *
## Comp.4 -3.6079 1.2481 -2.891 0.01263 *
## Comp.6 5.7067 2.0158 2.831 0.01416 *
## Comp.8 9.0212 4.0680 2.218 0.04502 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 4.788 on 13 degrees of freedom
## Multiple R-squared: 0.9521, Adjusted R-squared: 0.9301
## F-statistic: 43.11 on 6 and 13 DF, p-value: 7.696e-08
# Note that the order of the principal components does not correspond
# exactly to its importance in the regression!
# To perform prediction we need to compute first the scores associated to the
# new values of the predictors, conveniently preprocessed
# Predictions for FCB and RMA (although they are part of the training sample)
newPredictors <- laligaRed[1:2, ]
newPredictors <- scale(newPredictors, center = pcaLaligaRed$center,
scale = pcaLaligaRed$scale) # Centered and scaled
newScores <- t(apply(newPredictors, 1,
function(x) t(pcaLaligaRed$loadings) %*% x))
# We need a data frame for prediction
newScores <- data.frame("Comp" = newScores)
predict(modPCABIC, newdata = newScores, interval = "prediction")
## fit lwr upr
## Barcelona 93.64950 80.35115 106.9478
## Real Madrid 90.05622 77.11876 102.9937
# Reality
laliga[1:2, 1]
## [1] 91 90The second approach employs the function pls::pcr and is more direct, yet less connected with the techniques we have seen so far. It employs a model object that is different from the lm object and, as a consequence, functions like summary, BIC, MASS::stepAIC, or plot will not work properly. This implies that inference, model selection, and model diagnostics are not so straightforward. In exchange, pls::pcr allows for model fitting in an easier way and model selection through the use of cross-validation. In overall, this is a more pure predictive approach than predictive and explicative.
# Create a dataset without the problematic predictors and with the response
laligaRed2 <- subset(laliga, select = -c(Wins, Draws, Loses,
Matches.without.conceding))
# Simple call to pcr
library(pls)
modPcr <- pcr(Points ~ ., data = laligaRed2, scale = TRUE)
# Notice we do not need to create a data.frame with PCA, it is automatically
# done within pcr. We also have flexibility to remove predictors from the PCA
# scale = TRUE means that the predictors are scaled internally before computing
# PCA
# The summary of the model is different
summary(modPcr)
## Data: X dimension: 20 12
## Y dimension: 20 1
## Fit method: svdpc
## Number of components considered: 12
## TRAINING: % variance explained
## 1 comps 2 comps 3 comps 4 comps 5 comps 6 comps 7 comps 8 comps 9 comps 10 comps 11 comps 12 comps
## X 62.73 79.13 86.11 92.24 95.84 98.19 99.11 99.69 99.87 100.00 100 100.00
## Points 80.47 84.23 87.38 90.45 90.99 93.94 94.36 96.17 96.32 96.84 97 97.95
# First row: percentage of variance explained of the predictors
# Second row: percentage of variance explained of Y (the R^2)
# Note that we have the same R^2 for 3 and 12 components as in the previous
# approach
# Slots of information in the model -- most of them as 3-dim arrays with the
# third dimension indexing the number of components considered
names(modPcr)
## [1] "coefficients" "scores" "loadings" "Yloadings" "projection" "Xmeans" "Ymeans"
## [8] "fitted.values" "residuals" "Xvar" "Xtotvar" "fit.time" "ncomp" "method"
## [15] "center" "scale" "call" "terms" "model"
# The coefficients of the original predictors (gammas), not of the components!
modPcr$coefficients[, , 12]
## Goals.scored Goals.conceded Percentage.scored.goals Percentage.conceded.goals
## -602.85050765 -383.07010184 600.61255371 374.38729000
## Shots Shots.on.goal Penalties.scored Assistances
## -8.27239221 0.88174787 -2.14313238 24.42240486
## Fouls.made Yellow.cards Red.cards Offsides
## -2.96044265 5.51983512 1.20945331 -0.07231723
# pcr() computes up to ncomp (in this case, 12) linear models, each one
# considering one extra principal component. $coefficients returns in a
# 3-dim array the coefficients of all the linear models
# Prediction is simpler and can be done for different number of components
predict(modPcr, newdata = laligaRed2[1:2, ], ncomp = 12)
## , , 12 comps
##
## Points
## Barcelona 92.01244
## Real Madrid 91.38026
# Selecting the number of components to retain. All the components up to ncomp
# are selected, no further flexibility is possible
modPcr2 <- pcr(Points ~ ., data = laligaRed2, scale = TRUE, ncomp = 3)
summary(modPcr2)
## Data: X dimension: 20 12
## Y dimension: 20 1
## Fit method: svdpc
## Number of components considered: 3
## TRAINING: % variance explained
## 1 comps 2 comps 3 comps
## X 62.73 79.13 86.11
## Points 80.47 84.23 87.38
# Selecting the number of components to retain by Leave-One-Out
# cross-validation
modPcrCV1 <- pcr(Points ~ ., data = laligaRed2, scale = TRUE,
validation = "LOO")
summary(modPcrCV1)
