3.2 Essential dplyr commands
Assuming that the dplyr package (Wickham, François, et al., 2023) is a toolbox for tackling various Rwars challenges, we can ask: Which specific tools does it provide and which tasks are addressed by them? In the context of this chapter, the following dplyr functions are essential for reshaping and reducing data:
arrange()sorts cases (rows);filter()andslice()select cases (rows) by logical conditions or number;select()selects and reorders variables (columns);mutate()computes new variables (columns) and adds them to the existing ones;summarise()collapses multiple values of a variable (rows of a column) to a single one;
group_by()andungroup()change the unit of aggregation (in combination withmutate()andsummarise()).
The following sections illustrate each of these commands in the context of examples.
To keep things simple and entertaining, we use the toy dataset of sw <- dplyr::starwars to introduce the commands, but will use additional datasets in the exercises (in Section 3.5):
3.2.1 arrange() sorts rows
Using arrange() sorts cases (rows) by putting specific variables (columns) in specific orders (e.g., ascending or descending).
For instance, we could want to arrange cases (rows) by the name of individuals (in alphabetical order).
The dplyr function arrange() let’s us do this by calling:
Before we proceed, two simple observations will facilitate our future life a lot:
- In R, we can generally omit argument names (as long as the order of arguments makes it clear what is meant). Thus, we can write the same command more easily as:
- In dplyr and other tidyverse packages, we can rewrite commands by using the so-called pipe (written by the symbols
%>%) of the magrittr package (Bache & Wickham, 2025):
Think of the pipe as passing whatever is on its left (here: sw) to the first argument of the function on its right (here: .data).
(More details about using the pipe operator are provided below in Section 3.3.)
In other words, the last three commands (a), (b), and (c) are identical and yield the same output:
#> # A tibble: 87 × 14
#> name height mass hair_…¹ skin_…² eye_c…³ birth…⁴ sex gender homew…⁵
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 Ackbar 180 83 none brown … orange 41 male mascu… Mon Ca…
#> 2 Adi Gallia 184 50 none dark blue NA fema… femin… Corusc…
#> 3 Anakin Sky… 188 84 blond fair blue 41.9 male mascu… Tatooi…
#> 4 Arvel Cryn… NA NA brown fair brown NA male mascu… <NA>
#> 5 Ayla Secura 178 55 none blue hazel 48 fema… femin… Ryloth
#> 6 Bail Prest… 191 NA black tan brown 67 male mascu… Aldera…
#> 7 Barriss Of… 166 50 black yellow blue 40 fema… femin… Mirial
#> 8 BB8 NA NA none none black NA none mascu… <NA>
#> 9 Ben Quadin… 163 65 none grey, … orange NA male mascu… Tund
#> 10 Beru White… 165 75 brown light blue 47 fema… femin… Tatooi…
#> # … with 77 more rows, 4 more variables: species <chr>, films <list>,
#> # vehicles <list>, starships <list>, and abbreviated variable names
#> # ¹hair_color, ²skin_color, ³eye_color, ⁴birth_year, ⁵homeworldThis output contains the tibble sw, but arranged the rows alphabetically by the variable name, which is exactly what we wanted.
Although this is neat, two immediate questions are:
How can we arrange rows in different (e.g., descending, rather than ascending) orders?
How can we arrange rows by more than one variable?
Both of these tasks are solved rather intuitively by adjusting our calls to arrange():
#> # A tibble: 87 × 14
#> name height mass hair_…¹ skin_…² eye_c…³ birth…⁴ sex gender homew…⁵
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 Zam Wesell 168 55 blonde fair, … yellow NA fema… femin… Zolan
#> 2 Yoda 66 17 white green brown 896 male mascu… <NA>
#> 3 Yarael Poof 264 NA none white yellow NA male mascu… Quermia
#> 4 Wilhuff Ta… 180 NA auburn… fair blue 64 male mascu… Eriadu
#> 5 Wicket Sys… 88 20 brown brown brown 8 male mascu… Endor
#> 6 Wedge Anti… 170 77 brown fair hazel 21 male mascu… Corell…
#> 7 Watto 137 NA black blue, … yellow NA male mascu… Toydar…
#> 8 Wat Tambor 193 48 none green,… unknown NA male mascu… Skako
#> 9 Tion Medon 206 80 none grey black NA male mascu… Utapau
#> 10 Taun We 213 NA none grey black NA fema… femin… Kamino
#> # … with 77 more rows, 4 more variables: species <chr>, films <list>,
#> # vehicles <list>, starships <list>, and abbreviated variable names
#> # ¹hair_color, ²skin_color, ³eye_color, ⁴birth_year, ⁵homeworld#> # A tibble: 87 × 14
#> name height mass hair_c…¹ skin_…² eye_c…³ birth…⁴ sex gender homew…⁵
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 Taun We 213 NA none grey black NA fema… femin… Kamino
#> 2 Shaak Ti 178 57 none red, b… black NA fema… femin… Shili
#> 3 Lama Su 229 88 none grey black NA male mascu… Kamino
#> 4 Tion Medon 206 80 none grey black NA male mascu… Utapau
#> 5 Kit Fisto 196 87 none green black NA male mascu… Glee A…
#> 6 Plo Koon 188 80 none orange black 22 male mascu… Dorin
#> 7 Greedo 173 74 <NA> green black 44 male mascu… Rodia
#> 8 Nien Nunb 160 68 none grey black NA male mascu… Sullust
#> 9 Gasgano 122 NA none white,… black NA male mascu… Troiken
#> 10 BB8 NA NA none none black NA none mascu… <NA>
#> # … with 77 more rows, 4 more variables: species <chr>, films <list>,
#> # vehicles <list>, starships <list>, and abbreviated variable names
#> # ¹hair_color, ²skin_color, ³eye_color, ⁴birth_year, ⁵homeworldSee ?dplyr::arrange for more help and additional examples.
