13 Transforming data
Anyone regularly working with data is aware that transforming data (aka. “data munging” or “data wrangling”) is an essential pre-requisite for any successful data analysis.
The task of transforming data can be further decomposed into reducing or reshaping data. Our main focus in the current chapter lies on reducing data (by creating chains of functions that allow answering questions about the contents of a dataset). By contrast, the next Chapter 14 will primarily focus on reshaping data (by turning “messy” into “tidy” tables).
The key sections and topics (and corresponding R packages) of this chapter are:
Section 13.2: The pipe operator from magrittr (Bache & Wickham, 2025)
Section 13.3: Transforming tables with dplyr (Wickham, François, et al., 2023)
Both these sections and packages are designed for manipulating data structures (mostly vectors or tables) into other data structures (more vectors or tables). Although transforming data can be viewed as a challenge and a task in itself, our primary goal usually consists in gaining insights into the contents of our data.
Preparation
Recommended readings for this chapter include
Chapter 3: Transforming data of the ds4psy book (Neth, 2025a)
Chapter 5: Data transformation of the r4ds book (Wickham & Grolemund, 2017)
Preflections
Before reading, please take some time to reflect upon the following questions:
Assuming we had all the data required for answering our question, which additional obstacles would we face?
In which data structures are the answers to our data-related questions represented?
Do answers to our data-related questions always contain the entire data? Or do we typically select and reduce data, so that it becomes impossible to reconstruct the data from our answers?
13.1 Introduction
In Section 1.2.5, we introduced the notion of ecological rationality and quoted Herbert A. Simon:
Solving a problem simply means representing it so as to make the solution transparent.
(Simon, 1981, p. 153)
The deep wisdom expressed by this quote also applies to the efforts of data science in general, and to the task of transforming data in particular. In both cases, we aim and hope for some answer or insight that may be hidden in the data. If we succeed in re-representing the data in a way that makes this solution evident, our original problem disappears.
Transforming data is probably the single most important skill of data scientists. While flashy visualizations or fancy statistical models may be the most obvious fruits of our efforts, our skills for transforming data typically lead a more clandestine and silent life. But just as a group of star actors do not yet make a great movie, any successful data science project depends on solid data transformation skills.
One reason for the lack of attention to data transformation is that tutorials on statistical or visualization functions typically use data that just happens to be in the right format. While this conveniently allows demonstrating the power of those tools, it obscures the need and efforts required for bringing data into this form.
Any student of a foreign language will attest that understanding or repeating a well-formed sentence is easier than producing it. Thus, to become fluent in data science, we must be able to recognize different forms of data and learn how we can transform data into them. Ideally, we will eventually become so proficient in these skills that they will become automatic and invisible again. Thus, expert data scientists often fail to be aware of the difficulty that simple data transformation problems provide to novices. For them, solving a problem of transforming data simply means representing it so as to make the solution transparent.
As we have seen for previous challenges, our skills of transforming data are boosted by mastering a mix of concepts and tools.
13.1.1 Key concepts
In addition to introducing two popular tidyverse packages, this chapter introduces some terminology for talking about data transformations:
The current section will distinguish between reshaping and reducing data as two types of data transformation;
Key concepts of Section 13.2 are piping and the use of linear pipes in R (using the pipe operator
%>%of the magrittr package or the base R operator|>);Section 13.3 introduces functions to filter, select, and mutate variables, and group and summarize data (from the dplyr package). Each function denotes a corresponding concept.
13.1.2 Reshaping vs. reducing data
Data transformations and the corresponding functions can be classified into two general types:
Reshaping operations modify the shape or structure of data without changing its contents. A motivation for re-shaping data is that some shapes are more suitable for further analysis than others. Typical examples for reshaping data include transforming the values of a vector into an array or matrix, or re-arranging the rows or columns of a table. (A particular form of tabular data will be described as tidy in Chapter 14 on Tidying data.)
Reducing operations are data transformations that typically modify both the shape and content of data. Examples of data reductions include sampling values from a population, describing a set of values by some measure (e.g., its mean or median), or computing some summary measure out of a set of values (e.g., to learn about a test’s sensitivity or positive predictive value).
Importantly, reshaping data is less invasive than reducing data. Whereas reshaping operations can be reversed, reducing data usually goes beyond mere “trans-formation” by being uni-directional: We typically cannot reconstruct the original data from reduced data.
13.1.3 Practice: Reshape or reduce?
Assuming the following vectors v and a:
do the following operations reshape or reduce these data structures? Why?
Hint: Evaluate the expressions in order to see their results.
matrix(v, nrow = 3)
mean(v)
data.frame(nr = v, freq = v) %>%
group_by(nr) %>%
count()
rev(a)
table(a)
a == "A"Having distinguished between two types of transforming data, we can ask:
- Which data structures are being transformed?
- Which data structures are they being transformed into?
While both types of data transformations can be demonstrated with vectors, we mostly operate on data tables.
Working with a table (as inputs to functions) typically yields new tables (as outputs of functions).
This is where the pipe operator %>% from the magrittr package (Bache & Wickham, 2025) or the native pipe |> come into play:
Both operators pass (or “pipe”) the result of one operation (i.e., the output of a function) as an input to another operation (i.e., as the first argument of another function).
13.2 The pipe operator
The dplyr R package is awesome.
Pipes from the magrittr R package are awesome.
Put the two together and you have one of the most exciting things to happen to R in a long time.Sean C. Anderson (2014)
The pipe operator from the magrittr package (Bache & Wickham, 2025) is a simple tool for data manipulation.
Essentially, its so-called pipe operator %>% allows chaining commands in which the current result is passed to the next command (from left to right).
With such forward pipes, a sequence of simple commands can be chained into a compound command.
The pipe operator will be particularly useful when transforming tables with dplyr (in Section 13.3) and tidyr (in Section 14.2 of Chapter 14). But before diving into the depths data transformation, we show that the pipe is an interesting tool in itself.
13.2.1 Using pipes

Figure 13.1: Ceci est un pipe: %>% (from magrittr). In base R, |> represents a pipe operator.
What is a pipe? While some people see pipes primarily as smoking devices, others may recall representational issues in art history (see Wikipedia: The treachery of images). But a better analogy for the use of pipes in R is more pragmatic and profane: In the applied sub-domain of physics known as plumbing, a pipe is a device for directing fluid and solid substances from one location to another.
In R, the pipe is an operator that allows re-writing a nested call to multiple functions as a chain of individual functions.
Historically, the native forward pipe operator |> of base R (introduced in R version 4.1.0, published on 2021-05-18) was preceded by the %>% operator of the magrittr package (Bache & Wickham, 2025).
Despite some differences, both pipes allow turning a nested expression of function calls into a linear sequence of simple processing steps.
This sequence is easier to understand and avoids the need for saving intermediate results.
An indicator of the pipe’s popularity is the existence of a corresponding keyboard shortcut.
When using the the RStudio IDE, typing the key combination Cmd + Shift + M inserts the %>% operator (or the |> operator, when selecting “Select native pipe operator” in the Code section of Global options).
In the following examples, we mostly use the %>% operator of magrittr, but most examples would be the same for the native pipe operator |>.
Basic usage
For our present purposes, it is sufficient to think of the pipe operator %>% as passing whatever is on its left (or left-hand-side, abbreviated as lhs) to the first argument of the function on its right (or right-hand-side, rhs):
Here, lhs is an expression (i.e., typically an R function) that yields some output object (e.g., a number, vector, or table),
and rhs is an expression that uses this object as an input (i.e., an argument of an R function).
This description of an interaction between R expressions sounds more complicated than it is in practice. Actually, we are quite familiar with R expressions combine multiple steps. But so far, we have been nesting them in arithmetic formulas (with parentheses indicating operator precedence) or in hierarchical function calls, as in:
Using the pipe operator %>% of magrittr allows us to re-write the nested function calls into a linear chain of steps:
Thus, given three functions a(), b() and c(), the following pipe would compute the result of the compound expression c(b(a(x))):
As the intermediate steps get longer and more complicated, we typically re-write the same pipe sequence as follows:
Assigning pipe results
Importantly, the pipe function of passing values differs from the assignment of a value to a name or variable, as achieved by R’s assignment operator <- (see Section 13.2.2 for details and examples).
Thus, to assign the result of a pipe to some object y, we use the assignment operator <- at the (top) left of the pipe:
13.2.2 Example pipes
When transforming data, we mostly use pipes for manipulating tables into other tables. However, the following examples illustrate that pipes can be used for other tasks as well.
Arithmetic pipes
Whereas a description of the pipe operator may sound complicated, the underlying idea is quite simple: We often want to perform several operations in a row or sequence. This is familiar to most of us when computing arithmetic expressions. For instance, consider the following step-by-step instruction:
- Start with a number
x(e.g., 3). Then,
- multiply it by 4,
- add 20 to the result,
- subtract~7 from the result, and finally
- take the square root of the result.
When translating this sequence of steps into arithmetic notation, we could express its first step as \(x = 3\) and combine the next four steps into an expression \(\sqrt{x \cdot 4 + 20 - 7}\). Beyond knowing basic mathematical facts and operations, understanding this expression requires familiarity with arithmetic conventions and notations that determine the order of operations (e.g., reading from left to right, and rules like multiplying before adding or subtracting). To explicate the order of the elementary computations, we could use parentheses (e.g., \(\sqrt{((x \cdot 4) + 20) - 7}\)).
When translating an arithmetic expression into R code, similar conventions and rules govern the way we need to form our expression.
In our example, we can first assign an object x to 3 (i.e., using assignment to create a numeric vector of length 1).
The following arithmetic operations can then be invoked by using a mix of functions, arithmetic operators, and precedence rules:
x <- 3
sqrt(x * 4 + 20 - 7)
#> [1] 5Here, we can immediately spot the sqrt() function, but the arithmetic operators *, + and - also implement arithmetic functions, and the order of their execution is determined by conventions (or explicated by parentheses).
Avoiding the infix operators * and +, we can re-write the expression as a nested combination of R functions:
When combining multiple functions, the order of their evaluation is determined by their level of encapsulation in parentheses.
In R expressions, inner functions are evaluated before outer functions (e.g., prod() is evaluated first and sqrt() is evaluated last).
As this order of evaluation conflicts with our habit of reading and writing from left to right, expressions containing multiple functions are difficult to parse and understand.
The R pipe operators |> or %>% allow us re-writing the sequence of functions as a chain:
# Arithmetic pipe (using base R):
x |> prod(4) |> sum(20) |> sum(-7) |> sqrt()
#> [1] 5
# OR (using magrittr):
x %>% prod(4) %>% sum(20) %>% sum(-7) %>% sqrt()
#> [1] 5Note that the pipe notation corresponds closely to the step-by-step instructions above, particularly when we re-format the pipe to span multiple lines:
Thus, the pipe operator aligns the notation of functional expressions more closely with the order of their evaluation. The pipe may only provide a new notation, but allows us express chains of R functions in a way that matches their natural language description. Overall, piped expressions are powerful by aligning how we think with what is being computed in R.