## Data: X dimension: 20 12
## Y dimension: 20 1
## Fit method: svdpc
## Number of components considered: 12
##
## VALIDATION: RMSEP
## Cross-validated using 20 leave-one-out segments.
## (Intercept) 1 comps 2 comps 3 comps 4 comps 5 comps 6 comps 7 comps 8 comps 9 comps 10 comps 11 comps
## CV 18.57 8.505 8.390 8.588 7.571 7.688 6.743 7.09 6.224 6.603 7.547 8.375
## adjCV 18.57 8.476 8.356 8.525 7.513 7.663 6.655 7.03 6.152 6.531 7.430 8.236
## 12 comps
## CV 7.905
## adjCV 7.760
##
## TRAINING: % variance explained
## 1 comps 2 comps 3 comps 4 comps 5 comps 6 comps 7 comps 8 comps 9 comps 10 comps 11 comps 12 comps
## X 62.73 79.13 86.11 92.24 95.84 98.19 99.11 99.69 99.87 100.00 100 100.00
## Points 80.47 84.23 87.38 90.45 90.99 93.94 94.36 96.17 96.32 96.84 97 97.95
# View cross-validation Mean Squared Error in Prediction
validationplot(modPcrCV1, val.type = "MSEP") # l = 8 gives the minimum CV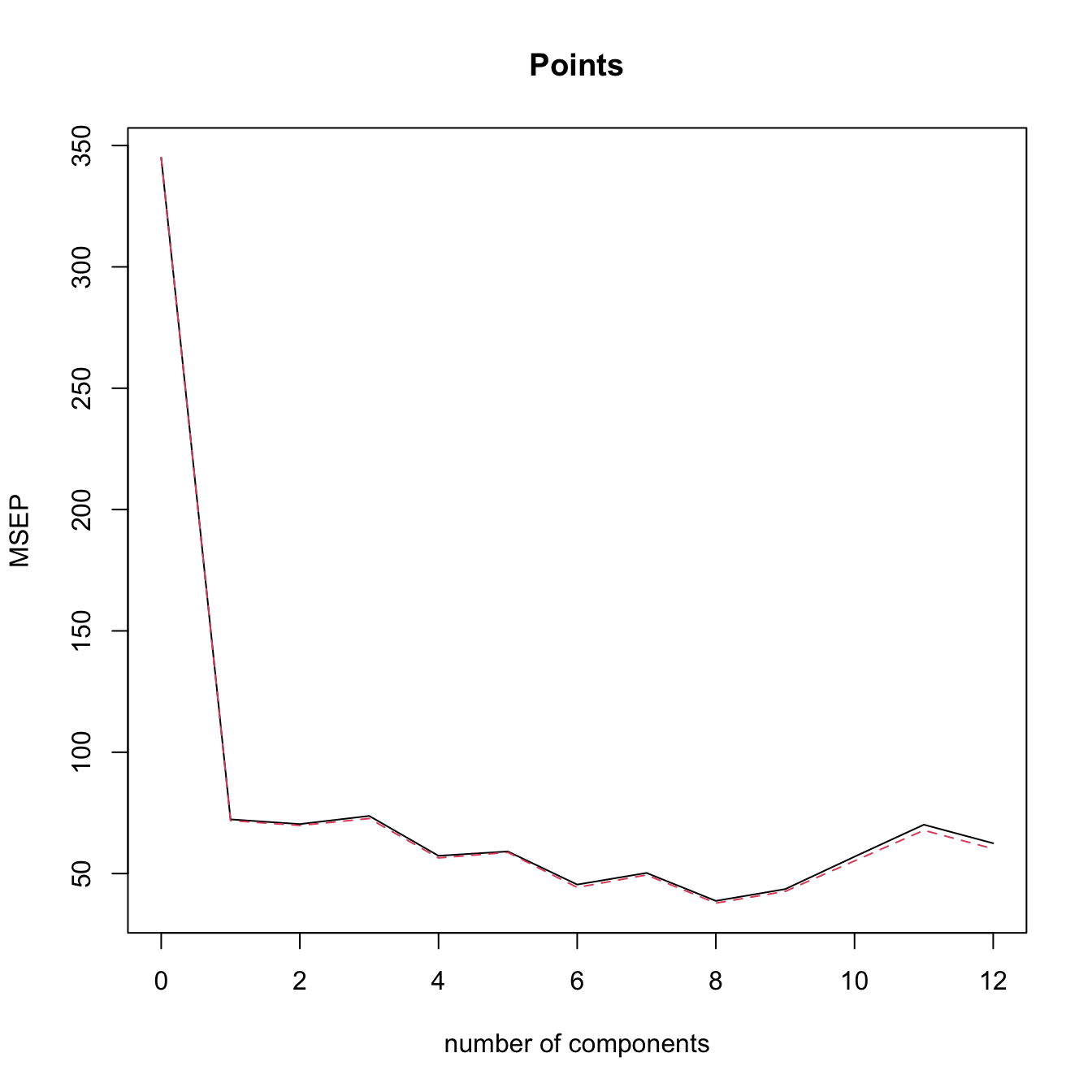
# The black is the CV loss, the dashed red line is the adjCV loss, a bias
# corrected version of the MSEP (not described in the notes)
# Selecting the number of components to retain by 10-fold Cross-Validation
# (k = 10 is the default, this can be changed with the argument segments)
modPcrCV10 <- pcr(Points ~ ., data = laligaRed2, scale = TRUE,
validation = "CV")
summary(modPcrCV10)
## Data: X dimension: 20 12
## Y dimension: 20 1
## Fit method: svdpc
## Number of components considered: 12
##
## VALIDATION: RMSEP
## Cross-validated using 10 random segments.
## (Intercept) 1 comps 2 comps 3 comps 4 comps 5 comps 6 comps 7 comps 8 comps 9 comps 10 comps 11 comps
## CV 18.57 8.520 8.392 8.911 7.187 7.124 6.045 6.340 5.728 6.073 7.676 9.300
## adjCV 18.57 8.464 8.331 8.768 7.092 7.043 5.904 6.252 5.608 5.953 7.423 8.968
## 12 comps
## CV 9.151
## adjCV 8.782
##
## TRAINING: % variance explained
## 1 comps 2 comps 3 comps 4 comps 5 comps 6 comps 7 comps 8 comps 9 comps 10 comps 11 comps 12 comps
## X 62.73 79.13 86.11 92.24 95.84 98.19 99.11 99.69 99.87 100.00 100 100.00
## Points 80.47 84.23 87.38 90.45 90.99 93.94 94.36 96.17 96.32 96.84 97 97.95
validationplot(modPcrCV10, val.type = "MSEP") # l = 8 gives the minimum CV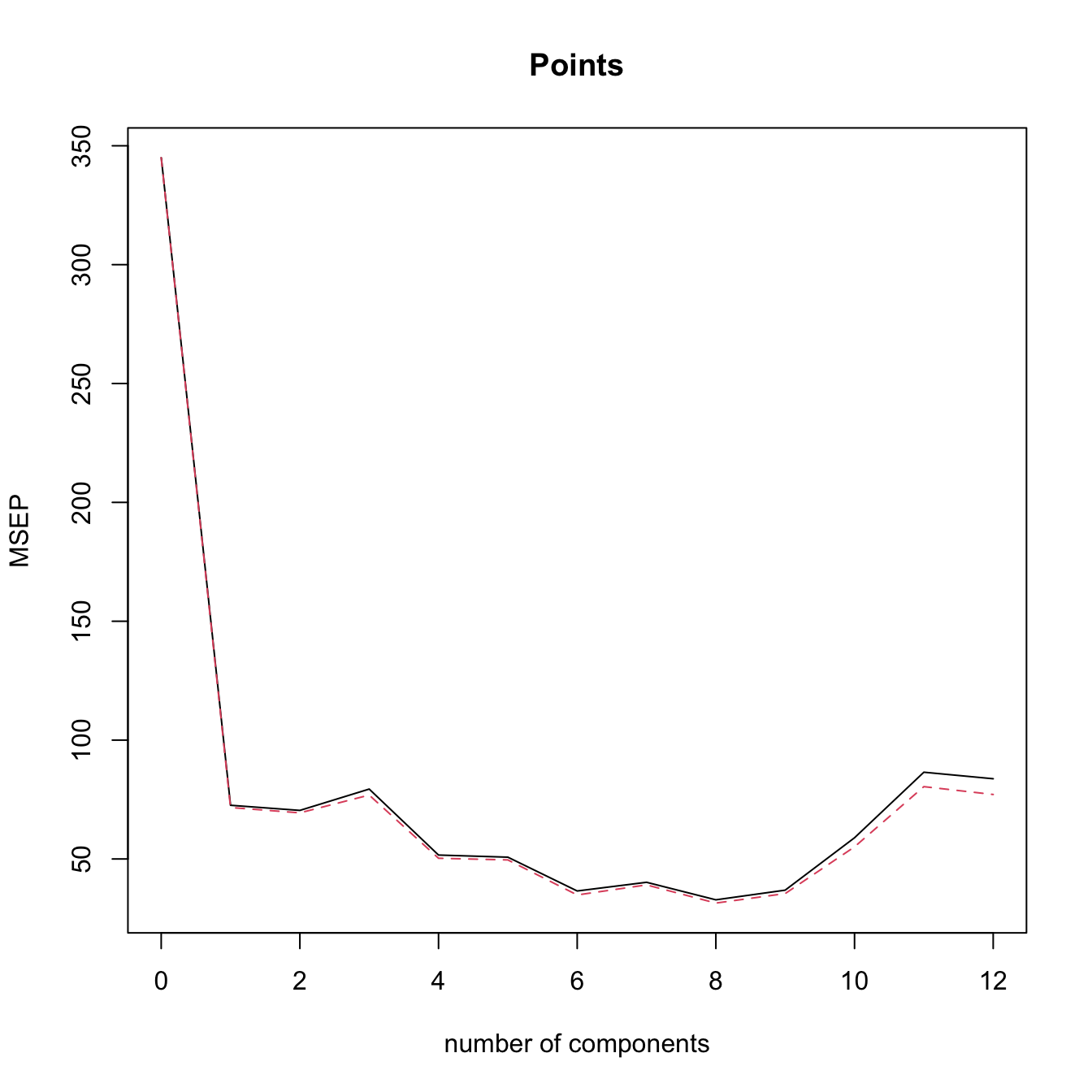
pcr() does an internal scaling of the predictors by their quasi-standard deviations. This means that each variable is divided by \(\frac{1}{\sqrt{n-1}},\) when in princomp a scaling of \(\frac{1}{\sqrt{n}}\) is applied (the standard deviations are employed). This results in a minor discrepancy in the scores object of both methods that is easily patchable. The scores of princomp() are the ones of pcr() multiplied by \(\sqrt{\frac{n}{n-1}}.\) This problem is inherited to the coefficients, which assume scores divided by \(\frac{1}{\sqrt{n-1}}.\) Therefore, the \(\hat{\boldsymbol{\gamma}}\) coefficients described in (3.16) are obtained by dividing the coefficients of pcr() by \(\sqrt{\frac{n}{n-1}}.\)
The next chunk of code illustrates the previous warning.
# Equality of loadings from princomp() and pcr()
max(abs(abs(pcaLaligaRed$loadings[, 1:3]) - abs(modPcr$loadings[, 1:3])))
## [1] 2.664535e-15
# Equality of scores from princomp() and pcr() (with the same standardization)
max(abs(abs(pcaLaligaRed$scores[, 1:3]) -
abs(modPcr$scores[, 1:3] * sqrt(n / (n - 1)))))
## [1] 9.298118e-15
# Equality of the gamma coefficients obtained previously for 3 PCA
# (with the same standardization)
modPcr$coefficients[, , 3] / sqrt(n / (n - 1))