Details
Note some details on using arrange() in the above examples:
All basic dplyr commands can be called as
verb(.data, ...)or — by using the pipe operator from magrittr — as.data %>% verb(...)(seevignette("magrittr")for details). Importantly, the pipe operator%>%is different from the+operator used inggplotcalls.In contrast to base R commands, sequences of multiple variables in tidyverse commands can be written as comma-separated variables, rather than as vectors of variable names (e.g.,
c("gender", "height")) and are unquoted.When specifying multiple variables in
arrange, their order (x, y, ...) specifies the order or priority of operations (first byx, then byy, etc.).
3.2.2 filter() selects rows
Using filter() selects cases (rows) by logical conditions or a criterion.
It keeps all rows for which the criterion is TRUE and drops all rows for which the criterion is FALSE or NA.
For instance, two identical ways to extract all humans from sw are:
# Filter to keep all humans:
filter(sw, species == "Human")
# The same command using the pipe:
sw %>% # Note: %>% is NOT + (used in ggplot)
filter(species == "Human")and result in:
#> # A tibble: 35 × 14
#> name height mass hair_…¹ skin_…² eye_c…³ birth…⁴ sex gender homew…⁵
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 Luke Skywa… 172 77 blond fair blue 19 male mascu… Tatooi…
#> 2 Darth Vader 202 136 none white yellow 41.9 male mascu… Tatooi…
#> 3 Leia Organa 150 49 brown light brown 19 fema… femin… Aldera…
#> 4 Owen Lars 178 120 brown,… light blue 52 male mascu… Tatooi…
#> 5 Beru White… 165 75 brown light blue 47 fema… femin… Tatooi…
#> 6 Biggs Dark… 183 84 black light brown 24 male mascu… Tatooi…
#> 7 Obi-Wan Ke… 182 77 auburn… fair blue-g… 57 male mascu… Stewjon
#> 8 Anakin Sky… 188 84 blond fair blue 41.9 male mascu… Tatooi…
#> 9 Wilhuff Ta… 180 NA auburn… fair blue 64 male mascu… Eriadu
#> 10 Han Solo 180 80 brown fair brown 29 male mascu… Corell…
#> # … with 25 more rows, 4 more variables: species <chr>, films <list>,
#> # vehicles <list>, starships <list>, and abbreviated variable names
#> # ¹hair_color, ²skin_color, ³eye_color, ⁴birth_year, ⁵homeworldTo filter by some criterion (here: a test that determines whether species == "Human" is TRUE or FALSE), we needed to know both the variable by which we wanted to filter (here: species) and its value of interest (here: "Human"). Note that the output of applying filter to sw is a new tibble, but this tibble only contains 35 cases (i.e., the humans from sw).
Filtering by more than one condition can be very effective, but requires some knowledge about logical operators:
# Filter by multiple (additive) conditions:
sw %>%
filter(height > 180, mass <= 75) # tall and light individuals#> # A tibble: 3 × 14
#> name height mass hair_…¹ skin_…² eye_c…³ birth…⁴ sex gender homew…⁵
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 Jar Jar Bin… 196 66 none orange orange 52 male mascu… Naboo
#> 2 Adi Gallia 184 50 none dark blue NA fema… femin… Corusc…
#> 3 Wat Tambor 193 48 none green,… unknown NA male mascu… Skako
#> # … with 4 more variables: species <chr>, films <list>, vehicles <list>,
#> # starships <list>, and abbreviated variable names ¹hair_color, ²skin_color,
#> # ³eye_color, ⁴birth_year, ⁵homeworld# The same command using the logical operator (&):
sw %>%
filter(height > 180 & mass <= 75) # tall and light individuals#> # A tibble: 3 × 14
#> name height mass hair_…¹ skin_…² eye_c…³ birth…⁴ sex gender homew…⁵
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 Jar Jar Bin… 196 66 none orange orange 52 male mascu… Naboo
#> 2 Adi Gallia 184 50 none dark blue NA fema… femin… Corusc…
#> 3 Wat Tambor 193 48 none green,… unknown NA male mascu… Skako
#> # … with 4 more variables: species <chr>, films <list>, vehicles <list>,
#> # starships <list>, and abbreviated variable names ¹hair_color, ²skin_color,
#> # ³eye_color, ⁴birth_year, ⁵homeworld# Filter for a range of a specific variable:
sw %>%
filter(height >= 150, height <= 165) # (a) using height twice#> # A tibble: 9 × 14
#> name height mass hair_…¹ skin_…² eye_c…³ birth…⁴ sex gender homew…⁵
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 Leia Organa 150 49 brown light brown 19 fema… femin… Aldera…
#> 2 Beru Whites… 165 75 brown light blue 47 fema… femin… Tatooi…
#> 3 Mon Mothma 150 NA auburn fair blue 48 fema… femin… Chandr…
#> 4 Nien Nunb 160 68 none grey black NA male mascu… Sullust
#> 5 Shmi Skywal… 163 NA black fair brown 72 fema… femin… Tatooi…
#> 6 Ben Quadina… 163 65 none grey, … orange NA male mascu… Tund
#> 7 Cordé 157 NA brown light brown NA fema… femin… Naboo
#> 8 Dormé 165 NA brown light brown NA fema… femin… Naboo
#> 9 Padmé Amida… 165 45 brown light brown 46 fema… femin… Naboo
#> # … with 4 more variables: species <chr>, films <list>, vehicles <list>,
#> # starships <list>, and abbreviated variable names ¹hair_color, ²skin_color,
#> # ³eye_color, ⁴birth_year, ⁵homeworld#> # A tibble: 9 × 14
#> name height mass hair_…¹ skin_…² eye_c…³ birth…⁴ sex gender homew…⁵
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 Leia Organa 150 49 brown light brown 19 fema… femin… Aldera…
#> 2 Beru Whites… 165 75 brown light blue 47 fema… femin… Tatooi…