The dot notation (of magrittr)
If we find the lack of an explicit representation of each step’s result on the right hand side of %>% confusing, we can re-write the magrittr pipe as follows:
# Arithmetic pipe (using the dot notation of magrittr):
x %>% prod(., 4) %>% sum(., 20, -7) %>% sqrt(.)
#> [1] 5Here, the dot . of the magrittr pipe operator is a placeholder for entering the result of the left (lhs) on the right (rhs).
Thus, the . represents whatever was passed (or “piped”) from the left to the right (here: the current value of x).
The key benefit of using the dot placeholder notation is that we may occasionally want the result of the left expression to be passed to a non-first argument of the right expression. When using R for statistics, a typical example consists in passing data to a linear model:
# The linear model:
lm(Sepal.Length ~ Petal.Length, data = iris)
# can be re-written as a pipe (with dot notation):
iris %>%
lm(Sepal.Length ~ Petal.Length, data = .)As both of these expressions effectively call the same lm() function, they also yield the same result:
#>
#> Call:
#> lm(formula = Sepal.Length ~ Petal.Length, data = .)
#>
#> Coefficients:
#> (Intercept) Petal.Length
#> 4.3066 0.4089By contrast, the base R pipe operator |> does not support the dot notation:
# ERROR: No dot notation in the base R pipe:
x |> prod(., 4) |> sum(., 20, -7) |> sqrt(.)
#> Error: object '.' not found
iris |>
lm(Sepal.Length ~ Petal.Length, data = .)
#> Error in eval(mf, parent.frame()): object '.' not foundAnother difference between both operators is that the magrittr pipe %>% allows dropping the parentheses when calling a function with no other arguments, whereas the native pipe |> always requires the parentheses.
(See Differences between the base R and magrittr pipes (by Hadley Wickham, 2023-04-21) for additional details.)
The pipe vs. assignment
As mentioned above, we must not confuse the pipe with R’s assignment operator <-.
Although %>% or |> may look and seem somewhat similar to the assignment operator <- (which also works as = or ->), they are and provide different functions.
Importantly, the pipe does not assign new objects, but rather apply a function to an existing object that then may serve as an input of another function.
The concrete input object changes every time a function is being applied and eventually results in an output object.
What happens if we tried to use a pipe to assign something to a new object \(y\)?
Assuming there is no function y() defined in our current environment, the following code does not assign anything to y, but instead yields an error:
# The pipe does NOT assign objects:
# ERROR with base R pipe:
x |> y
# ERROR with magrittr pipe:
x %>% y
#> Error in y: The pipe operator requires a function call as RHS (<input>:4:6)Thus, for actually assigning the result of a pipe to an object y, we need to use our standard assignment operator on the left (or at the beginning) of the pipe:
# Pipe and assignment by `<-`:
y <- x %>%
prod(4) %>%
sum(20, -7) %>%
sqrt()
y # evaluates to:
#> [1] 5The following also works, but using <- (above) is clearer than using ->:
13.2.3 Evaluation
Overall, the effects of the pipe operators %>% and |> are similar to introducing a new mathematical notation:
They do not allow us to do anything we could not do before, but allow us re-writing chains of commands in a more natural, sequential fashion.
Essentially, embedded or nested calls of functions within functions are untangled into a linear chain of processing steps.
This only re-writes R expressions, but in a way that corresponds more closely to how we think about them (i.e., making them easier to parse and understand).
Piped expressions are particularly useful when generating and transforming data objects (e.g., vectors or tables) by a series of functions that all share the same type of inputs and outputs (e.g., vectors or tables). As using the pipe avoids the need for saving intermediate objects, it makes complex sequences of function calls easier to construct, write, and understand.
While using pipes can add convenience and reduce complexity, these benefits also have some costs.
From an applied perspective, a key requirement for using the pipe is that we must be aware of the data structures serving as inputs and outputs at each step.
More importantly, piping functions implies that we do not need a record of all intermediate results.
Whenever the results of intermediate steps are required later, we still have to assign them to corresponding objects.
From a programming perspective, enabling piped expressions requires that R functions accept some key input as their first argument (unless we use the . notation of magrittr).
Fortunately, most R functions are written in just this way, but it is something to keep in mind when writing our own functions.
13.2.4 Piping color palettes
When using R for data analysis and statistics, piped expressions are primarily used for transforming data tables into other data tables.
However, the pipe operator %>% can be used in other contexts as well, provided functions are written in ways that enable their combination into pipes.
As a concrete and colorful example, we will illustrate piped expressions in common visualization tasks.
For instance, consider the usecol() and seecol() functions of the unikn package (Neth & Gradwohl, 2024):
Besides defining some custom colors (like Seeblau or Pinky), the unikn package provides two main functions for creating and viewing color palettes:
The
usecol()function uses an input argumentpalto define a color palette (e.g., as a vector of color names) and extends this palette tonvalues. Its output is a color palette (as a vector of color codes).The
seecol()function shows and provides detail information on a given color palette.
A typical task when selecting colors is to define a new color palette and then visually inspecting them.
As the first input argument of seecol() matches the output of usecol(), we can use the pipe operator to chain both commands:
A more traditional (and explicit) version of the same commands would first use usecol() for defining a color palette as an R object (e.g., by assigning it to my_col) and then use this as the first argument of the seecol() function:
my_col <- usecol(c(Seeblau, "white", Pinky), n = 7)
seecol(pal = my_col, main = "My new color palette")Note that both of these code snippets call the same functions and thus create identical visualizations.
However, using the pipe did not define the color palette my_col as a separate object in our environment.
The piped chain solution is more compact and immediate, but only the second solution would allow us to re-use my_col later.
Again, we see that the benefits and costs of piped expressions depend on what we want to do with the objects that are being created and manipulated.
13.2.5 Practice pipes
Native pipes
Assume the following objects:
v <- 1:3
x <- v[1]; y <- v[2]; z <- v[3]and re-write the following magrittr pipes by only using the base R pipe |>:
Solution
The following assume an R version 4.1.0 (published on 2021-05-18) or newer:
Note:
As the native R pipe operator |> does not support the . (dot or placeholder) notation, the last expression transformed the function '^'(., z) into the arithmetic infix operator .^z.
Colorful pipes
The grepal() function of unikn searches all R color names (in colors()) for some term (e.g., “gold”, “orange”, or “white”) and returns the corresponding colors.
Construct some color pipes that finds different versions of key colors and displays the corresponding colors.
Are there more “black” or more “white” colors in R?
How many shades of grey are provided by
colors()?
Solution
A pipe that shows all “orange” colors (i.e., colors with “orange” in their name) would be:
There is only one “black”, but many different types of “white” in R:
grepal("black") %>% seecol()
grepal("white") %>% seecol()
# Note:
grepal("grey") %>% seecol()
grepal("dark") %>% seecol()
grepal("light") %>% seecol()Overall, these examples have shown that pipe operators facilitate writing R expressions in arithmetic and visualization contexts. However, the primary use of pipes in R consists in transforming data.
Transforming data with pipes
As we have seen, R pipe operators are a tool for providing data inputs to functions. This functionality is particularly useful when modifying tables of data (i.e., data frames or tibbles) by R functions that are designed to re-shape or reduce data. In the rest of this and in the next chapter, we will be using pipes to illustrate the tools provided by two popular tidyverse packages:
Section 13.3 of this chapter: Transforming tables with dplyr (Wickham, François, et al., 2023)
Section 14.2 of Chapter 14: Transforming tables with tidyr (Wickham & Girlich, 2025)
A good question to occasionally ask ourselves is: If both these packages transform data tables, what is the difference between dplyr and tidyr? We will revisit this question after introducing the essential commands of both packages (in Section 14.3.1).
13.3 Transforming tables with dplyr
The dplyr package (Wickham, François, et al., 2023) is a core component of the tidyverse. Like ggplot2, dplyr is widely used by people who otherwise do not reside within the tidyverse. But as dplyr is a package that is both immensely useful and embodies many of the tidyverse principles in paradigmatic form, we can think of it as the primary citizen of the tidyverse.
dplyr provides a set of functions — best thought of as verbs — that allow slicing and dicing rectangular datasets and computing many summary statistics. While each individual function is simple, they can be combined (e.g., by pipes) into a powerful language of data manipulation. In combination with other functions, using dplyr provides us with quick overviews of datasets that amount to what psychologists often call descriptive statistics. Although a descriptive understanding of data is no substitute for statistical testing or scientific insight, it is an important pre-requisite of both.
The following sections merely provide a summary of essential dplyr functions. More extensive resources for this section include:
Chapter 3: Transforming data of the ds4psy book (Neth, 2025a).
Chapter 5: Data transformation of the r4ds book (Wickham & Grolemund, 2017).
The documentation of the dplyr package (Wickham, François, et al., 2023).
13.3.1 The function of pliers
The name of the dplyr package is inspired by the notion of “pliers”:

Figure 13.2: Pliers are tools for pulling out things or twisting their shapes. (Image by Evan-Amos, via Wikimedia Commons.)
Pliers are tools for pulling out parts and tugging, tweaking, or twisting things into different shapes (see Figure 13.2). In our current context, the thing to tweak is a rectangular set of data (as an R data frame or tibble) and the dplyr tool allows manipulating this table into other tables that contain parts, additional or fewer variables, or provide summary information.
dplyr = MS Excel + control
For users that are familiar with basic spreadsheet in MS Excel: The dplyr functions allow similar manipulations of tabular data in R. However, spreadsheet users are typically solving many tasks by clicking interface buttons, entering simple formulas, and many copy-and-paste operations. While this can be simple and engaging, it is terribly error prone. The main problem with spreadsheets is that the process, typically consisting of many small interactive steps, remains transient and is lost, as only the resulting data table is stored. If a sequence of 100 steps included a minor error on step 29, we often need to start from scratch. Thus, it is very easy to make mistakes and almost impossible to recover from them if they are not noticed immediately.
By contrast, dplyr provides a series of functions for solving (sequences of) simple data manipulation tasks (e.g., arranging or selecting rows or columns, categorizing variables into groups, and computing simple summary tables). Rather than incrementally constructing a spreadsheet and many implicit cut-and-paste operations, dplyr uses sequences of simple functions that explicate the entire process. In the spirit of reproducible research (see Section 1.3), this documents precisely what is being done and allows making corrections later.