## Goals.scored Goals.conceded Percentage.scored.goals Percentage.conceded.goals
## 1.7305124 -3.4048178 1.7416378 -3.3944235
## Shots Shots.on.goal Penalties.scored Assistances
## 0.2347716 0.8782162 2.6044699 1.4548813
## Fouls.made Yellow.cards Red.cards Offsides
## -0.1171732 -1.7826488 -2.6423211 1.3755697
gamma[-1]
## Goals.scored Goals.conceded Percentage.scored.goals Percentage.conceded.goals
## 1.7305124 -3.4048178 1.7416378 -3.3944235
## Shots Shots.on.goal Penalties.scored Assistances
## 0.2347716 0.8782162 2.6044699 1.4548813
## Fouls.made Yellow.cards Red.cards Offsides
## -0.1171732 -1.7826488 -2.6423211 1.3755697
# Coefficients associated to the principal components -- same as in modPCA3
lm(Points ~ ., data = data.frame("Points" = laliga$Points,
modPcr$scores[, 1:3] * sqrt(n / (n - 1))))
##
## Call:
## lm(formula = Points ~ ., data = data.frame(Points = laliga$Points,
## modPcr$scores[, 1:3] * sqrt(n/(n - 1))))
##
## Coefficients:
## (Intercept) Comp.1 Comp.2 Comp.3
## 52.400 -5.769 2.438 -3.422
modPCA3
##
## Call:
## lm(formula = Points ~ Comp.1 + Comp.2 + Comp.3, data = laligaPCA)
##
## Coefficients:
## (Intercept) Comp.1 Comp.2 Comp.3
## 52.400 5.769 -2.438 3.422
# Of course, flipping of signs is always possible with PCAThe selection of \(\ell\) by cross-validation attempts to minimize the Mean Squared Error in Prediction (MSEP) or, equivalently, the Root MSEP (RMSEP) of the model.97 This is a Swiss army knife method valid for the selection of any tuning parameter \(\lambda\) that affects the form of the estimate \(\hat{m}_\lambda\) of the regression function \(m\) (remember (1.1)). Given the sample \(\{(\mathbf{X}_i,Y_i)\}_{i=1}^n,\) leave-one-out cross-validation considers the tuning parameter
\[\begin{align} \hat{\lambda}_\text{CV}:=\arg\min_{\lambda\geq0} \sum_{i=1}^n (Y_i-\hat{m}_{\lambda,-i}(\mathbf{X}_i))^2,\tag{3.17} \end{align}\]
where \(\hat{m}_{\lambda,-i}\) represents the fit of the model \(\hat{m}_\lambda\) without the \(i\)-th observation \((\mathbf{X}_i,Y_i).\)
A less computationally expensive variation on leave-one-out cross-validation is \(k\)-fold cross-validation, which partitions the data into \(k\) folds \(F_1,\ldots, F_k\) of approximately equal size, trains the model \(\hat{m}_\lambda\) in the aggregation of \(k-1\) folds, and evaluates its MSEP in the remaining fold:
\[\begin{align} \hat{\lambda}_{k\text{-CV}}:=\arg\min_{\lambda\geq0} \sum_{j=1}^k \sum_{i\in F_j} (Y_i-\hat{m}_{\lambda,-F_{j}}(\mathbf{X}_i))^2,\tag{3.18} \end{align}\]
where \(\hat{m}_{\lambda,-F_{j}}\) represents the fit of the model \(\hat{m}_\lambda\) excluding the data from the \(j\)-th fold \(F_{j}.\) Recall that \(k\)-fold cross-validation is more general than leave-one-out cross-validation, since the latter is a particular case of the former with \(k=n.\)
summary() function on the output of lm(). The coefficients \(\boldsymbol{\alpha}_{0:\ell}\) are the ones estimated by least squares when considering the scores of the \(\ell\) principal components as the predictors. This transformation of the predictors does not affect the validity of the inferential results of Section 2.4 (derived conditionally on the predictors). But recall that this inference is not about \(\boldsymbol{\gamma}.\)
Inference in PCR is based on the assumptions of the linear model being satisfied for the considered principal components. The evaluation of the assumptions can be done using the exact same tools described in Section 3.5. However, keep in mind that PCA is a linear transformation of the data. Therefore:
- If the linearity assumption fails for the predictors \(X_1,\ldots,X_p,\) then it will also likely fail for \(\Gamma_1,\ldots,\Gamma_\ell,\) since the transformation will not introduce nonlinearities able to capture the nonlinear effects.
- Similarly, if the homoscedasticity, normality, or independence assumptions fail for \(X_1,\ldots,X_p,\) then they will also likely fail for \(\Gamma_1,\ldots,\Gamma_\ell.\)
Exceptions to the previous common implications are possible, and may involve the association of one or several problematic predictors (e.g., have nonlinear effects on the response) to the principal components that are excluded from the model. Up to which extent the failure of the assumptions in the original predictors can be mitigated by PCR depends on each application.
3.6.3 Partial least squares regression
PCR works by replacing the predictors \(X_1,\ldots,X_p\) by a set of principal components \(\Gamma_1,\ldots,\Gamma_\ell,\) under the hope that these directions, that explain most of the variability of the predictors, are also the best directions for predicting the response \(Y.\) While this is a reasonable belief, it is not a guaranteed fact. Partial Least Squares Regression (PLSR) precisely tackles this point with the idea of regressing the response \(Y\) on a set of new variables \(P_1,\ldots,P_\ell\) that are constructed with the objective of predicting \(Y\) from \(X_1,\ldots,X_p\) in the best linear way.
As with PCA, the idea is to find linear combinations of the predictors \(X_1,\ldots,X_p,\) that is, to have:
\[\begin{align} P_1:=\sum_{j=1}^pa_{1j}X_j,\,\ldots,\,P_p:=\sum_{j=1}^pa_{pj}X_j.\tag{3.19} \end{align}\]
These new predictors are called PLS components.98
The question is how to choose the coefficients \(a_{kj},\) \(j,k=1,\ldots,p.\) PLSR does it by placing the most weight in the predictors that are most strongly correlated with \(Y\) and in such a way that the resulting \(P_1,\ldots,P_p\) are uncorrelated between them. After standardizing the variables, for \(P_1\) this is achieved by setting \(a_{1j}\) equal to the theoretical slope coefficient of regressing \(Y\) into \(X_j,\) that is:99
\[\begin{align*} a_{1j}:=\frac{\mathrm{Cov}[X_j,Y]}{\mathbb{V}\mathrm{ar}[X_j]}, \end{align*}\]
where \(a_{1j}\) stems from \(Y=a_{0}+a_{1j}X_j+\varepsilon.\)
The second partial least squares direction, \(P_2,\) is computed in a similar way, but once the linear effects of \(P_1\) on \(X_1,\ldots,X_p\) are removed. This is achieved by:
- Regressing each of \(X_1,\ldots,X_p\) on \(P_1.\) That is, fit the \(p\) simple linear models
\[\begin{align*} X_j=\alpha_0+\alpha_{1j}P_1+\varepsilon_j,\quad j=1,\ldots,p. \end{align*}\]
Regress \(Y\) on each of the \(p\) random errors \(\varepsilon_{1},\ldots,\varepsilon_{p}\;\)100 from the above regressions, yielding
\[\begin{align*} a_{2j}:=\frac{\mathrm{Cov}[\varepsilon_j,Y]}{\mathbb{V}\mathrm{ar}[\varepsilon_j]}, \end{align*}\]
where \(a_{2j}\) stems from \(Y=a_{0}+a_{2j}\varepsilon_j+\varepsilon.\)
The coefficients for \(P_j,\) \(j>2,\) are computed101 by iterating the former process. Once \(P_1,\ldots,P_\ell\) are obtained, then PLSR proceeds as PCR and fits the model
\[\begin{align*} Y=\beta_0+\beta_1P_1+\cdots+\beta_\ell P_\ell+\varepsilon. \end{align*}\]
The implementation of PLSR can be done by the function pls::plsr, which has an analogous syntax to pls::pcr.