#> 3 Mon Mothma 150 NA auburn fair blue 48 fema… femin… Chandr…
#> 4 Nien Nunb 160 68 none grey black NA male mascu… Sullust
#> 5 Shmi Skywal… 163 NA black fair brown 72 fema… femin… Tatooi…
#> 6 Ben Quadina… 163 65 none grey, … orange NA male mascu… Tund
#> 7 Cordé 157 NA brown light brown NA fema… femin… Naboo
#> 8 Dormé 165 NA brown light brown NA fema… femin… Naboo
#> 9 Padmé Amida… 165 45 brown light brown 46 fema… femin… Naboo
#> # … with 4 more variables: species <chr>, films <list>, vehicles <list>,
#> # starships <list>, and abbreviated variable names ¹hair_color, ²skin_color,
#> # ³eye_color, ⁴birth_year, ⁵homeworld# Filter by multiple (alternative) conditions:
sw %>%
filter(homeworld == "Kashyyyk" | skin_color == "green")#> # A tibble: 8 × 14
#> name height mass hair_…¹ skin_…² eye_c…³ birth…⁴ sex gender homew…⁵
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 Chewbacca 228 112 brown unknown blue 200 male mascu… Kashyy…
#> 2 Greedo 173 74 <NA> green black 44 male mascu… Rodia
#> 3 Yoda 66 17 white green brown 896 male mascu… <NA>
#> 4 Bossk 190 113 none green red 53 male mascu… Trando…
#> 5 Rugor Nass 206 NA none green orange NA male mascu… Naboo
#> 6 Kit Fisto 196 87 none green black NA male mascu… Glee A…
#> 7 Poggle the … 183 80 none green yellow NA male mascu… Geonos…
#> 8 Tarfful 234 136 brown brown blue NA male mascu… Kashyy…
#> # … with 4 more variables: species <chr>, films <list>, vehicles <list>,
#> # starships <list>, and abbreviated variable names ¹hair_color, ²skin_color,
#> # ³eye_color, ⁴birth_year, ⁵homeworldA common criterion for filtering is that we want to (a) only obtain cases with missing values on some variable(s), or (b) only keep cases without missing values on some variable(s):
#> # A tibble: 4 × 14
#> name height mass hair_…¹ skin_…² eye_c…³ birth…⁴ sex gender homew…⁵
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 Ric Olié 183 NA brown fair blue NA <NA> <NA> Naboo
#> 2 Quarsh Pana… 183 NA black dark brown 62 <NA> <NA> Naboo
#> 3 Sly Moore 178 48 none pale white NA <NA> <NA> Umbara
#> 4 Captain Pha… NA NA unknown unknown unknown NA <NA> <NA> <NA>
#> # … with 4 more variables: species <chr>, films <list>, vehicles <list>,
#> # starships <list>, and abbreviated variable names ¹hair_color, ²skin_color,
#> # ³eye_color, ⁴birth_year, ⁵homeworld# (b) Filter cases with existing (non-NA) values on specific variables:
sw %>%
filter(!is.na(mass), !is.na(birth_year)) #> # A tibble: 36 × 14
#> name height mass hair_…¹ skin_…² eye_c…³ birth…⁴ sex gender homew…⁵
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 Luke Skywa… 172 77 blond fair blue 19 male mascu… Tatooi…
#> 2 C-3PO 167 75 <NA> gold yellow 112 none mascu… Tatooi…
#> 3 R2-D2 96 32 <NA> white,… red 33 none mascu… Naboo
#> 4 Darth Vader 202 136 none white yellow 41.9 male mascu… Tatooi…
#> 5 Leia Organa 150 49 brown light brown 19 fema… femin… Aldera…
#> 6 Owen Lars 178 120 brown,… light blue 52 male mascu… Tatooi…
#> 7 Beru White… 165 75 brown light blue 47 fema… femin… Tatooi…
#> 8 Biggs Dark… 183 84 black light brown 24 male mascu… Tatooi…
#> 9 Obi-Wan Ke… 182 77 auburn… fair blue-g… 57 male mascu… Stewjon
#> 10 Anakin Sky… 188 84 blond fair blue 41.9 male mascu… Tatooi…
#> # … with 26 more rows, 4 more variables: species <chr>, films <list>,
#> # vehicles <list>, starships <list>, and abbreviated variable names
#> # ¹hair_color, ²skin_color, ³eye_color, ⁴birth_year, ⁵homeworldAs filter() selects cases, its result should typically be a table with the same number of columns as the original one, but fewer rows.
See ?dplyr::filter for more help and additional examples.
Details
Note some details on using filter:
Separating multiple conditions by commas is the same as using the logical AND (
&).As seen with
arrange(), variable names are unquoted.A comma between conditions or tests (
x, y, ...) means the same as&(logical AND), as each test results in a vector of Boolean values.Unlike in base R, rows for which the condition evaluates to
NAare dropped.Additional filter functions include
near()for testing numerical (near-)identity.
Practice
Here are some exercises to practice the combination of filter() and arrange() in dplyr pipes:
- Verify for an example that filtering by two criteria yields the same result as filtering twice (once for each criterion).
# (a) Filtering by 2 criteria:
tb1 <- sw %>%
filter(height >= 150, height <= 165) %>%
arrange(name)
# (b) Filtering first by criterion 1, then by criterion 2:
tb2 <- sw %>%
filter(height >= 150) %>%
filter(height <= 165) %>%
arrange(name)
# (c) Filtering first by criterion 2, then by criterion 1:
tb3 <- sw %>%
filter(height <= 165) %>%
filter(height >= 150) %>%
arrange(name)
# Verify equality (of name variable):
all.equal(tb1$name, tb2$name)#> [1] TRUE#> [1] TRUECan you explain why we added arrange(name) to the end of each filter pipe?
- Use
filter()on theswdata to select some either diverse or narrow subset of individuals. For instance,
- which individual with blond hair and blue eyes has an unknown mass?
- of which species are individuals that are over 2m tall and have brown hair?
- which individuals from Tatooine are not male (according to
sex) but of masculinegender?