13.3.2 Essential dplyr functions
The following sections will briefly illustrate essential dplyr functions and their corresponding tasks:
-
arrange()sorts cases (rows); -
filter()andslice()select cases (rows) by logical conditions; -
select()selects and reorders variables (columns); -
mutate()andtransmute()compute new variables (columns) out of existing ones; -
summarise()collapses multiple values of a variable (rows of a column) to a single one;
-
group_by()changes the unit of aggregation (in combination withmutate()andsummarise()).
Learning dplyr essentially consists in learning and applying these functions as the verbs of a new language. And just as the words of a language are typically combined into longer sequences (e.g., sentences, paragraphs, etc.) dplyr functions are often combined into longer data transformation pipes. Studying a few examples of each function will make it easy and straightforward to combine them into powerful pipes that allow slicing, dicing, and summarizing large data tables.
Example data
Section 3.2: Essential dplyr commands uses the starwars data from the dplyr package:
The starwars data is a tibble containing 87 cases (rows) and 14 variables (columns).
See ?dplyr::starwars for a description of the data and its variables.
To provide additional examples, we use the storms data from the dplyr package in this chapter:
The storms data is a tibble containing 11859 cases (rows) and 13 variables (columns).
See ?dplyr::storms for a description of the data and its variables.
A key difference between the storms and the starwars data lies in their units of observations:
Each row of starwars corresponds to an individual creature, so that each individual creature appears in exactly one row.
By contrast, each row in storms describes a particular observation of a storm, but the same storm can be observed at multiple times (i.e., appears in multiple lines).
As a consequence, working with the storms data is slightly more challenging than working with the starwars data.
The following examples use the storms data.
If you find any of them difficult to understand, check out
Section 3.2: Essential dplyr commands of the ds4psy book (Neth, 2025a), which provide corresponding examples based on the starwars data.
1. Using arrange() to sort cases (rows)
The arrange() function keeps all data, but arranges its cases (rows) by the variable (column) mentioned:
#> # A tibble: 11,859 × 13
#> name year month day hour lat long status categ…¹ wind press…²
#> <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <chr> <ord> <int> <int>
#> 1 Zeta 2006 1 1 0 25.6 -38.3 tropical sto… 0 50 997
#> 2 Zeta 2006 1 1 6 25.4 -38.4 tropical sto… 0 50 997
#> 3 Zeta 2006 1 1 12 25.2 -38.5 tropical sto… 0 50 997
#> 4 Zeta 2006 1 1 18 25 -38.6 tropical sto… 0 55 994
#> 5 Zeta 2006 1 2 0 24.6 -38.9 tropical sto… 0 55 994
#> 6 Zeta 2006 1 2 6 24.3 -39.7 tropical sto… 0 50 997
#> 7 Zeta 2006 1 2 12 23.8 -40.4 tropical sto… 0 45 1000
#> 8 Zeta 2006 1 2 18 23.6 -40.8 tropical sto… 0 50 997
#> 9 Zeta 2006 1 3 0 23.4 -41 tropical sto… 0 55 994
#> 10 Zeta 2006 1 3 6 23.3 -41.3 tropical sto… 0 55 994
#> # … with 11,849 more rows, 2 more variables:
#> # tropicalstorm_force_diameter <int>, hurricane_force_diameter <int>, and
#> # abbreviated variable names ¹category, ²pressureArranging rows by multiple variables is also possible:
#> # A tibble: 11,859 × 13
#> name year month day hour lat long status categ…¹ wind press…²
#> <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <chr> <ord> <int> <int>
#> 1 Isidore 1990 9 4 0 7.2 -23.4 tropical d… -1 25 1010
#> 2 Isidore 1990 9 4 6 7.4 -25.1 tropical d… -1 25 1010
#> 3 Kirk 2018 9 22 6 7.7 -21.8 tropical d… -1 30 1007
#> 4 Kirk 2018 9 22 12 8.1 -22.9 tropical s… 0 35 1005
#> 5 Pablo 1995 10 4 18 8.3 -31.4 tropical d… -1 30 1009
#> 6 Pablo 1995 10 5 0 8.4 -32.8 tropical d… -1 30 1009
#> 7 Isidore 1990 9 4 12 8.4 -26.7 tropical d… -1 25 1009
#> 8 Kirk 2018 9 22 18 8.5 -24.1 tropical s… 0 35 1005
#> 9 Isidore 1996 9 24 12 8.6 -23.3 tropical d… -1 25 1008
#> 10 Arthur 1990 7 22 6 8.8 -41.9 tropical d… -1 25 1010
#> # … with 11,849 more rows, 2 more variables:
#> # tropicalstorm_force_diameter <int>, hurricane_force_diameter <int>, and
#> # abbreviated variable names ¹category, ²pressureThe arrange() function sorts text variables (of type “character”) in alphabetical and numeric variables (of type “integer” or “double”) in ascending order. Use desc() to sort a variable in the opposite order:
#> # A tibble: 11,859 × 13
#> name year month day hour lat long status categ…¹ wind press…²
#> <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <chr> <ord> <int> <int>
#> 1 Zeta 2020 10 29 12 35.3 -83.6 tropical sto… 0 45 990
#> 2 Zeta 2020 10 29 6 32.8 -87.5 tropical sto… 0 60 986
#> 3 Zeta 2020 10 29 0 30.2 -89.9 hurricane 2 85 973
#> 4 Zeta 2020 10 28 21 29.2 -90.6 hurricane 3 100 970
#> 5 Zeta 2020 10 28 18 28 -91.1 hurricane 2 95 973
#> 6 Zeta 2020 10 28 12 26 -91.7 hurricane 1 80 978
#> 7 Zeta 2005 12 31 6 25.7 -37.6 tropical sto… 0 50 997
#> 8 Zeta 2005 12 31 12 25.7 -37.9 tropical sto… 0 50 997
#> 9 Zeta 2005 12 31 18 25.7 -38.1 tropical sto… 0 45 1000
#> 10 Zeta 2005 12 31 0 25.6 -37.3 tropical sto… 0 45 1000
#> # … with 11,849 more rows, 2 more variables:
#> # tropicalstorm_force_diameter <int>, hurricane_force_diameter <int>, and
#> # abbreviated variable names ¹category, ²pressureNote that the variable names specified in arrange() — or in other dplyr functions — are not enclosed in quotation marks.
This may seem a bit strange at first, but becomes totally intuitive after typing a few commands.
2. Using filter() or slice() to select cases (rows)
Many questions concern only a subset of the cases (rows) of our data.
In these instances, a typical first step consists in filtering rows for particular values on one or more variables.
For instance, the following command reduces the 11859 rows of the st data quite drastically by only including rows in which the wind speed exceeds 150 knots:
#> # A tibble: 12 × 13
#> name year month day hour lat long status category wind pressure
#> <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <chr> <ord> <int> <int>
#> 1 Gilbert 1988 9 14 0 19.7 -83.8 hurricane 5 160 888
#> 2 Gilbert 1988 9 14 6 19.9 -85.3 hurricane 5 155 889
#> 3 Mitch 1998 10 26 18 16.9 -83.1 hurricane 5 155 905
#> 4 Mitch 1998 10 27 0 17.2 -83.8 hurricane 5 155 910
#> 5 Rita 2005 9 22 3 24.7 -87.3 hurricane 5 155 895
#> 6 Rita 2005 9 22 6 24.8 -87.6 hurricane 5 155 897
#> 7 Wilma 2005 10 19 12 17.3 -82.8 hurricane 5 160 882
#> 8 Dorian 2019 9 1 12 26.5 -76.5 hurricane 5 155 927
#> 9 Dorian 2019 9 1 16 26.5 -77 hurricane 5 160 910
#> 10 Dorian 2019 9 1 18 26.5 -77.1 hurricane 5 160 910
#> 11 Dorian 2019 9 2 0 26.6 -77.7 hurricane 5 155 914
#> 12 Dorian 2019 9 2 2 26.6 -77.8 hurricane 5 155 914
#> # … with 2 more variables: tropicalstorm_force_diameter <int>,
#> # hurricane_force_diameter <int>As before, filter() can use multiple variables and include numeric and character variables:
# Select cases (rows) based on several conditions:
st %>%
filter(year > 2014, month == 9, status == "hurricane")#> # A tibble: 234 × 13
#> name year month day hour lat long status category wind pressure
#> <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <chr> <ord> <int> <int>
#> 1 Fred 2015 9 1 0 17.4 -24.9 hurricane 1 65 991
#> 2 Joaquin 2015 9 30 6 25.4 -71.8 hurricane 1 65 978
#> 3 Joaquin 2015 9 30 12 24.9 -72.2 hurricane 1 70 971
#> 4 Joaquin 2015 9 30 18 24.4 -72.5 hurricane 1 80 961
#> 5 Gaston 2016 9 1 0 35.5 -46.3 hurricane 2 90 969
#> 6 Gaston 2016 9 1 6 36.3 -44.3 hurricane 2 85 973
#> 7 Gaston 2016 9 1 12 37.1 -42 hurricane 1 80 976
#> 8 Gaston 2016 9 1 18 37.8 -39.5 hurricane 1 75 981
#> 9 Gaston 2016 9 2 0 38.2 -37 hurricane 1 70 985
#> 10 Gaston 2016 9 2 6 38.5 -35 hurricane 1 65 988
#> # … with 224 more rows, and 2 more variables:
#> # tropicalstorm_force_diameter <int>, hurricane_force_diameter <int>A variant of filter() is slice(), which is used to select particular rows, which are either described by some number or some combination of a property and a number:
# Select cases (rows):
st %>% slice_head(n = 3) # select the first 3 cases#> # A tibble: 3 × 13
#> name year month day hour lat long status categ…¹ wind press…² tropi…³
#> <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <chr> <ord> <int> <int> <int>
#> 1 Amy 1975 6 27 0 27.5 -79 tropi… -1 25 1013 NA
#> 2 Amy 1975 6 27 6 28.5 -79 tropi… -1 25 1013 NA
#> 3 Amy 1975 6 27 12 29.5 -79 tropi… -1 25 1013 NA
#> # … with 1 more variable: hurricane_force_diameter <int>, and abbreviated
#> # variable names ¹category, ²pressure, ³tropicalstorm_force_diameter
st %>% slice_tail(n = 3) # select the last 3 cases#> # A tibble: 3 × 13
#> name year month day hour lat long status categ…¹ wind press…² tropi…³
#> <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <chr> <ord> <int> <int> <int>
#> 1 Iota 2020 11 18 0 13.8 -86.7 tropi… 0 40 1000 140
#> 2 Iota 2020 11 18 6 13.8 -87.8 tropi… 0 35 1005 140
#> 3 Iota 2020 11 18 12 13.7 -89 tropi… -1 25 1006 0
#> # … with 1 more variable: hurricane_force_diameter <int>, and abbreviated
#> # variable names ¹category, ²pressure, ³tropicalstorm_force_diameter#> # A tibble: 3 × 13
#> name year month day hour lat long status categ…¹ wind press…² tropi…³
#> <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <chr> <ord> <int> <int> <int>
#> 1 AL07… 2003 7 26 12 32.3 -82 tropi… -1 20 1022 NA
#> 2 AL07… 2003 7 26 18 32.8 -82.6 tropi… -1 15 1022 NA
#> 3 AL07… 2003 7 27 0 33 -83 tropi… -1 15 1022 NA
#> # … with 1 more variable: hurricane_force_diameter <int>, and abbreviated
#> # variable names ¹category, ²pressure, ³tropicalstorm_force_diameter#> # A tibble: 3 × 13
#> name year month day hour lat long status categ…¹ wind press…² tropi…³
#> <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <chr> <ord> <int> <int> <int>
#> 1 Wilma 2005 10 19 12 17.3 -82.8 hurri… 5 160 882 265
#> 2 Gilb… 1988 9 14 0 19.7 -83.8 hurri… 5 160 888 NA
#> 3 Gilb… 1988 9 14 6 19.9 -85.3 hurri… 5 155 889 NA
#> # … with 1 more variable: hurricane_force_diameter <int>, and abbreviated
#> # variable names ¹category, ²pressure, ³tropicalstorm_force_diameter
st %>% slice_sample(n = 3) # select 3 random cases#> # A tibble: 3 × 13
#> name year month day hour lat long status categ…¹ wind press…² tropi…³
#> <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <chr> <ord> <int> <int> <int>
#> 1 Jose… 2008 9 3 18 13.8 -29.2 tropi… 0 55 994 180
#> 2 Bonn… 1992 9 20 12 36.5 -56 hurri… 2 85 974 NA
#> 3 Nico… 2016 10 12 0 27.4 -66.6 hurri… 1 75 976 180
#> # … with 1 more variable: hurricane_force_diameter <int>, and abbreviated
#> # variable names ¹category, ²pressure, ³tropicalstorm_force_diameterNote that filter() and slice() reduced the number of cases (rows), but left the number of variables (columns) intact.