# Simple call to plsr -- very similar to pcr
modPlsr <- plsr(Points ~ ., data = laligaRed2, scale = TRUE)
# The summary of the model
summary(modPlsr)
## Data: X dimension: 20 12
## Y dimension: 20 1
## Fit method: kernelpls
## Number of components considered: 12
## TRAINING: % variance explained
## 1 comps 2 comps 3 comps 4 comps 5 comps 6 comps 7 comps 8 comps 9 comps 10 comps 11 comps 12 comps
## X 62.53 76.07 84.94 88.29 92.72 95.61 98.72 99.41 99.83 100.00 100.00 100.00
## Points 83.76 91.06 93.93 95.43 95.94 96.44 96.55 96.73 96.84 96.84 97.69 97.95
# First row: percentage of variance explained of the predictors
# Second row: percentage of variance explained of Y (the R^2)
# Note we have the same R^2 for 12 components as in the linear model
# Slots of information in the model
names(modPlsr)
## [1] "coefficients" "scores" "loadings" "loading.weights" "Yscores" "Yloadings"
## [7] "projection" "Xmeans" "Ymeans" "fitted.values" "residuals" "Xvar"
## [13] "Xtotvar" "fit.time" "ncomp" "method" "center" "scale"
## [19] "call" "terms" "model"
# PLS scores
head(modPlsr$scores)
## Comp 1 Comp 2 Comp 3 Comp 4 Comp 5 Comp 6 Comp 7 Comp 8 Comp 9
## Barcelona 7.36407868 -0.6592285 -0.4262822 -0.1357589 0.8668796 -0.52701770 -0.1547704 -0.3104798 -0.15841110
## Real Madrid 6.70659816 -1.2848902 0.7191143 0.5679398 -0.5998745 0.30663290 0.5382173 0.2110337 0.15242836
## Atlético Madrid 2.45577740 2.6469837 0.4573647 0.6179274 -0.4307465 0.13368949 0.2951566 -0.1597672 -0.15232469
## Villarreal 0.06913485 2.0275895 0.6548014 -0.2358589 0.8350426 0.61305618 -0.6684330 0.0556628 0.07919102
## Athletic 0.96513341 0.3860060 -0.2718465 -0.1580349 -0.2649106 0.01822319 -0.1899969 0.3846156 -0.28926416
## Celta -0.39588401 -0.5296417 0.6148093 0.1494496 0.5817902 0.69106828 0.1586472 0.2396975 0.41575546
## Comp 10 Comp 11 Comp 12
## Barcelona -0.18952142 -0.0001005279 0.0011993985
## Real Madrid 0.15826286 -0.0011752690 -0.0030013550
## Atlético Madrid 0.05857664 0.0028149603 0.0012838990
## Villarreal -0.05343433 0.0012705829 -0.0001652195
## Athletic 0.03702926 -0.0008077855 0.0052995290
## Celta -0.03488514 0.0003208900 0.0022585552
# Also uncorrelated
head(cov(modPlsr$scores))
## Comp 1 Comp 2 Comp 3 Comp 4 Comp 5 Comp 6 Comp 7 Comp 8
## Comp 1 7.393810e+00 3.973430e-16 -2.337312e-16 -7.304099e-18 9.115515e-16 1.149665e-15 3.272236e-16 7.011935e-17
## Comp 2 3.973430e-16 1.267859e+00 -2.980072e-16 -1.840633e-16 -2.103580e-16 -1.920978e-16 -2.337312e-17 -9.495329e-17
## Comp 3 -2.337312e-16 -2.980072e-16 9.021126e-01 -8.399714e-18 1.256305e-16 1.003401e-16 -2.921640e-18 5.843279e-18
## Comp 4 -7.304099e-18 -1.840633e-16 -8.399714e-18 3.130984e-01 -5.660677e-17 -4.660928e-17 -3.652049e-19 2.282531e-17
## Comp 5 9.115515e-16 -2.103580e-16 1.256305e-16 -5.660677e-17 2.586132e-01 9.897054e-17 8.180591e-17 3.432926e-17
## Comp 6 1.149665e-15 -1.920978e-16 1.003401e-16 -4.660928e-17 9.897054e-17 2.792408e-01 1.132135e-17 1.314738e-17
## Comp 9 Comp 10 Comp 11 Comp 12
## Comp 1 -4.090295e-17 -2.571043e-16 -1.349090e-16 -3.928806e-16
## Comp 2 -1.358562e-16 -3.652049e-17 -1.000234e-16 2.990686e-17
## Comp 3 -1.168656e-17 -2.848599e-17 8.478176e-18 -9.205732e-18
## Comp 4 2.246010e-17 2.757297e-17 -2.658007e-17 -7.249889e-18
## Comp 5 4.382459e-18 1.679943e-17 -5.937433e-18 7.983152e-18
## Comp 6 1.150396e-17 2.191230e-18 -2.137412e-17 -5.567770e-18
# The coefficients of the original predictors, not of the components!
modPlsr$coefficients[, , 2]
## Goals.scored Goals.conceded Percentage.scored.goals Percentage.conceded.goals
## 1.8192870 -4.4038213 1.8314760 -4.4045722
## Shots Shots.on.goal Penalties.scored Assistances
## 0.4010902 0.9369002 -0.2006251 2.3688050
## Fouls.made Yellow.cards Red.cards Offsides
## 0.2807601 -1.6677725 -2.4952503 1.2187529
# Obtaining the coefficients of the PLS components
lm(formula = Points ~., data = data.frame("Points" = laliga$Points,
modPlsr$scores[, 1:3]))
##
## Call:
## lm(formula = Points ~ ., data = data.frame(Points = laliga$Points,
## modPlsr$scores[, 1:3]))
##
## Coefficients:
## (Intercept) Comp.1 Comp.2 Comp.3
## 52.400 6.093 4.341 3.232
# Prediction
predict(modPlsr, newdata = laligaRed2[1:2, ], ncomp = 12)
## , , 12 comps
##
## Points
## Barcelona 92.01244
## Real Madrid 91.38026
# Selecting the number of components to retain
modPlsr2 <- plsr(Points ~ ., data = laligaRed2, scale = TRUE, ncomp = 2)
summary(modPlsr2)
## Data: X dimension: 20 12
## Y dimension: 20 1
## Fit method: kernelpls
## Number of components considered: 2
## TRAINING: % variance explained
## 1 comps 2 comps
## X 62.53 76.07
## Points 83.76 91.06
# Selecting the number of components to retain by Leave-One-Out cross-validation
modPlsrCV1 <- plsr(Points ~ ., data = laligaRed2, scale = TRUE,
validation = "LOO")
summary(modPlsrCV1)