- which individuals do neither identify as masculine nor as feminine (according to
gender) OR are heavier than 150kg?
sw %>%
filter(hair_color == "blond", eye_color == "blue")
sw %>%
filter(height > 200, hair_color == "brown")
sw %>%
filter(homeworld == "Tatooine", sex != "male", gender == "masculine")
sw %>%
filter((gender != "masculine" & gender != "feminine") | mass > 150)Note that the sw data distinguishes between an individual’s sex and gender.
slice() selects rows by number or value
If we want to select specific rows of a data table and already know their row number, we can use the slice() command of dplyr:
#> # A tibble: 1 × 14
#> name height mass hair_color skin_color eye_co…¹ birth…² sex gender homew…³
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 C-3PO 167 75 <NA> gold yellow 112 none mascu… Tatooi…
#> # … with 4 more variables: species <chr>, films <list>, vehicles <list>,
#> # starships <list>, and abbreviated variable names ¹eye_color, ²birth_year,
#> # ³homeworld#> # A tibble: 3 × 14
#> name height mass hair_…¹ skin_…² eye_c…³ birth…⁴ sex gender homew…⁵
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 Luke Skywal… 172 77 blond fair blue 19 male mascu… Tatooi…
#> 2 C-3PO 167 75 <NA> gold yellow 112 none mascu… Tatooi…
#> 3 R2-D2 96 32 <NA> white,… red 33 none mascu… Naboo
#> # … with 4 more variables: species <chr>, films <list>, vehicles <list>,
#> # starships <list>, and abbreviated variable names ¹hair_color, ²skin_color,
#> # ³eye_color, ⁴birth_year, ⁵homeworld#> # A tibble: 2 × 14
#> name height mass hair_…¹ skin_…² eye_c…³ birth…⁴ sex gender homew…⁵
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 Luke Skywal… 172 77 blond fair blue 19 male mascu… Tatooi…
#> 2 Padmé Amida… 165 45 brown light brown 46 fema… femin… Naboo
#> # … with 4 more variables: species <chr>, films <list>, vehicles <list>,
#> # starships <list>, and abbreviated variable names ¹hair_color, ²skin_color,
#> # ³eye_color, ⁴birth_year, ⁵homeworldSeveral variants of slice() allow selecting a number n or proportion prop of all cases from the head or tail of a table, draw random samples of rows, or select the case(s) with the highest or lowest values on some variable:
#> # A tibble: 4 × 14
#> name height mass hair_…¹ skin_…² eye_c…³ birth…⁴ sex gender homew…⁵
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 Luke Skywal… 172 77 blond fair blue 19 male mascu… Tatooi…
#> 2 C-3PO 167 75 <NA> gold yellow 112 none mascu… Tatooi…
#> 3 R2-D2 96 32 <NA> white,… red 33 none mascu… Naboo
#> 4 Darth Vader 202 136 none white yellow 41.9 male mascu… Tatooi…
#> # … with 4 more variables: species <chr>, films <list>, vehicles <list>,
#> # starships <list>, and abbreviated variable names ¹hair_color, ²skin_color,
#> # ³eye_color, ⁴birth_year, ⁵homeworld#> # A tibble: 3 × 14
#> name height mass hair_…¹ skin_…² eye_c…³ birth…⁴ sex gender homew…⁵
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 BB8 NA NA none none black NA none mascu… <NA>
#> 2 Captain Pha… NA NA unknown unknown unknown NA <NA> <NA> <NA>
#> 3 Padmé Amida… 165 45 brown light brown 46 fema… femin… Naboo
#> # … with 4 more variables: species <chr>, films <list>, vehicles <list>,
#> # starships <list>, and abbreviated variable names ¹hair_color, ²skin_color,
#> # ³eye_color, ⁴birth_year, ⁵homeworld#> # A tibble: 3 × 14
#> name height mass hair_…¹ skin_…² eye_c…³ birth…⁴ sex gender homew…⁵
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 Saesee Tiin 188 NA none pale orange NA male mascu… Iktotch
#> 2 Cliegg Lars 183 NA brown fair blue 82 male mascu… Tatooi…
#> 3 Ki-Adi-Mundi 198 82 white pale yellow 92 male mascu… Cerea
#> # … with 4 more variables: species <chr>, films <list>, vehicles <list>,
#> # starships <list>, and abbreviated variable names ¹hair_color, ²skin_color,
#> # ³eye_color, ⁴birth_year, ⁵homeworld#> # A tibble: 1 × 14
#> name height mass hair_…¹ skin_…² eye_c…³ birth…⁴ sex gender homew…⁵
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 Jabba Desil… 175 1358 <NA> green-… orange 600 herm… mascu… Nal Hu…
#> # … with 4 more variables: species <chr>, films <list>, vehicles <list>,
#> # starships <list>, and abbreviated variable names ¹hair_color, ²skin_color,
#> # ³eye_color, ⁴birth_year, ⁵homeworld#> # A tibble: 1 × 14
#> name height mass hair_color skin_color eye_co…¹ birth…² sex gender homew…³
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 Yoda 66 17 white green brown 896 male mascu… <NA>
#> # … with 4 more variables: species <chr>, films <list>, vehicles <list>,
#> # starships <list>, and abbreviated variable names ¹eye_color, ²birth_year,
#> # ³homeworldAn R purist could point out that all functions of slice() and its variants can be replaced by base R indexing (or subsetting) functions. For instance, the expression sw[(nrow(sw) - 2):nrow(sw), ] would also select the final three rows of sw (see Section 1.5.3).
Alternatively, we can replace many slice() commands by corresponding filter() expressions. For instance, we can create a column that contains the number of the corresponding row and then filter() for the numeric values of this column:
tb <- sw # copy data (to keep sw)
# Add a column that contains the row number:
tb$row_nr <- 1:nrow(sw)
# Apply filters (to the new column):
filter(tb, row_nr == 2) # the 2nd row
filter(tb, row_nr == 1:3) # the first 3 rows
filter(tb, row_nr == 1 | row_nr == nrow(sw)) # the 1st and last rows
filter(tb, row_nr %in% sample(1:nrow(tb), size = 3)) # 3 random rowsHowever, replacing slice() commands by their alternatives shows that the corresponding expressions can often get cumbersome and cryptic.
As we will see throughout this book, R often provides many alternative ways of getting things done — and there is nothing wrong with having dedicated tools for solving simple tasks. Hence, slice() and its variants are a welcome addition to our dplyr vocabulary.