The complement is select(), which has the opposite effects.
3. Using select() to select variables (columns)
The select() function provides an easy way of selecting and re-arranging the variables (columns) of tables:
#> # A tibble: 11,859 × 3
#> name pressure wind
#> <chr> <int> <int>
#> 1 Amy 1013 25
#> 2 Amy 1013 25
#> 3 Amy 1013 25
#> 4 Amy 1013 25
#> 5 Amy 1012 25
#> 6 Amy 1012 25
#> 7 Amy 1011 25
#> 8 Amy 1006 30
#> 9 Amy 1004 35
#> 10 Amy 1002 40
#> # … with 11,849 more rowsImportant assets of select() are its additional features:
-
va:vxselects a range of variables (e.g., fromvatovx); -
!vyallows negative selections (e.g., selecting all variables exceptvy); -
&or|selects the intersection or union of two sets of variables; -
starts_with("abc")andends_with("xyz")selects all variables whose names start or end with some characters (e.g., “abc” or “xyz”); -
everything()selects all variables not selected yet (e.g., to re-order all variables).
Here are some typical examples for corresponding selections (adding slice_sample(n = 3) for showing only three random rows of the resulting table):
# Select a range of variables:
st %>% select(name, year:day, lat:long) %>% slice_sample(n = 3) #> # A tibble: 3 × 6
#> name year month day lat long
#> <chr> <dbl> <dbl> <int> <dbl> <dbl>
#> 1 Harvey 2005 8 6 33.5 -56.7
#> 2 Debby 1982 9 14 22.4 -71.8
#> 3 Lisa 2010 9 25 22.3 -28.3
# Select an intersection of negated variables:
st %>% select(!status & !ends_with("diameter")) %>% slice_sample(n = 3) #> # A tibble: 3 × 10
#> name year month day hour lat long category wind pressure
#> <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <ord> <int> <int>
#> 1 Marilyn 1995 9 20 0 34.2 -66.8 1 80 974
#> 2 Harvey 1981 9 15 0 28.4 -62.6 4 115 946
#> 3 Hermine 1980 9 25 0 17.7 -95.5 0 45 1000
# Re-order the columns of a table (selecting everything):
st %>% select(year, name, lat:long, everything()) %>% slice_sample(n = 3) #> # A tibble: 3 × 13
#> year name lat long month day hour status categ…¹ wind press…² tropi…³
#> <dbl> <chr> <dbl> <dbl> <dbl> <int> <dbl> <chr> <ord> <int> <int> <int>
#> 1 1996 Lili 33.2 -53.8 10 23 6 hurri… 1 65 985 NA
#> 2 2012 Sandy 29.7 -75.6 10 27 18 hurri… 1 70 960 690
#> 3 2018 Lesl… 34.2 -57.6 10 5 0 tropi… 0 55 984 410
#> # … with 1 more variable: hurricane_force_diameter <int>, and abbreviated
#> # variable names ¹category, ²pressure, ³tropicalstorm_force_diameterWhen using a pipe for quickly answering some descriptive question, it is common practice to first apply some combination of filter() and slice() (for removing non-needed rows), and select() (for removing non-needed columns).
4. Using mutate() for computing new variables
A frequent task in data analysis consists in computing some new variable out of existing ones.
Metaphorically, this can be viewed as “mutating” some table’s current information into slightly different form.
The mutate() function of dplyr first names a new variable (e.g., var_new =) and then uses existing R expressions (e.g., arithmetic operators, functions, etc.) for computing the values of the new variable.
As an example, let’s combine some variables into a new date variable:
# Compute and add a new variable:
st %>%
select(name, year, month, day) %>% # select/remove variables
mutate(date = paste(year, month, day, sep = "-")) %>%
slice_sample(n = 5) # show 5 random cases (rows)#> # A tibble: 5 × 5
#> name year month day date
#> <chr> <dbl> <dbl> <int> <chr>
#> 1 Danielle 1998 8 30 1998-8-30
#> 2 Alex 1998 7 28 1998-7-28
#> 3 Klaus 1984 11 11 1984-11-11
#> 4 Gabrielle 2019 9 8 2019-9-8
#> 5 Hugo 1989 9 17 1989-9-17In this example, we used the R function paste() to combine the three variables year, month, and day, into a single character variable.
Note that the assignment to a new variable was signaled by the operator =, rather than R’s typical assignment operator <-.
To make our example clear, we used the select() function to remove all variables not needed for this illustration. In most real-world settings, we would not remove all other variables, but rather add new variables to the existing ones (and occasionally use select() to re-order them).
Note also that, when actually working with dates and times later, we would not transform date values into text objects. Instead, we use R functions for parsing date and time variables into dedicated variables that represent dates or date-times (see Chapter 10: Dates and times).
As with the other dplyr verbs, we can compute several new variables in one mutate() command by separating them by commas.
If we wanted to get rid of the old variables, we could immediately remove them from the data by using transmute():
# Compute several new variables (replacing the old ones):
st %>%
transmute(name = paste0(name, " (", status, ")"),
date = paste(year, month, day, sep = "-"),
loc = paste0("(", lat, "; ", long, ")")) %>%
slice_sample(n = 5) # show 5 random cases (rows)#> # A tibble: 5 × 3
#> name date loc
#> <chr> <chr> <chr>
#> 1 Jeanne (tropical storm) 2004-9-15 (17.1; -64)
#> 2 Nine (tropical depression) 2015-9-17 (15.6; -45)
#> 3 Humberto (hurricane) 2007-9-13 (29.5; -94.4)
#> 4 Marco (tropical storm) 1996-11-24 (16; -78)
#> 5 Ana (tropical depression) 1979-6-23 (14; -61.3)However, it is usually a bad idea to throw away source data. Especially since any act of computing new variables can be error-prone, we generally do not recommend using transmute(). Instead, a careful data analyst prefers mutate() for creating new variables and immediately checks whether the newly created variables are correct. And keeping all original variables is rarely a problem, as we always can remove unwanted variables (by selecting only the ones needed) later.
As both our examples so far have used mutate() or transmute() to create new character variables, here’s an example on numerical data.
If we wanted to add a variable that rounds the values of pressure to the nearest multiple of 10, we could first divide these values by 10, round the result to the nearest integer (using the R function round(x, 0)), before multiplying by 10 again:
# Compute and add a new numeric variable:
st %>%
mutate(press_10 = round(pressure/10, 0) * 10) %>%
select(name, pressure, press_10) %>%
slice_sample(n = 5) # show 5 random cases (rows)#> # A tibble: 5 × 3
#> name pressure press_10
#> <chr> <int> <dbl>
#> 1 Mitch 923 920
#> 2 AL042000 1011 1010
#> 3 Gabrielle 993 990
#> 4 Floyd 967 970
#> 5 Debby 995 1000The immense power of mutate() lies in its use of functions for computing new variables out of the existing ones.
As any R function can be used, the possibilities for creating new variables are limitless.
However, note that the computations of each mutate() command are typically constrained to each individual case (row).
This changes with the following two dplyr commands.
5. Using summarise() for aggregating over values of variables
Whereas mutate() computes new variables out of exiting ones for each case (i.e., by row), summarise() (and summarize()) computes summaries for individual variables (i.e., by column).
Each summary is assigned to a new variable, using the same var_new = ... syntax as mutate().
The type of summary is indicated by applying a function to one or more variables.
Useful functions for potential summaries include:
Count:
n(),n_distinct()Range:
min(),max(),quantile()
When using summary functions, it is important to choose sensible names for the variables to which their results are being assigned. Here is a bad and a better example for naming summary variables:
#> # A tibble: 1 × 4
#> n name mean max
#> <int> <int> <dbl> <int>
#> 1 11859 214 53.6 160
# better names:
st %>%
summarise(nr = n(),
nr_names = n_distinct(name),
mean_wind = mean(wind),
max_wind = max(wind))#> # A tibble: 1 × 4
#> nr nr_names mean_wind max_wind
#> <int> <int> <dbl> <int>
#> 1 11859 214 53.6 160Fortunately, if we fail to provide variable names, dplyr provides sensible default names:
#> # A tibble: 1 × 4
#> `n()` `n_distinct(name)` `mean(wind)` `max(wind)`
#> <int> <int> <dbl> <int>
#> 1 11859 214 53.6 160Notice that using summarise() on a data table yields a modified — and typically much smaller — data table (as a tibble). Thus, summarise() is a function primarily used for reducing data.