## Data: X dimension: 20 12
## Y dimension: 20 1
## Fit method: kernelpls
## Number of components considered: 12
##
## VALIDATION: RMSEP
## Cross-validated using 20 leave-one-out segments.
## (Intercept) 1 comps 2 comps 3 comps 4 comps 5 comps 6 comps 7 comps 8 comps 9 comps 10 comps 11 comps
## CV 18.57 8.307 7.221 6.807 6.254 6.604 6.572 6.854 7.348 7.548 7.532 7.854
## adjCV 18.57 8.282 7.179 6.742 6.193 6.541 6.490 6.764 7.244 7.430 7.416 7.717
## 12 comps
## CV 7.905
## adjCV 7.760
##
## TRAINING: % variance explained
## 1 comps 2 comps 3 comps 4 comps 5 comps 6 comps 7 comps 8 comps 9 comps 10 comps 11 comps 12 comps
## X 62.53 76.07 84.94 88.29 92.72 95.61 98.72 99.41 99.83 100.00 100.00 100.00
## Points 83.76 91.06 93.93 95.43 95.94 96.44 96.55 96.73 96.84 96.84 97.69 97.95
# View cross-validation Mean Squared Error Prediction
validationplot(modPlsrCV1, val.type = "MSEP") # l = 4 gives the minimum CV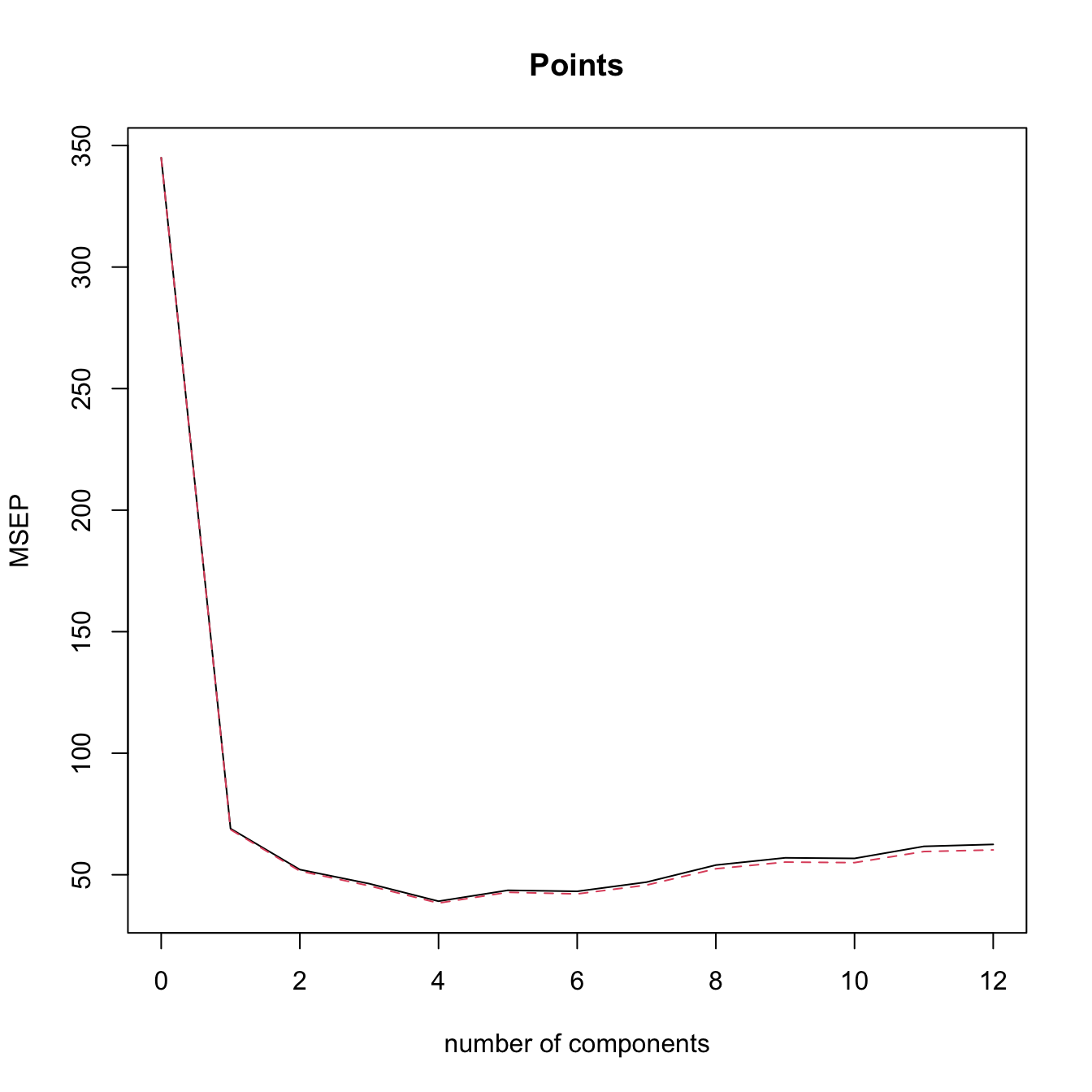
# Selecting the number of components to retain by 10-fold Cross-Validation
# (k = 10 is the default)
modPlsrCV10 <- plsr(Points ~ ., data = laligaRed2, scale = TRUE,
validation = "CV")
summary(modPlsrCV10)