3.2.3 select() selects columns
Using select() selects variables (columns) by their names or numbers.
As it works exactly like filter() (but selects columns, rather than rows), we can immediately select not just one, but multiple variables.
Actually, there are many ways of achieving the same result:
#> # A tibble: 87 × 4
#> name species birth_year gender
#> <chr> <chr> <dbl> <chr>
#> 1 Luke Skywalker Human 19 masculine
#> 2 C-3PO Droid 112 masculine
#> 3 R2-D2 Droid 33 masculine
#> 4 Darth Vader Human 41.9 masculine
#> 5 Leia Organa Human 19 feminine
#> 6 Owen Lars Human 52 masculine
#> 7 Beru Whitesun lars Human 47 feminine
#> 8 R5-D4 Droid NA masculine
#> 9 Biggs Darklighter Human 24 masculine
#> 10 Obi-Wan Kenobi Human 57 masculine
#> # … with 77 more rows# The same when using the pipe:
sw %>% # Note: %>% is NOT + (used in ggplot)
select(name, species, birth_year, gender)#> # A tibble: 87 × 4
#> name species birth_year gender
#> <chr> <chr> <dbl> <chr>
#> 1 Luke Skywalker Human 19 masculine
#> 2 C-3PO Droid 112 masculine
#> 3 R2-D2 Droid 33 masculine
#> 4 Darth Vader Human 41.9 masculine
#> 5 Leia Organa Human 19 feminine
#> 6 Owen Lars Human 52 masculine
#> 7 Beru Whitesun lars Human 47 feminine
#> 8 R5-D4 Droid NA masculine
#> 9 Biggs Darklighter Human 24 masculine
#> 10 Obi-Wan Kenobi Human 57 masculine
#> # … with 77 more rows# The same when providing a vector of variable names:
sw %>%
select(c(name, species, birth_year, gender)) #> # A tibble: 87 × 4
#> name species birth_year gender
#> <chr> <chr> <dbl> <chr>
#> 1 Luke Skywalker Human 19 masculine
#> 2 C-3PO Droid 112 masculine
#> 3 R2-D2 Droid 33 masculine
#> 4 Darth Vader Human 41.9 masculine
#> 5 Leia Organa Human 19 feminine
#> 6 Owen Lars Human 52 masculine
#> 7 Beru Whitesun lars Human 47 feminine
#> 8 R5-D4 Droid NA masculine
#> 9 Biggs Darklighter Human 24 masculine
#> 10 Obi-Wan Kenobi Human 57 masculine
#> # … with 77 more rows#> # A tibble: 87 × 4
#> name homeworld birth_year sex
#> <chr> <chr> <dbl> <chr>
#> 1 Luke Skywalker Tatooine 19 male
#> 2 C-3PO Tatooine 112 none
#> 3 R2-D2 Naboo 33 none
#> 4 Darth Vader Tatooine 41.9 male
#> 5 Leia Organa Alderaan 19 female
#> 6 Owen Lars Tatooine 52 male
#> 7 Beru Whitesun lars Tatooine 47 female
#> 8 R5-D4 Tatooine NA none
#> 9 Biggs Darklighter Tatooine 24 male
#> 10 Obi-Wan Kenobi Stewjon 57 male
#> # … with 77 more rows#> # A tibble: 87 × 4
#> name homeworld birth_year sex
#> <chr> <chr> <dbl> <chr>
#> 1 Luke Skywalker Tatooine 19 male
#> 2 C-3PO Tatooine 112 none
#> 3 R2-D2 Naboo 33 none
#> 4 Darth Vader Tatooine 41.9 male
#> 5 Leia Organa Alderaan 19 female
#> 6 Owen Lars Tatooine 52 male
#> 7 Beru Whitesun lars Tatooine 47 female
#> 8 R5-D4 Tatooine NA none
#> 9 Biggs Darklighter Tatooine 24 male
#> 10 Obi-Wan Kenobi Stewjon 57 male
#> # … with 77 more rowsWhen selecting ranges of variables, the : operator allows selecting ranges of variables:
#> # A tibble: 87 × 6
#> name height mass gender homeworld species
#> <chr> <int> <dbl> <chr> <chr> <chr>
#> 1 Luke Skywalker 172 77 masculine Tatooine Human
#> 2 C-3PO 167 75 masculine Tatooine Droid
#> 3 R2-D2 96 32 masculine Naboo Droid
#> 4 Darth Vader 202 136 masculine Tatooine Human
#> 5 Leia Organa 150 49 feminine Alderaan Human
#> 6 Owen Lars 178 120 masculine Tatooine Human
#> 7 Beru Whitesun lars 165 75 feminine Tatooine Human
#> 8 R5-D4 97 32 masculine Tatooine Droid
#> 9 Biggs Darklighter 183 84 masculine Tatooine Human
#> 10 Obi-Wan Kenobi 182 77 masculine Stewjon Human
#> # … with 77 more rowsSelecting can also be used to re-arrange variables. In this case, the function everything() is useful, and refers to every variable not already specified:
# Select to re-order variables (columns) with everything():
sw %>%
select(species, name, gender, everything())#> # A tibble: 87 × 14
#> species name gender height mass hair_…¹ skin_…² eye_c…³ birth…⁴ sex
#> <chr> <chr> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr>
#> 1 Human Luke Skywa… mascu… 172 77 blond fair blue 19 male
#> 2 Droid C-3PO mascu… 167 75 <NA> gold yellow 112 none
#> 3 Droid R2-D2 mascu… 96 32 <NA> white,… red 33 none
#> 4 Human Darth Vader mascu… 202 136 none white yellow 41.9 male
#> 5 Human Leia Organa femin… 150 49 brown light brown 19 fema…
#> 6 Human Owen Lars mascu… 178 120 brown,… light blue 52 male
#> 7 Human Beru White… femin… 165 75 brown light blue 47 fema…
#> 8 Droid R5-D4 mascu… 97 32 <NA> white,… red NA none
#> 9 Human Biggs Dark… mascu… 183 84 black light brown 24 male
#> 10 Human Obi-Wan Ke… mascu… 182 77 auburn… fair blue-g… 57 male
#> # … with 77 more rows, 4 more variables: homeworld <chr>, films <list>,
#> # vehicles <list>, starships <list>, and abbreviated variable names
#> # ¹hair_color, ²skin_color, ³eye_color, ⁴birth_yearA number of additional helper functions allow more sophisticated selections by testing variable names:
#> # A tibble: 87 × 4