Interestingly, we can immediately use a computed variable (like mean_wind or sd_wind) in a subsequent computation:
st %>%
summarise(nr = n(),
mean_wind = mean(wind),
sd_wind = sd(wind),
mean_minus_sd = mean_wind - sd_wind,
mean_plus_sd = mean_wind + sd_wind)
#> # A tibble: 1 × 5
#> nr mean_wind sd_wind mean_minus_sd mean_plus_sd
#> <int> <dbl> <dbl> <dbl> <dbl>
#> 1 11859 53.6 26.2 27.4 79.8Summaries of columns are certainly nice to have, but nothing for which we needed a new function for.
Instead, we could have simply computed the same summaries directly for the vectors of the st data:
nrow(st)
#> [1] 11859
n_distinct(st$name)
#> [1] 214
mean(st$wind)
#> [1] 53.63774
max(st$wind)
#> [1] 160Thus, the true value of the summarise() function lies in the fact that it aggregates not only over all values of a variable (i.e., entire columns), but also over the levels of grouped variables, or all combinations of grouped variables.
To use this feature, we need to precede a summarise() function by a group_by() function.
6. Using group_by() for changing the unit of aggregation
The group_by() function does very little by itself, but becomes immensely powerful in combination with other dplyr commands.
To see how this works, let’s select only four variables from our st data and examine the effects of group_by():
#> # A tibble: 11,859 × 4
#> # Groups: name [214]
#> name year wind pressure
#> <chr> <dbl> <int> <int>
#> 1 Amy 1975 25 1013
#> 2 Amy 1975 25 1013
#> 3 Amy 1975 25 1013
#> 4 Amy 1975 25 1013
#> 5 Amy 1975 25 1012
#> 6 Amy 1975 25 1012
#> 7 Amy 1975 25 1011
#> 8 Amy 1975 30 1006
#> 9 Amy 1975 35 1004
#> 10 Amy 1975 40 1002
#> # … with 11,849 more rowsThe resulting tibble contains all 11859 cases (rows) of st and the four selected variables.
So what did the group_by(name) command do?
Inspecting the output more closely shows the message:
“Groups: name [198]”.
This suggests that something has changed, even though we do not see any effects. Interestingly, the number of groups mentioned (i.e., 214) matches the number of distinct storm names that we obtained above by evaluating n_distinct(st$name).
Let’s try a second command:
#> # A tibble: 11,859 × 4
#> # Groups: name, year [512]
#> name year wind pressure
#> <chr> <dbl> <int> <int>
#> 1 Amy 1975 25 1013
#> 2 Amy 1975 25 1013
#> 3 Amy 1975 25 1013
#> 4 Amy 1975 25 1013
#> 5 Amy 1975 25 1012
#> 6 Amy 1975 25 1012
#> 7 Amy 1975 25 1011
#> 8 Amy 1975 30 1006
#> 9 Amy 1975 35 1004
#> 10 Amy 1975 40 1002
#> # … with 11,849 more rowsThe resulting tibble seems unaltered, but the message below the tibble dimensions now reads:
“Groups: name, year [426]”.
As there are 198 different values of name and the range of year values varies from 1975 to 2020, the number of 426 groups is not immediately obvious. As we will see momentarily, it results from the fact that some, but not all instances of name occur in more than one year.
The easiest way to identify the specific groups in both cases is to follow the last two statements by the count() function.
This returns the groups (as the rows of a tibble) together with a variable n that counts the number of observations in each group:
# Group st by name and count (n of) observations per group:
st %>%
select(name, year, wind, pressure) %>%
group_by(name) %>%
count() %>%
filter(name == "Felix") # show only one group#> # A tibble: 1 × 2
#> # Groups: name [1]
#> name n
#> <chr> <int>
#> 1 Felix 178
# Group st by name, year and count (n of) observations per group:
st %>%
select(name, year, wind, pressure) %>%
group_by(name, year) %>%
count() %>%
filter(name == "Felix") # show only one group#> # A tibble: 4 × 3
#> # Groups: name, year [4]
#> name year n
#> <chr> <dbl> <int>
#> 1 Felix 1989 57
#> 2 Felix 1995 59
#> 3 Felix 2001 40
#> 4 Felix 2007 22Rather than returning the entire tibble, we added filter(name == "Felix") at the end to only return lines with this particular name value. When group_by(name), there is only one such group (containing 178 observations).
By contrast, when group_by(name, year), there are four such groups (with a sum of 178 observations). Thus, a storm with the name Felix was observed in four distinct years.
ungroup() removes group()
The ungroup() function removes existing grouping factors.
This is occasionally necessary for applying additional dplyr commands.
For instance, if we first wanted to group storms by name and later draw a random sample of size n = 10 , we would need to add an intermediate ungroup() before applying slice_sample(n = 10) to the tibble of groups:
#> # A tibble: 5 × 13
#> name year month day hour lat long status categ…¹ wind press…² tropi…³
#> <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <chr> <ord> <int> <int> <int>
#> 1 Jean… 1980 11 12 0 24.1 -87.4 hurri… 2 85 988 NA
#> 2 Erin 2007 8 16 12 28.1 -97.1 tropi… -1 30 1006 0
#> 3 Patty 2012 10 13 0 25.4 -71.9 tropi… 0 35 1007 30
#> 4 Evel… 1977 10 14 6 30.9 -64.9 tropi… 0 35 1005 NA
#> 5 AL02… 2003 6 11 12 9.7 -44.2 tropi… -1 30 1008 NA
#> # … with 1 more variable: hurricane_force_diameter <int>, and abbreviated
#> # variable names ¹category, ²pressure, ³tropicalstorm_force_diameterWhy do we want to group() data tables?
The benefits of grouping become obvious by combining a group_by() function with a subsequent mutate() or summarise() function.
In both cases, the unit of aggregation is changed from all cases to those within each group.
The following sections provide examples for both combinations.
Grouped mutates
To illustrate the effects of a sequence of group_by() and mutate(), let’s compute the mean wind speed twice:
the 1st use of
mutate()computes the mean wind speedmn_wind_1over all cases. The number of cases over which the mean is aggregated can be counted by then()function (assigned to a variablemn_n_1).the 2nd use of
mutate()also computes the mean wind speedmn_wind_2and uses the samen()function (assigned to a variablemn_n_2). The key difference is not in the content of themutate()function, but the fact that the 2ndmutate()is located after thegroup_by(name)expression. This changed the unit of aggregation to this particular group.
st %>%
select(name, year, wind) %>% # select some variables
mutate(mn_wind_1 = mean(wind), # compute mean 1
mn_n_1 = n()) %>% # compute nr 1
group_by(name) %>% # group by name
mutate(mn_wind_2 = mean(wind), # compute mean 2
mn_n_2 = n()) %>% # compute n 2
ungroup() %>% # ungroup
slice_sample(n = 10)#> # A tibble: 10 × 7
#> name year wind mn_wind_1 mn_n_1 mn_wind_2 mn_n_2
#> <chr> <dbl> <int> <dbl> <int> <dbl> <int>
#> 1 Luis 1995 110 53.6 11859 96.7 52
#> 2 Georges 1998 90 53.6 11859 67.3 92
#> 3 Bonnie 1986 30 53.6 11859 52.4 209
#> 4 Ivan 2004 45 53.6 11859 79.3 141
#> 5 Frederic 1979 115 53.6 11859 52.6 66
#> 6 Lisa 2010 50 53.6 11859 45.0 122
#> 7 Michael 2012 50 53.6 11859 69.8 66
#> 8 Humberto 2001 25 53.6 11859 64.2 132
#> 9 Emily 2005 120 53.6 11859 59.6 217
#> 10 Arthur 2002 50 53.6 11859 39.0 119To better inspect the resulting tibble, we followed the 2nd mutate() function with ungroup(). This removes any existing grouping operations and allows manipulating the table with the new variables. In this case, we used slice_sample(n = 10) to draw 10 random rows (out of the 11859 rows from st). These 10 rows show the difference in results of both mutate() commands. The first variable mn_wind_1 shows an identical value for all rows (as it computed the means over all 11859 rows). The second variable mn_wind_2 shows a different value for each name, as it was computed over those rows that shared the same name.
Beware that the results of grouped mutate() commands are easily misinterpreted:
For instance, the values of mn_wind_2 could be misinterpreted as the mean wind speed of a particular storm.
However, mn_wind_2 was only computed over those observations that shared the same name value.
As some names occur repeatedly (in different years), the values do not necessarily refer to the same storm.
Aggregating by storm would require a unique identifier for each storm (e.g., some combination of its name and date).
Grouped summaries
Perhaps even more frequent than following a group() function by mutate() is following it by summarise().
In this case, we aggregate the specified summaries over each group, rather than all data values.
To illustrate the effects of grouped summaries, compare the following three pipes of dplyr commands: All three contain the same summarise() part, which computes three new variables that report the number of cases in each summary n_cases, the mean wind speed mn_wind, and the maximum wind speed max_wind. The difference between the three versions lies in the group_by() statements prior to the summarise() command:
- the 1st pipe computes the summary for all of
st(without grouping):
#> # A tibble: 1 × 3
#> n_cases mn_wind max_wind
#> <int> <dbl> <int>
#> 1 11859 53.6 160- the 2nd pipe computes the summary for each
yearofst:
# Pipe 2:
st %>%
group_by(year) %>%
summarise(n_cases = n(),
mn_wind = mean(wind),
max_wind = max(wind))#> # A tibble: 46 × 4
#> year n_cases mn_wind max_wind
#> <dbl> <int> <dbl> <int>
#> 1 1975 86 50.9 100
#> 2 1976 52 59.9 105
#> 3 1977 53 54.0 150
#> 4 1978 54 40.5 80
#> 5 1979 301 48.7 150
#> 6 1980 161 53.7 90
#> # ℹ 40 more rows- the 3rd pipe computes the summary for each
yearandmonthofst:
# Pipe 3:
st %>%
group_by(year, month) %>%
summarise(n_cases = n(),
mn_wind = mean(wind),
max_wind = max(wind))#> # A tibble: 227 × 5
#> # Groups: year [46]
#> year month n_cases mn_wind max_wind
#> <dbl> <dbl> <int> <dbl> <int>
#> 1 1975 6 16 37.5 60
#> 2 1975 7 14 56.8 60
#> 3 1975 8 40 45 100
#> 4 1975 9 16 73.8 95
#> 5 1976 8 18 68.1 105
#> 6 1976 9 18 56.4 90
#> # ℹ 221 more rowsNote that each pipe results in a tibble, but their dimensions differ considerably.
Whereas the first pipe yielded a compact 1 by 3 data structure,
the 2nd one contained 48 rows and 4 columns, and the 3rd one contained 259 rows and 5 columns.
As we can use all kinds of R functions in the mutate() and summarise() parts, preceding them by group_by() allows computing all kinds of new variables and descriptive summary statistics.
In the following, we illustrate how we can answer quite interesting questions by appropriate dplyr pipes.