## Data: X dimension: 20 12
## Y dimension: 20 1
## Fit method: kernelpls
## Number of components considered: 12
##
## VALIDATION: RMSEP
## Cross-validated using 10 random segments.
## (Intercept) 1 comps 2 comps 3 comps 4 comps 5 comps 6 comps 7 comps 8 comps 9 comps 10 comps 11 comps
## CV 18.57 7.895 6.944 6.571 6.330 6.396 6.607 7.018 7.487 7.612 7.607 8.520
## adjCV 18.57 7.852 6.868 6.452 6.201 6.285 6.444 6.830 7.270 7.372 7.368 8.212
## 12 comps
## CV 8.014
## adjCV 7.714
##
## TRAINING: % variance explained
## 1 comps 2 comps 3 comps 4 comps 5 comps 6 comps 7 comps 8 comps 9 comps 10 comps 11 comps 12 comps
## X 62.53 76.07 84.94 88.29 92.72 95.61 98.72 99.41 99.83 100.00 100.00 100.00
## Points 83.76 91.06 93.93 95.43 95.94 96.44 96.55 96.73 96.84 96.84 97.69 97.95
validationplot(modPlsrCV10, val.type = "MSEP")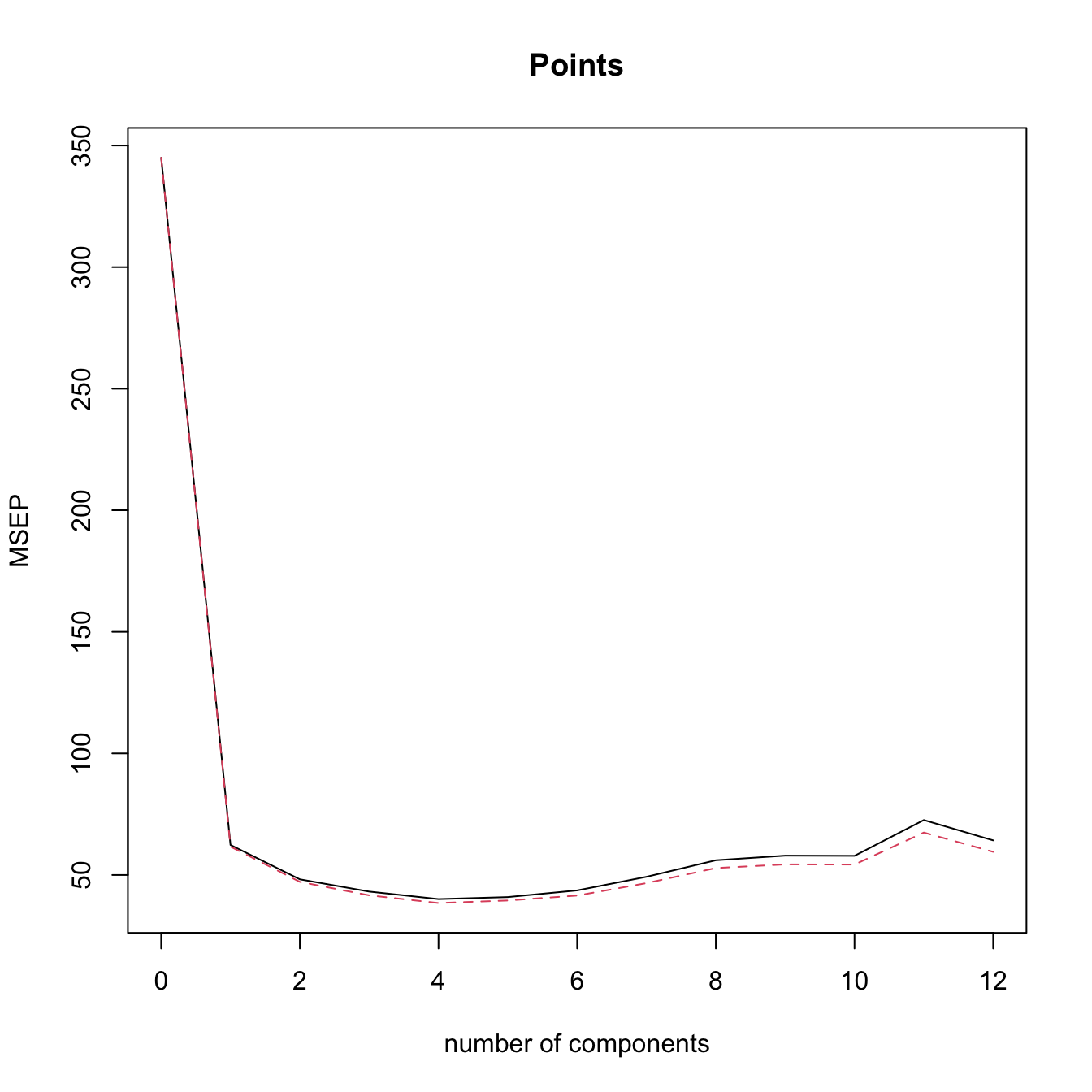
# l = 4 is close to the minimum CV
# Regress manually Points in the scores, in order to have an lm object
# Create a new dataset with the response + PLS components
laligaPLS <- data.frame("Points" = laliga$Points, cbind(modPlsr$scores))
# Regression on the first two PLS
modPlsr <- lm(Points ~ Comp.1 + Comp.2, data = laligaPLS)
summary(modPlsr) # Predictors clearly significative -- same R^2 as in modPlsr2
##
## Call:
## lm(formula = Points ~ Comp.1 + Comp.2, data = laligaPLS)
##
## Residuals:
## Min 1Q Median 3Q Max
## -9.3565 -3.6157 0.4508 2.3288 12.3116
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 52.4000 1.2799 40.941 < 2e-16 ***
## Comp.1 6.0933 0.4829 12.618 4.65e-10 ***
## Comp.2 4.3413 1.1662 3.723 0.00169 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 5.724 on 17 degrees of freedom
## Multiple R-squared: 0.9106, Adjusted R-squared: 0.9
## F-statistic: 86.53 on 2 and 17 DF, p-value: 1.225e-09
car::vif(modPlsr) # No problems at all
## Comp.1 Comp.2
## 1 1Let’s perform PCR and PLSR in the iris dataset. Recall that this dataset contains a factor variable, which can not be treated directly by princomp but can be used in PCR/PLSR (it is transformed internally to two dummy variables). Do the following:
- Compute the PCA of
iris, excludingSpecies. What is the percentage of variability explained with two components? - Draw the biplot and look for interpretations of the principal components.
- Plot the PCA scores in a
car::scatterplotMatrixplot such that the scores are colored by the levels inSpecies(you can use thegroupsargument). - Compute the PCR of
Petal.Widthwith \(\ell=2\) (excludeSpecies). Inspect by leave-one-out cross-validation the best selection of \(\ell.\) - Repeat the previous point with PLSR.
- Plot the PLS scores of the data in a
car::scatterplotMatrixplot such that the scores are colored by the levels inSpecies. Compare with the PCA scores. Is there any interesting interpretation? (A quick way of converting thescoresto a matrix is withrbind.)
We conclude by illustrating the differences between PLSR and PCR with a small numerical example. For that, we consider two correlated predictors \[\begin{align} \begin{pmatrix}X_1\\X_2\end{pmatrix}\sim \mathcal{N}_2\left(\begin{pmatrix}0\\0\end{pmatrix},\begin{pmatrix}1&\rho\\\rho&1\end{pmatrix}\right), \quad -1\leq\rho\leq1,\tag{3.20} \end{align}\] and the linear model \[\begin{align} Y=\beta_1X_1+\beta_2X_2+\varepsilon,\quad \boldsymbol{\beta}=\begin{pmatrix}\cos(\theta)\\\sin(\theta)\end{pmatrix},\tag{3.21} \end{align}\] where \(\theta\in[0,2\pi)\) and \(\varepsilon\sim\mathcal{N}(0,1).\) Therefore:
- The correlation \(\rho\) controls the linear dependence between \(X_1\) and \(X_2\). This parameter is the only one that affects the two principal components \(\Gamma_1=a_{11}X_1+a_{21}X_2\) and \(\Gamma_2=a_{12}X_1+a_{22}X_2\) (recall (3.9)). The loadings \((\hat{a}_{11},\hat{a}_{21})'\) and \((\hat{a}_{12},\hat{a}_{22})'\) are the (sample) PC1 and PC2 directions.