#> skin_color sex species starships
#> <chr> <chr> <chr> <list>
#> 1 fair male Human <chr [2]>
#> 2 gold none Droid <chr [0]>
#> 3 white, blue none Droid <chr [0]>
#> 4 white male Human <chr [1]>
#> 5 light female Human <chr [0]>
#> 6 light male Human <chr [0]>
#> 7 light female Human <chr [0]>
#> 8 white, red none Droid <chr [0]>
#> 9 light male Human <chr [1]>
#> 10 fair male Human <chr [5]>
#> # … with 77 more rows#> # A tibble: 87 × 5
#> mass species films vehicles starships
#> <dbl> <chr> <list> <list> <list>
#> 1 77 Human <chr [5]> <chr [2]> <chr [2]>
#> 2 75 Droid <chr [6]> <chr [0]> <chr [0]>
#> 3 32 Droid <chr [7]> <chr [0]> <chr [0]>
#> 4 136 Human <chr [4]> <chr [0]> <chr [1]>
#> 5 49 Human <chr [5]> <chr [1]> <chr [0]>
#> 6 120 Human <chr [3]> <chr [0]> <chr [0]>
#> 7 75 Human <chr [3]> <chr [0]> <chr [0]>
#> 8 32 Droid <chr [1]> <chr [0]> <chr [0]>
#> 9 84 Human <chr [1]> <chr [0]> <chr [1]>
#> 10 77 Human <chr [6]> <chr [1]> <chr [5]>
#> # … with 77 more rows#> # A tibble: 87 × 4
#> hair_color skin_color eye_color birth_year
#> <chr> <chr> <chr> <dbl>
#> 1 blond fair blue 19
#> 2 <NA> gold yellow 112
#> 3 <NA> white, blue red 33
#> 4 none white yellow 41.9
#> 5 brown light brown 19
#> 6 brown, grey light blue 52
#> 7 brown light blue 47
#> 8 <NA> white, red red NA
#> 9 black light brown 24
#> 10 auburn, white fair blue-gray 57
#> # … with 77 more rows#> # A tibble: 87 × 4
#> hair_color skin_color eye_color homeworld
#> <chr> <chr> <chr> <chr>
#> 1 blond fair blue Tatooine
#> 2 <NA> gold yellow Tatooine
#> 3 <NA> white, blue red Naboo
#> 4 none white yellow Tatooine
#> 5 brown light brown Alderaan
#> 6 brown, grey light blue Tatooine
#> 7 brown light blue Tatooine
#> 8 <NA> white, red red Tatooine
#> 9 black light brown Tatooine
#> 10 auburn, white fair blue-gray Stewjon
#> # … with 77 more rowsThe helper function where() allows applying a function to all variables and returns only those for which the function returns TRUE. For instance, if we wanted to only select numeric columns of sw, we could use:
#> # A tibble: 87 × 3
#> height mass birth_year
#> <int> <dbl> <dbl>
#> 1 172 77 19
#> 2 167 75 112
#> 3 96 32 33
#> 4 202 136 41.9
#> 5 150 49 19
#> 6 178 120 52
#> 7 165 75 47
#> 8 97 32 NA
#> 9 183 84 24
#> 10 182 77 57
#> # … with 77 more rowsThe same could be achieved by using the select_if() variant of select():
#> # A tibble: 87 × 3
#> height mass birth_year
#> <int> <dbl> <dbl>
#> 1 172 77 19
#> 2 167 75 112
#> 3 96 32 33
#> 4 202 136 41.9
#> 5 150 49 19
#> 6 178 120 52
#> 7 165 75 47
#> 8 97 32 NA
#> 9 183 84 24
#> 10 182 77 57
#> # … with 77 more rowsAs select() selects variables, its result should typically be a table with the same number of cases as the original one, but fewer columns, unless the variables specified include everything().
(See ?dplyr::select for more help and additional examples, as well as ?dplyr::select_if for conditional variants.)
rename()` renames variables
A dplyr function that is often useful when selecting variables is rename(), which does exactly what it says:
#> # A tibble: 87 × 14
#> creature height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 Luke Sk… 172 77 blond fair blue 19 male mascu…
#> 2 C-3PO 167 75 <NA> gold yellow 112 none mascu…
#> 3 R2-D2 96 32 <NA> white, bl… red 33 none mascu…
#> 4 Darth V… 202 136 none white yellow 41.9 male mascu…
#> 5 Leia Or… 150 49 brown light brown 19 fema… femin…
#> 6 Owen La… 178 120 brown, gr… light blue 52 male mascu…
#> 7 Beru Wh… 165 75 brown light blue 47 fema… femin…
#> 8 R5-D4 97 32 <NA> white, red red NA none mascu…
#> 9 Biggs D… 183 84 black light brown 24 male mascu…
#> 10 Obi-Wan… 182 77 auburn, w… fair blue-gray 57 male mascu…
#> # ℹ 77 more rows
#> # ℹ 5 more variables: is_from <chr>, species <chr>, films <list>,
#> # vehicles <list>, starships <list>In its basic form, rename() uses paired combinations of new_name = old_name to rename variables, but does keep all other variables.
To permanently drop the old and keep only the new names, we would need to re-assign the altered sw (to sw itself).
Note also that the (new and old) variable names do not need to be enclosed in quotation marks.
For changing all variable names of a table in the same fashion, the rename_with() function allows directly applying a function to variable names.
For instance, to change all uppercase characters in variable names to lowercase characters, we could use:
#> [1] "Sepal.Length" "Sepal.Width" "Petal.Length" "Petal.Width" "Species"# Rename variables by applying a function:
iris |>
# Apply tolower() to all names:
rename_with(tolower) |>
# Print only the names:
names()#> [1] "sepal.length" "sepal.width" "petal.length" "petal.width" "species"More complicated re-naming scenarios can specify sets of variables in combination with quantifiers (e.g., see the examples of ?rename() using a lookup vector and the any_of() or all_of() modifiers).