13.3.3 Answering questions by data transformation
Using dplyr pipes to reshape or reduce tables and then either inspecting the resulting tibble or visualizing it with ggplot2 provides powerful combinations for data exploration. As we will address the topic of Exploring data more explicitly in Chapter 15, this section only provides some more advanced examples.
To illustrate the typical workflow, we will continue to work with the dplyr storms data (copied to st).
So let’s ask some non-trivial questions and then provide a descriptive answer to it by summary tables and visualizations (i.e., a combination of dplyr and ggplot2 expressions).
Answer
We can translate this question into the following one: What was the maximal wind speed that was recorded for each (named) storm?
st <- dplyr::storms # copy data
st_top10_wind <- st %>%
group_by(name) %>%
summarise(max_wind = max(wind)) %>%
arrange(desc(max_wind)) %>%
slice(1:10)
st_top10_wind#> # A tibble: 10 × 2
#> name max_wind
#> <chr> <int>
#> 1 Dorian 160
#> 2 Gilbert 160
#> 3 Wilma 160
#> 4 Mitch 155
#> 5 Rita 155
#> 6 Andrew 150
#> 7 Anita 150
#> 8 David 150
#> 9 Dean 150
#> 10 Felix 150Answer
We arrange the rows by (descending) wind speed, then group it by the name of storms, and select only the top row of each group:
#> [1] 214 13The resulting table st_max_wind contains only 214 rows.
Does this correspond to the number of unique storm names in st?
Let’s check:
Note a key difference between the two tables st_top10_wind and st_max_wind:
st_top10_wind is only a small summary table that answers our question from above, whereas st_max_wind is a much larger subset of the original data (and includes the same variables as st).
In fact, our summary information of st_top10_wind should be contained within st_max_wind.
Let’s check this: Inspecting wind_top10 shows four storms with wind speeds of at least 150 knots.
Do we obtain the same storms when filtering the table st_max_wind for these values?
The following expressions verify this by re-computing the top-10 storm names from the data in st_max_wind:
top10_2 <- st_max_wind %>%
filter(wind >= 150) %>%
select(name, wind) %>%
arrange(desc(wind))
top10_2#> # A tibble: 12 × 2
#> # Groups: name [12]
#> name wind
#> <chr> <int>
#> 1 Dorian 160
#> 2 Gilbert 160
#> 3 Wilma 160
#> 4 Mitch 155
#> 5 Rita 155
#> 6 Andrew 150
#> 7 Anita 150
#> 8 David 150
#> 9 Dean 150
#> 10 Felix 150
#> 11 Katrina 150
#> 12 Maria 150
all.equal(st_top10_wind$name, top10_2$name)#> [1] "Lengths (10, 12) differ (string compare on first 10)"Answer
Using a dplyr pipe to compute a grouped summary table t_w:
t_w <- st %>%
group_by(category) %>%
summarise(n = n(),
mn_wind = mean(wind))
knitr::kable(t_w, caption = "Mean wind speed of storms (from **dplyr**).")| category | n | mn_wind |
|---|---|---|
| -1 | 2898 | 27.49482 |
| 0 | 5347 | 45.66392 |
| 1 | 1934 | 70.95140 |
| 2 | 749 | 89.41923 |
| 3 | 434 | 104.48157 |
| 4 | 411 | 122.10462 |
| 5 | 86 | 145.58140 |
Using t_w to plot results with ggplot2:
ggplot(t_w, aes(x = category, y = mn_wind)) +
geom_point(aes(size = n), col = "firebrick") +
labs(tag = "A", title = "Wind speed by storm category",
x = "Storm category", y = "Wind speed (mean)") +
ylim(0, 150) +
theme_ds4psy()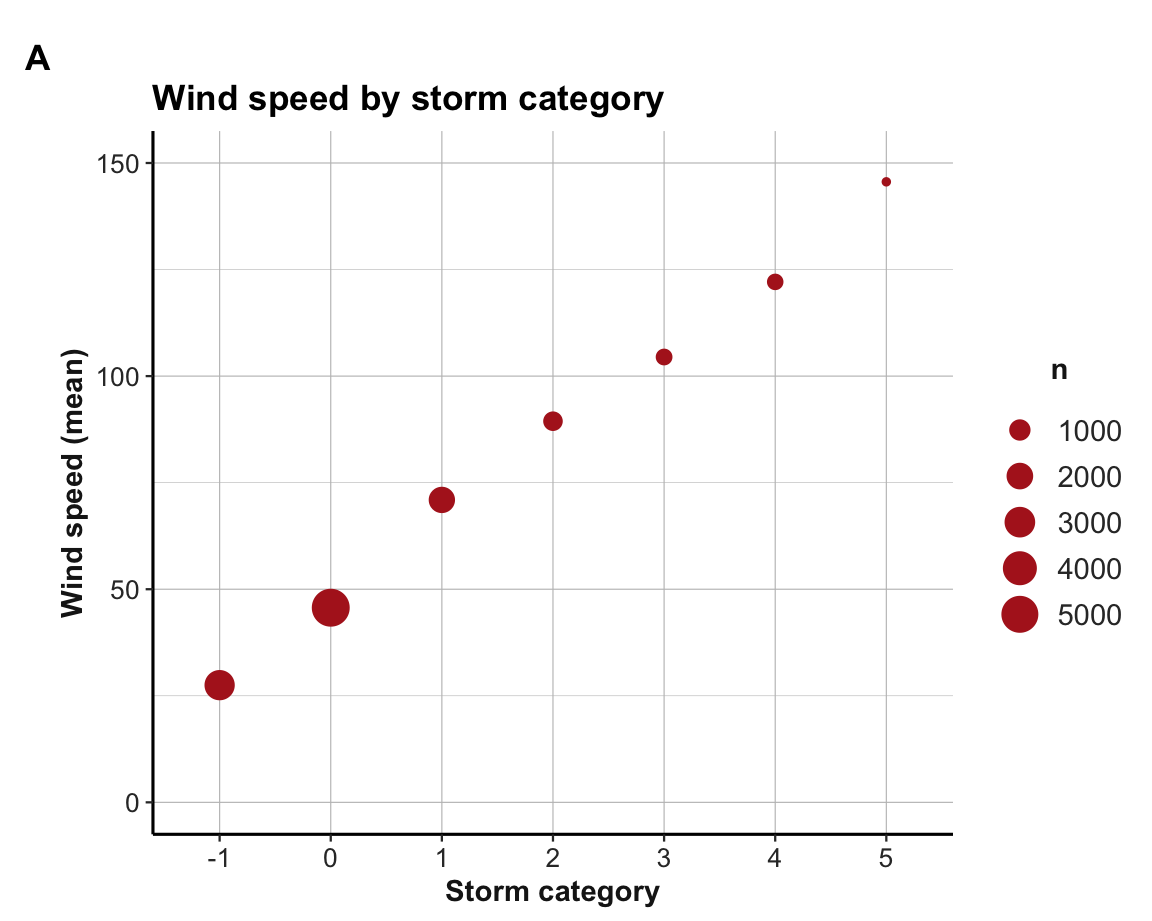
Note that the aesthetic mapping size = n expresses the frequency count as point size (i.e., visualizes a 2nd variable).
13.3.4 Practice: Using dplyr (and ggplot2)
The following exercises can be solved by pipes of dplyr and ggplot2 functions.
Exercise 1: Counting groups and visualizing two variables
Try answering the following questions by creating a dplyr pipe and a visualization that shows the solution:
- What was the average air pressure (in millibar) by storm category?
- How frequent is the corresponding storm category?
Solution
Using a dplyr pipe to compute a summary table t_p:
| category | n | mn_press |
|---|---|---|
| -1 | 2898 | 1007.5390 |
| 0 | 5347 | 999.2910 |
| 1 | 1934 | 981.1887 |
| 2 | 749 | 966.9359 |
| 3 | 434 | 953.9124 |
| 4 | 411 | 939.3942 |
| 5 | 86 | 917.4070 |
Using t_p to plot results with ggplot2:
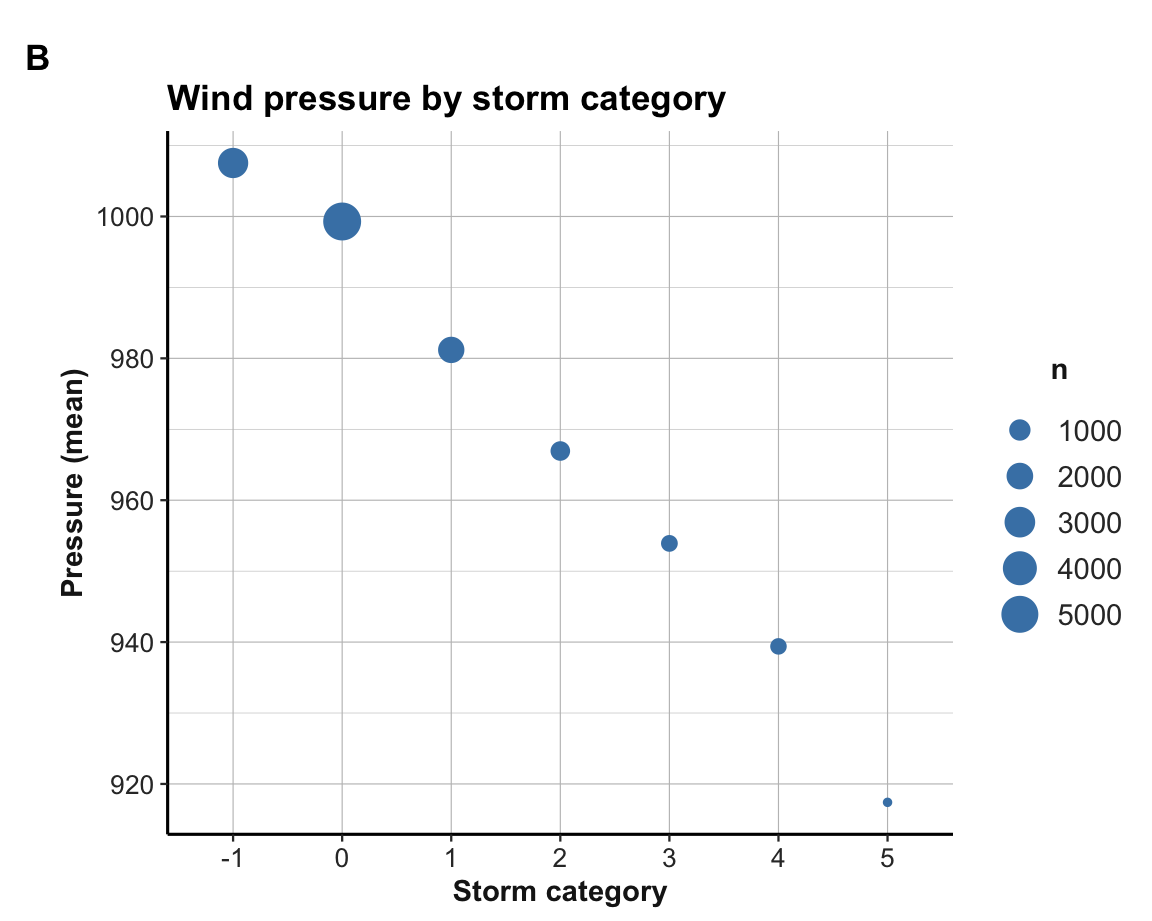
Exercise 2: Plotting storm counts per category and month
Use the data from dplyr::storms to show that there are specific storm seasons throughout the year.