- The angle \(\theta\) controls the direction of \(\boldsymbol{\beta},\) that is, the linear relation between \(Y\) and \((X_1,X_2)\). Consequently, \(\theta\) affects the PLS components \(P_1=b_{11}X_1+b_{21}X_2\) and \(P_2=b_{12}X_1+b_{22}X_2\) (recall (3.19), with adapted notation). The loadings \((\hat{b}_{11},\hat{b}_{21})'\) and \((\hat{b}_{12},\hat{b}_{22})'\) are the (sample) PLS1 and PLS2 directions.
The animation in Figure 3.35 contains samples from (3.20) colored according to their expected value under (3.21). It illustrates the following insights:
- PC directions are unaware of \(\boldsymbol{\beta};\) PLS directions are affected by \(\boldsymbol{\beta}.\)
- PC directions are always orthogonal; PLS directions are not.
- PLS1 aligns102 with \(\boldsymbol{\beta}\) when \(\rho = 0.\) This does not happen when \(\rho \neq 0\) (remember Figure 2.7).
- Under high correlation, the PLS and PC directions are very similar.
- Under low-moderate correlation, PLS1 better explains103 \(\boldsymbol{\beta}\) than PC1.
Figure 3.35: Illustration of the differences and similarities between PCR and PLSR. The points are sampled from (3.20) and colored according to their expected value under (3.21): yellow for larger values; dark violet for smaller. The color gradient is thus controlled by the direction of \(\boldsymbol{\beta}\) (black arrow) and informs on the position of the plane \(y=\beta_1x_1+\beta_2x_2.\) The PC/PLS directions are described in the text. Application available here.
That is, \(\mathbb{E}[X_j]=0,\) \(j=1,\ldots,p.\) This is important since PCA is sensitive to the centering of the data.↩︎
Recall that the covariance matrix is a real, symmetric, semi-positive definite matrix.↩︎
Therefore, \(\mathbf{A}^{-1}=\mathbf{A}'.\)↩︎
Since the variance-covariance matrix \(\mathbb{V}\mathrm{ar}[\boldsymbol{\Gamma}]\) is diagonal.↩︎
Up to now, the exposition has been focused exclusively on the population case.↩︎
Some care is needed here. The matrix \(\mathbf{S}\) is obtained from linear combinations of the \(n\) vectors \(\mathbf{X}_1-\bar{\mathbf{X}},\ldots,\mathbf{X}_n-\bar{\mathbf{X}}.\) Recall that these \(n\) vectors are not linearly independent, as they are guaranteed to add \(\mathbf{0},\) \(\sum_{i=1}^n[\mathbf{X}_i-\bar{\mathbf{X}}]=\mathbf{0},\) so it is possible to express one perfectly on the rest. That implies that the \(p\times p\) matrix \(\mathbf{S}\) has rank smaller or equal to \(n-1.\) If \(p\leq n-1,\) then the matrix has full rank \(p\) and it is invertible (excluding degenerate cases in which the \(p\) variables are collinear). But if \(p\geq n,\) then \(\mathbf{S}\) is singular and, as a consequence, \(\lambda_j=0\) for \(j\geq n.\) This implies that the principal components for those eigenvalues cannot be determined univocally.↩︎
Therefore, a sample of lengths measured in centimeters will have a variance \(10^4\) times larger than the same sample measured in meters – yet it is the same information!↩︎
The biplot is implemented in R by
biplot.princomp(or bybiplotapplied to aprincompobject). This function applies an internal scaling of the scores and variables to improve the visualization (see?biplot.princomp), which can be disabled with the argumentscale = 0.↩︎For the sample version, replace the variable \(X_j\) by its sample \(X_{1j},\ldots,X_{nj},\) the loadings \(a_{ji}\) by their estimates \(\hat a_{ji},\) and the principal component \(\Gamma_j\) by the scores \(\{\hat{\Gamma}_{ij}\}_{i=1}^n.\)↩︎
Observe that both \(X_j=a_{j1}\Gamma_{1} + a_{j2}\Gamma_{2} + \cdots + a_{jp}\Gamma_{p}\) and \(\Gamma_j=a_{1j}X_{1} + a_{2j}X_{2} + \cdots + a_{pj}X_{p},\) \(j=1,\ldots,p,\) hold simultaneously due to the orthogonality of \(\mathbf{A}.\)↩︎
The first two principal components of the dataset explain the \(78\%\) of the variability of the data. Hence, the insights obtained from the biplot should be regarded as “\(78\%\)-accurate”.↩︎
This is certainly not always true, but it is often the case. See Figure 3.35.↩︎
For the sake of simplicity, we considering the exposition the first \(\ell\) principal components, but obviously other combinations of \(\ell\) principal components are possible.↩︎
Since we know by (3.11) that \(\boldsymbol{\Gamma}=\mathbf{A}'(\mathbf{X}-\boldsymbol{\mu}),\) where \(\mathbf{A}\) is the \(p\times p\) matrix of loadings.↩︎
For example, thanks to \(\hat{\boldsymbol{\gamma}}\) we know that the estimated conditional response of \(Y\) precisely increases \(\hat{\gamma}_j\) units for a marginal unit increment in \(X_j.\)↩︎
If \(\ell=p,\) then PCR is equivalent to least squares estimation.↩︎
Alternatively, the “in Prediction” part of the latter terms is dropped and they are just referred to as the MSE and RMSE.↩︎
They play the role of \(X_1,\ldots,X_p\) in the computation of \(P_1\) and can be regarded as \(X_1,\ldots,X_p\) after filtering the linear information explained by \(P_1.\)↩︎
Of course, in practice, the computations need to be done in terms of the sample versions of the presented population versions.↩︎
Recall that we do not care about whether the directions of PLS1 and \(\boldsymbol{\beta}\) are close, but rather if the axes spanned by PLS1 and \(\boldsymbol{\beta}\) are close. The signs of \(\boldsymbol{\beta}\) are learned during the model fitting!↩︎
In the sense that the absolute value of the projection of \(\boldsymbol{\beta}\) along \((\hat{b}_{11},\hat{b}_{21})',\) \(|(\hat{b}_{11},\hat{b}_{21})\boldsymbol{\beta}|,\) is usually larger than that along \((\hat{a}_{11},\hat{a}_{21})'.\)↩︎