Details
Note some details on using select():
select()works both by specifying variable (column) names and by specifying column numbers.Again, variable names are unquoted.
The sequence of variable names (separated by commas) specifies the order of columns in the resulting tibble.
Selecting and adding
everything()allows re-ordering variables.Various helper functions (e.g.,
starts_with,ends_with,contains,matches,num_range) refer to (parts of) variable names or apply functions to variables (e.g.,where).
Similarly, the rename() function changes the names of the specified variables (typed without quotes) and keeps all other variables.
Practice
Here are some exercises to practice the dplyr verbs encountered so far:
- What is the result of
sw %>% select(height)? More specifically, how does it differ from the vectorsw$height?
- Use
select()orselect_if()on thedplyr::starwarsdata (sw) to select and re-order specific subsets of variables (e.g., all variables starting with “h”, all even columns, all character variables, etc.).
sw %>% select(starts_with("h"))
even_col <- ((1:ncol(sw) %% 2) == 0)
sw %>% select_if(even_col)
sw %>% select(where(is.character))- Advanced: The dplyr command
select(sw, where(is.numeric))selects all numeric columns of a tibblesw. How could the same be achieved by using subsetting in base R?
Hint: As this turns out to be quite difficult, we will only cover the commands used here much later (in Chapter 12 on Iteration). This illustrates that dplyr enables us to solve tasks that would otherwise be much harder.
3.2.4 mutate() computes new variables
Using mutate() computes new variables (columns) from scratch or existing ones.
For instance, the height variable is provided in centimeters (cm).
To transform this measure into feet, we need to divide it by 100 (to obtain height in meters) and multiply the result by a factor of 3.28084:
# Preparation: Save only a subset variables of sw as sws:
sws <- select(sw, name:mass, birth_year:species)
sws # => 87 cases (rows), but only 7 variables (columns)#> # A tibble: 87 × 8
#> name height mass birth_year sex gender homeworld species
#> <chr> <int> <dbl> <dbl> <chr> <chr> <chr> <chr>
#> 1 Luke Skywalker 172 77 19 male masculine Tatooine Human
#> 2 C-3PO 167 75 112 none masculine Tatooine Droid
#> 3 R2-D2 96 32 33 none masculine Naboo Droid
#> 4 Darth Vader 202 136 41.9 male masculine Tatooine Human
#> 5 Leia Organa 150 49 19 female feminine Alderaan Human
#> 6 Owen Lars 178 120 52 male masculine Tatooine Human
#> 7 Beru Whitesun lars 165 75 47 female feminine Tatooine Human
#> 8 R5-D4 97 32 NA none masculine Tatooine Droid
#> 9 Biggs Darklighter 183 84 24 male masculine Tatooine Human
#> 10 Obi-Wan Kenobi 182 77 57 male masculine Stewjon Human
#> # … with 77 more rows# Conversion factor (cm to feet):
factor_cm_2_feet <- 3.28084/100
# Compute 2 new variables and add them to existing ones:
mutate(sws, id = 1:nrow(sw),
height_feet = factor_cm_2_feet * height)#> # A tibble: 87 × 10
#> name height mass birth…¹ sex gender homew…² species id heigh…³
#> <chr> <int> <dbl> <dbl> <chr> <chr> <chr> <chr> <int> <dbl>
#> 1 Luke Skywalk… 172 77 19 male mascu… Tatooi… Human 1 5.64
#> 2 C-3PO 167 75 112 none mascu… Tatooi… Droid 2 5.48
#> 3 R2-D2 96 32 33 none mascu… Naboo Droid 3 3.15
#> 4 Darth Vader 202 136 41.9 male mascu… Tatooi… Human 4 6.63
#> 5 Leia Organa 150 49 19 fema… femin… Aldera… Human 5 4.92
#> 6 Owen Lars 178 120 52 male mascu… Tatooi… Human 6 5.84
#> 7 Beru Whitesu… 165 75 47 fema… femin… Tatooi… Human 7 5.41
#> 8 R5-D4 97 32 NA none mascu… Tatooi… Droid 8 3.18
#> 9 Biggs Darkli… 183 84 24 male mascu… Tatooi… Human 9 6.00
#> 10 Obi-Wan Keno… 182 77 57 male mascu… Stewjon Human 10 5.97
#> # … with 77 more rows, and abbreviated variable names ¹birth_year, ²homeworld,
#> # ³height_feet#> # A tibble: 87 × 10
#> name height mass birth…¹ sex gender homew…² species id heigh…³
#> <chr> <int> <dbl> <dbl> <chr> <chr> <chr> <chr> <int> <dbl>
#> 1 Luke Skywalk… 172 77 19 male mascu… Tatooi… Human 1 5.64
#> 2 C-3PO 167 75 112 none mascu… Tatooi… Droid 2 5.48
#> 3 R2-D2 96 32 33 none mascu… Naboo Droid 3 3.15
#> 4 Darth Vader 202 136 41.9 male mascu… Tatooi… Human 4 6.63
#> 5 Leia Organa 150 49 19 fema… femin… Aldera… Human 5 4.92
#> 6 Owen Lars 178 120 52 male mascu… Tatooi… Human 6 5.84
#> 7 Beru Whitesu… 165 75 47 fema… femin… Tatooi… Human 7 5.41
#> 8 R5-D4 97 32 NA none mascu… Tatooi… Droid 8 3.18
#> 9 Biggs Darkli… 183 84 24 male mascu… Tatooi… Human 9 6.00
#> 10 Obi-Wan Keno… 182 77 57 male mascu… Stewjon Human 10 5.97
#> # … with 77 more rows, and abbreviated variable names ¹birth_year, ²homeworld,
#> # ³height_feetA closely related dplyr verb is transmute(), which only keeps computed variables and drops all other ones:
# Transmute computes and only keeps new variables:
sws %>%
transmute(id = 1:nrow(sw),
height_feet = factor_cm_2_feet * height)#> # A tibble: 87 × 2
#> id height_feet
#> <int> <dbl>
#> 1 1 5.64
#> 2 2 5.48
#> 3 3 3.15
#> 4 4 6.63
#> 5 5 4.92
#> 6 6 5.84
#> 7 7 5.41
#> 8 8 3.18
#> 9 9 6.00
#> 10 10 5.97
#> # … with 77 more rowsAlthough mutate() and transmute() compute the same variables,
mutate() is of an incremental nature (by adding new variables to the existing table), whereas transmute() drastically changes the table (by only keeping new variables).