- How many storms of each category were recorded in each month?
- Is there an observable trend of storm observations over the calendar year?
- To begin addressing these tasks, we could use a dplyr pipe to compute a grouped summary table that shows the number of storms per category and month:
#> # A tibble: 37 × 3
#> # Groups: month [10]
#> month category n
#> <dbl> <dbl> <int>
#> 1 1 1 5
#> 2 1 NA 65
#> 3 4 NA 66
#> 4 5 NA 201
#> 5 6 1 18
#> 6 6 NA 791
#> 7 7 1 148
#> 8 7 2 35
#> 9 7 3 19
#> 10 7 4 18
#> # ℹ 27 more rows- Try using
t_2to plot results with ggplot2:
The following plot aims to visualize our summary table t_2:
ggplot(t_2, aes(x = factor(month))) +
geom_bar(aes(fill = factor(category))) +
labs(title = "Storm counts by category per month",
x = "Month", y = "Number of observations") +
theme_ds4psy()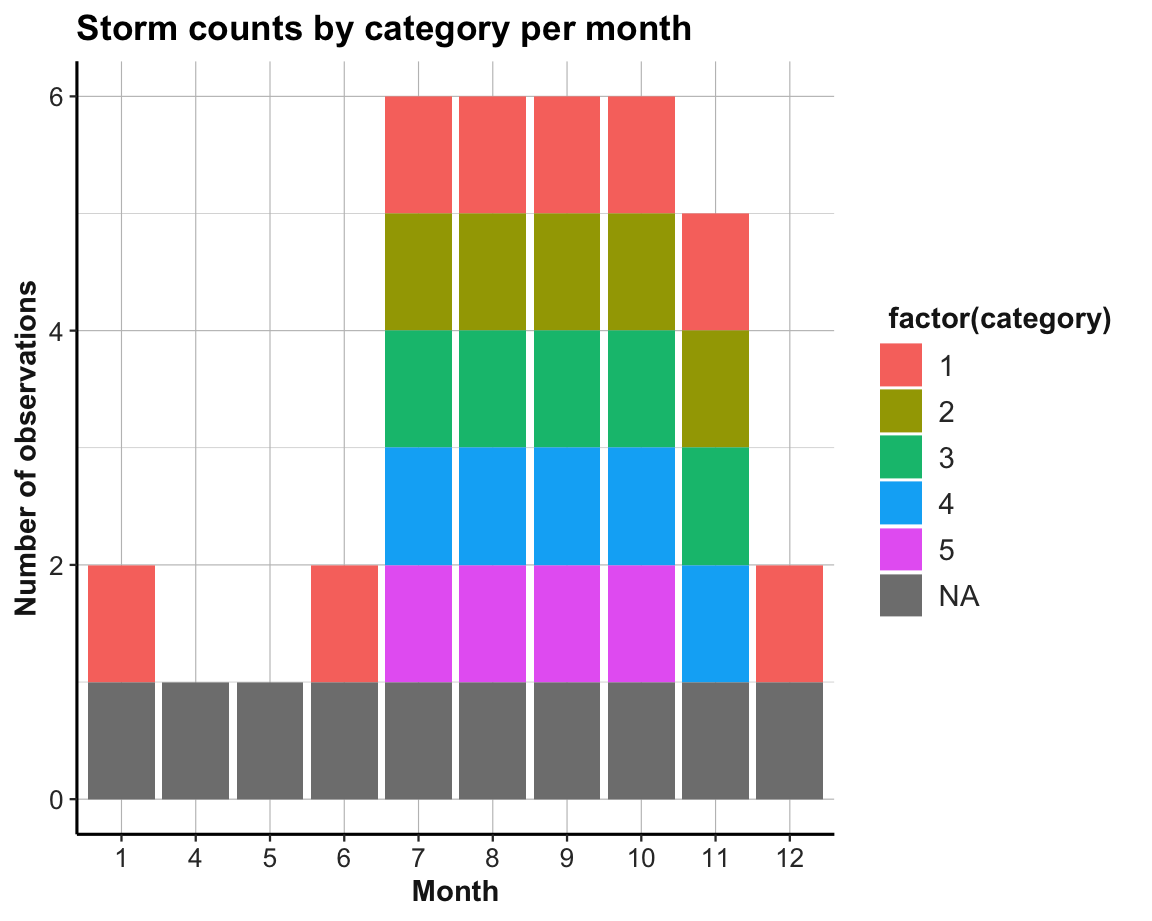
Figure 13.3: A misleading visualization: All storm category counts appear to be 0 or 1.
However, this plot seems wrong and is actually quite misleading.
- What’s the problem with it? How can it be fixed?
Solution
The problem is that the bar plot counted how often each category-month combination occurs in t_2 (with geom_bar() using its default stat = "count").
Thus, the only two possible counts for each combination of storm category and month are 0 (i.e., absence of a cell value) and 1 (i.e., presence of a cell value).
Here are two possible corrections of the misleading visualization:
- Use the
t_2data, butgeom_bar()usingy = nandstat = "identity":
ggplot(t_2, aes(x = factor(month))) +
geom_bar(aes(y = n, fill = category), stat = "identity") +
labs(title = "Storm counts by category per month",
x = "Month", y = "Number of observations") +
theme_ds4psy()- Use the raw data of
standgeom_bar()with default settings (i.e.,stat = "count"):
ggplot(st, aes(x = factor(month))) +
geom_bar(aes(fill = category, group = category)) +
labs(title = "Storm counts by category per month",
x = "Month", y = "Number of observations") +
theme_ds4psy()- An alternative way to address this task may be to compute a better summary table (showing the counts of storms by category and month). The following solution uses tidyr functions (discussed in Chapter 14) to first re-shape the summary table
t_2from its long to a wider format:
| month | NA | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|---|
| 1 | 65 | 5 | 0 | 0 | 0 | 0 |
| 4 | 66 | 0 | 0 | 0 | 0 | 0 |
| 5 | 201 | 0 | 0 | 0 | 0 | 0 |
| 6 | 791 | 18 | 0 | 0 | 0 | 0 |
| 7 | 1430 | 148 | 35 | 19 | 18 | 1 |
| 8 | 3404 | 581 | 198 | 113 | 114 | 32 |
| 9 | 5314 | 1145 | 581 | 359 | 309 | 70 |
| 10 | 2335 | 466 | 150 | 86 | 88 | 13 |
| 11 | 949 | 152 | 29 | 16 | 24 | 0 |
| 12 | 179 | 33 | 0 | 0 | 0 | 0 |
Interpretation
Both visualizations and the summary table suggest the same trend over the calendar year: The number of storms in all categories (including
NA) increase in spring, peak in late summer (September), and rapidly decrease afterwards.Curiously, there are zero observations in the data in months 2 and 3 (i.e., February and March).
A crucial limitation of all these solutions is that we were counting observations (i.e., rows) in our data, rather than storms. As some storms occur more often than others, the number of observations does not correspond to the number of storms per category.
Exercise 3: Names of re-occuring storms
Identify all storms in
stthat were observed in more than one year.There are two ways in which a storm
namecan appear in more than one year:
- A single storm may occur in multiple years (e.g., from December to January)
- A storm
nameis used repeatedly (i.e., to name different storms in different years)
Check how often each of these cases occurs.
Solution
ad 1.: Grouping by the name and year variables,
we first count the number of times a storm occurs per year.
We then group by name to count storms occurring in more than one year.
st %>%
select(name, year) %>%
group_by(name, year) %>%
count() %>%
# select(name, year) %>%
group_by(name) %>%
count() %>%
filter(n > 1) %>%
head() #> # A tibble: 6 × 2
#> # Groups: name [6]
#> name n
#> <chr> <int>
#> 1 Alberto 7
#> 2 Alex 4
#> 3 Allison 3
#> 4 Ana 7
#> 5 Andrew 2
#> 6 Arthur 7
# tail()Verify these results for some storms:
- Does “Alberto” really occur in 6 different years?
st %>%
filter(name == "Alberto") %>%
group_by(name, year) %>%
count()
#> # A tibble: 7 × 3
#> # Groups: name, year [7]
#> name year n
#> <chr> <dbl> <int>
#> 1 Alberto 1982 17
#> 2 Alberto 1988 11
#> 3 Alberto 1994 32
#> 4 Alberto 2000 79
#> 5 Alberto 2006 18
#> 6 Alberto 2012 13
#> # … with 1 more row
#> # ℹ Use `print(n = ...)` to see more rows- Does “Ana” really occur in 7 different years?
st %>%
filter(name == "Ana") %>%
group_by(name, year) %>%
count()
#> # A tibble: 7 × 3
#> # Groups: name, year [7]
#> name year n
#> <chr> <dbl> <int>
#> 1 Ana 1979 19
#> 2 Ana 1985 14
#> 3 Ana 1991 12
#> 4 Ana 1997 15
#> 5 Ana 2003 13
#> 6 Ana 2009 15
#> # … with 1 more row
#> # ℹ Use `print(n = ...)` to see more rows- Does “Zeta” really occur in 3 different years?
13.3.5 Other dplyr functions
Our introduction of dplyr in this section only focused on a small set of key functions. Although these functions will suffice for the majority of our daily data transformation tasks, some omissions will briefly be mentioned here.
Merging data tables with *join*()
A limitation of the dplyr functions that we have seen so far is that they all referred to one data table. When we need to combine or merge variables (columns) from two distinct data tables, additional two-table functions are required.
Whenever the two tables contain different cases (rows), simply pasting together the columns of two tables (e.g, by using cbind()) is no longer an option. Instead, we need to recruit some concepts from database management to describe which variables in one table correspond to those of the other table and must specify the particular way in which we want to join the pair of tables. Typical notions occurring in this context are “full join” or “left join”, and terms for set operations like “union”, “intersection”, or “set difference”.
Whereas the base R function merge() can handle many such cases, a dedicated set of dplyr functions containing the keyword join provides tailored solutions to many special cases.
See the dplyr vignette on Two-table verbs or
Chapter 8: Joining data for an overview of these functions.
Re-naming variables with rename()
When we need to change the names of variables (i.e., of the columns of a data frame or tibble), the rename() function is useful.
In its basic form, it uses combinations of new_name = old_name to rename variables.
For instance, we could change the status variable to a type variable and simplify two long variable names of the st data as follows:
st |>
# Rename 3 variables (by new_name = old_name pairs):
rename(type = status,
trop_diam = tropicalstorm_force_diameter,
hurr_diam = hurricane_force_diameter) |>
# Reduce and print table:
select(name, year, type, trop_diam, hurr_diam) |>
slice_sample(n = 10) |>
knitr::kable()| name | year | type | trop_diam | hurr_diam |
|---|---|---|---|---|
| Marilyn | 1995 | tropical storm | NA | NA |
| Danielle | 1998 | hurricane | NA | NA |
| Henri | 1979 | tropical depression | NA | NA |
| Leslie | 2018 | tropical storm | 380 | 0 |
| Humberto | 2013 | tropical storm | 180 | 0 |
| Sean | 2011 | tropical storm | 250 | 0 |
| Dean | 1995 | tropical depression | NA | NA |
| Chantal | 2001 | tropical storm | NA | NA |
| Bonnie | 1998 | hurricane | NA | NA |
| Alicia | 1983 | tropical depression | NA | NA |
Note that we would need to re-assign st (to st itself) to permanently drop the old and keep only the new names.
For changing all variable names of a table in the same fashion, the rename_with() function allows directly applying a function to variable names.
For instance, to change all uppercase characters in variable names to lowercase characters, we could use:
# Example:
names(iris) # contains uppercase names
#> [1] "Sepal.Length" "Sepal.Width" "Petal.Length" "Petal.Width" "Species"
# Rename variables by applying a function:
iris |>
# Apply tolower() to all names:
rename_with(tolower) |>
# Print only the names:
names()
#> [1] "sepal.length" "sepal.width" "petal.length" "petal.width" "species"More complicated re-naming scenarios can specify sets of variables in combination with quantifiers (e.g., see the examples of ?rename() using a lookup vector and the any_of() or all_of() modifiers).
13.3.6 Summary and evaluation
This concludes our summary of essential dplyr functions (Wickham, François, et al., 2023). To sum up:
Summary: Key dplyr functions (Wickham, François, et al., 2023) include:
-
arrange()sorts cases (rows); -
filter()andslice()select cases (rows) by logical conditions; -
select()selects and reorders variables (columns); -
mutate()andtransmute()compute new variables (columns) out of existing ones; -
summarise()collapses multiple values of a variable (rows of a column) to a single one;
-
group_by()changes the unit of aggregation (in combination withmutate()andsummarise()).
Other useful dplyr functions include variants of rename() and the two-table *join*() functions.
In our evaluation of the pipe operators %>% and |> (in Section 13.2.3 above), we compared their effects to introducing a new notation and emphasized that they provide no new functionality. Similarly, all tasks addressed by dplyr functions could also be performed by clever combinations of base R functions (e.g., by indexing, aggregate(), and tapply()).
With respect to the enthusiasm expressed in the quote above (in Section 13.2), this raises the question:
- How can two tools that provide nothing new create something awesome?
Just as we saw for the pipe operator, the beauty and power of dplyr functions derives from their ecological rationality (see Section 1.2.5). Being specifically designed to address simple data transformation tasks, they match the way we usually think about data. Thus, rather than requiring us to re-shape our thinking to the logic of base R, dplyr provides a set of handy tools that aim to do what we want with a minimum of fuzz. While many users find the dplyr functions easier and more natural to use than the corresponding base R functions, which tool we prefer is mostly a matter of experience or of familiarity.
Overall, combining dplyr functions into pipes provides very flexible and powerful tools for transforming (i.e., reshaping and reducing) data. Next, we will encounter additional functions for data transformation from the tidyr package (in Chapter 14). Whereas many of the dplyr pipes of this chapter reduced our data (e.g., when filtering, selecting, or summarizing variables), the primary purpose of tidyr functions is to re-shape data tables into “tidy” data.
13.4 Conclusion
13.4.1 Summary
This chapter first distinguished between two ways of transforming data: Whereas reshaping data changes the form of data, reducing data typically changes both form and content.
We then introduced the %>% operator of the magrittr package (Bache & Wickham, 2025) and its base R cousin |>.
Both operators allow chaining sequences of function calls into powerful pipes.
The dplyr package (Wickham, François, et al., 2023) provides a set of useful functions for reshaping and reducing data. In combination with ggplot2 and tidyr, dplyr is a key pillar of the tidyverse (Wickham et al., 2019).
Summary: Data transformation reshapes or reduces data.
The magrittr pipe operator %>% and its base R cousin |> turn nested expressions into sequential expressions (Bache & Wickham, 2025).
Key dplyr functions (Wickham, François, et al., 2023) include:
-
arrange()sorts cases (rows); -
filter()andslice()select cases (rows) by logical conditions; -
select()selects and reorders variables (columns); -
mutate()andtransmute()compute new variables (columns) out of existing ones; -
summarise()collapses multiple values of a variable (rows of a column) to a single one;
-
group_by()changes the unit of aggregation (in combination withmutate()andsummarise()).
Other useful dplyr functions include variants of rename() and the two-table *join*() functions.
13.4.2 Resources
Here are some pointers to related articles, cheatsheets, and additional links:
Different pipes
For details on and the differences between the magrittr pipe operator %>% and R’s native pipe operator |>, see
New features in R 4.1.0 (by Russ Hyde, 2021-05-17)
The new R pipe (by Elio Campitelli, 2021-05-25)
Differences between the base R and magrittr pipes (by Hadley Wickham, 2023-04-21)
Despite their differences, we can regard both pipe operators as interchangeable for most practical purposes.
On dplyr
- See the cheatsheet on transforming data with dplyr from Posit cheatsheets:
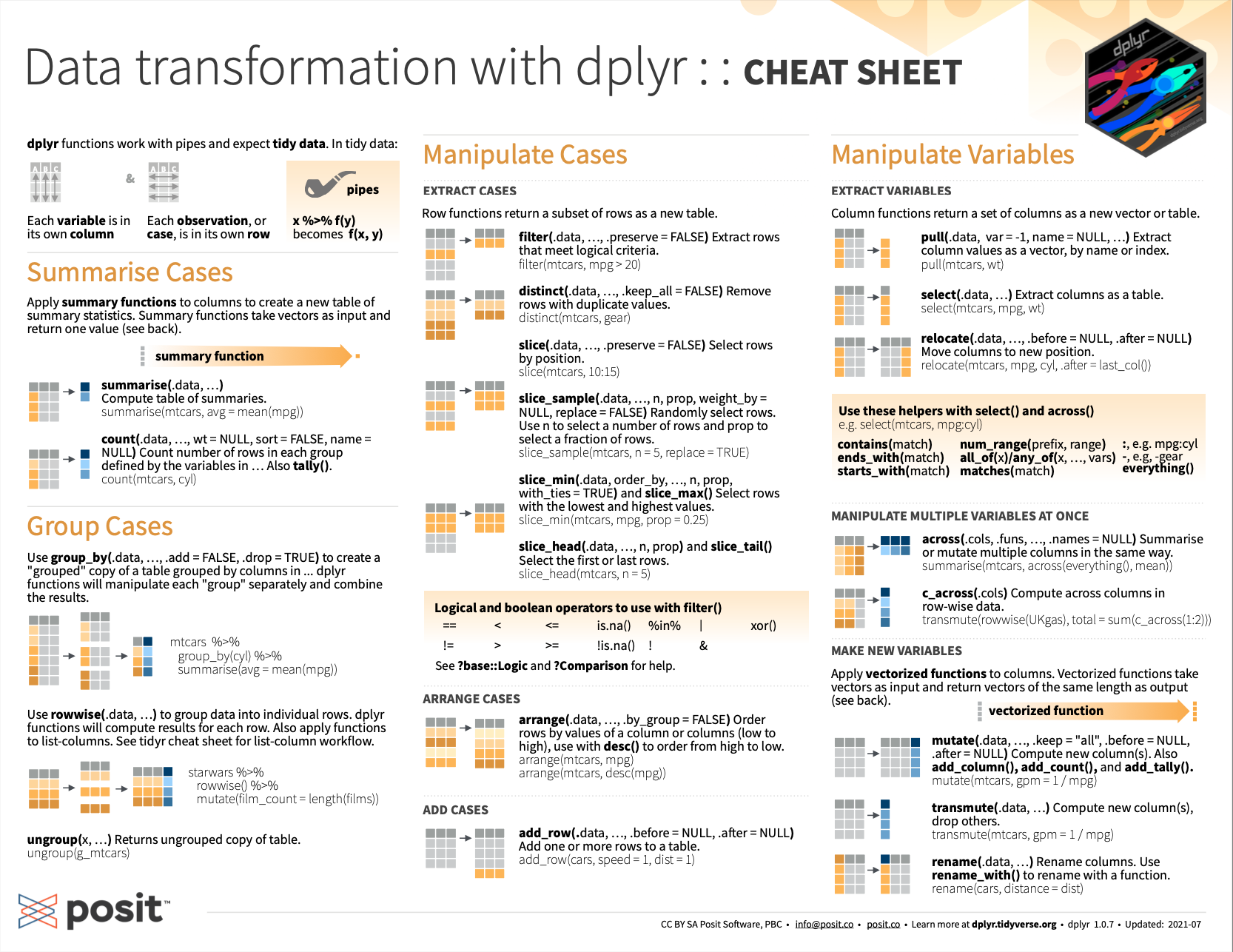
Figure 13.4: Data transformation with dplyr from Posit cheatsheets.
- Introduction on dplyr and pipes: The basics (by Sean C. Anderson, 2014-09-13)
13.4.3 Preview
Having learned how to create data transformation pipes by magrittr and dplyr, the next Chapter 14) on tidying data will expand these skills by adding tools from the tidyr package.
13.5 Exercises
To check our knowledge on dplyr functions and pipes for data transformation, we distinguish between conceptual and practical exercises.
13.5.1 Reshaping vs. reducing data
The essential dplyr functions perform data transformation tasks. Discuss the specific purpose of each function in terms of reshaping or reducing data (as introduced in Section 13.1).
13.5.2 Replacing dplyr functions by base R functionality
Discuss how each of the essential dplyr functions introduced in this chapter could be replaced by using base R functionality.
Hint: As there is no 1:1-correspondence between functions, identify the task performed by each function before thinking about alternative ways of tackling these tasks.
Practical exercises
The following exercises on transforming data with dplyr pipes (Wickham, François, et al., 2023) use a variety of datasets. The links point to corresponding exercises of Chapter 3: Transforming data of the ds4psy book (Neth, 2025a).