For most purposes, adding new columns to the existing table is perfectly fine.
Interestingly, a variable computed by mutate() or transmute() can immediately be used for computing another variable:
# Compute variables based on multiple others (including computed ones):
sws %>%
select(name, height, mass, species) %>%
mutate(BMI = mass / ((height / 100) ^ 2), # compute body mass index (kg/m^2)
BMI_low = BMI < 18.5, # classify low BMI values
BMI_high = BMI > 30, # classify high BMI values
BMI_norm = !BMI_low & !BMI_high # classify normal BMI values
)#> # A tibble: 87 × 8
#> name height mass species BMI BMI_low BMI_high BMI_norm
#> <chr> <int> <dbl> <chr> <dbl> <lgl> <lgl> <lgl>
#> 1 Luke Skywalker 172 77 Human 26.0 FALSE FALSE TRUE
#> 2 C-3PO 167 75 Droid 26.9 FALSE FALSE TRUE
#> 3 R2-D2 96 32 Droid 34.7 FALSE TRUE FALSE
#> 4 Darth Vader 202 136 Human 33.3 FALSE TRUE FALSE
#> 5 Leia Organa 150 49 Human 21.8 FALSE FALSE TRUE
#> 6 Owen Lars 178 120 Human 37.9 FALSE TRUE FALSE
#> 7 Beru Whitesun lars 165 75 Human 27.5 FALSE FALSE TRUE
#> 8 R5-D4 97 32 Droid 34.0 FALSE TRUE FALSE
#> 9 Biggs Darklighter 183 84 Human 25.1 FALSE FALSE TRUE
#> 10 Obi-Wan Kenobi 182 77 Human 23.2 FALSE FALSE TRUE
#> # ℹ 77 more rowsNote that this uses three variables for classifying each creature’s BMI value.
A better solution could immediate categorize the BMI values by using the base R function cut():
sws %>%
select(name, height, mass, species) %>%
mutate(BMI = mass / ((height / 100) ^ 2), # compute body mass index (kg/m^2)
BMI_cat = cut(BMI,
breaks = c(-Inf, 18.5, 30, Inf),
labels = c("low", "norm", "high"))
)#> # A tibble: 87 × 6
#> name height mass species BMI BMI_cat
#> <chr> <int> <dbl> <chr> <dbl> <fct>
#> 1 Luke Skywalker 172 77 Human 26.0 norm
#> 2 C-3PO 167 75 Droid 26.9 norm
#> 3 R2-D2 96 32 Droid 34.7 high
#> 4 Darth Vader 202 136 Human 33.3 high
#> 5 Leia Organa 150 49 Human 21.8 norm
#> 6 Owen Lars 178 120 Human 37.9 high
#> 7 Beru Whitesun lars 165 75 Human 27.5 norm
#> 8 R5-D4 97 32 Droid 34.0 high
#> 9 Biggs Darklighter 183 84 Human 25.1 norm
#> 10 Obi-Wan Kenobi 182 77 Human 23.2 norm
#> # ℹ 77 more rowsAs mutate() typically changes variables (by computing new ones), it seems appropriately named.
However, note that mutate() does typically not change the identity of the cases (rows) of a data table.
See ?dplyr::mutate for more help and additional examples.
Details
Let’s summarize some noteworthy details on mutate() and transmute():
mutate()computes new variables (columns) and adds them to existing ones, whiletransmute()drops existing ones.Each
mutate()command specifies a new variable name (without quotes), followed by=and a rule for computing the new variable from existing ones.Multiple
mutate()steps are separated by commas, each of which creates a new variable.Later variables may use earlier ones.
Again, variable names are unquoted.
See http://r4ds.had.co.nz/transform.html#mutate-funs for useful functions for creating new variables.
3.2.5 summarise() computes summaries
The dplyr verb summarise() computes a function for a specified variable and collapses the values of the specified variable (i.e., the rows of a specified columns) to a single value:
# Summarise allows computing a function for a variable (column):
summarise(sw, mn_mass = mean(mass, na.rm = TRUE)) # => 97.31 kg #> # A tibble: 1 × 1
#> mn_mass
#> <dbl>
#> 1 97.3#> # A tibble: 1 × 1
#> mn_mass
#> <dbl>
#> 1 97.3In most cases, we want to compute not just one summary statistic of a variable (e.g., mass), but several ones:
# Multiple summarise steps allow applying
# different functions for 1 dependent variable:
sw %>%
summarise(n_mass = sum(!is.na(mass)),
mn_mass = mean(mass, na.rm = TRUE),
md_mass = median(mass, na.rm = TRUE),
sd_mass = sd(mass, na.rm = TRUE),
max_mass = max(mass, na.rm = TRUE),
big_mass = any(mass > 1000)
)#> # A tibble: 1 × 6
#> n_mass mn_mass md_mass sd_mass max_mass big_mass
#> <int> <dbl> <dbl> <dbl> <dbl> <lgl>
#> 1 59 97.3 79 169. 1358 TRUESimilarly, we often want to obtain summary information about more than one variable.
For instance, we may want to know basic statistics about the height and weight variables in our sw data, and count some characteristics of character variables:
# Multiple summarise steps also allow applying
# different functions to different dependent variables:
sw %>%
summarise(# Descriptives of height:
n_height = sum(!is.na(height)),
mn_height = mean(height, na.rm = TRUE),
sd_height = sd(height, na.rm = TRUE),
# Descriptives of mass:
n_mass = sum(!is.na(mass)),
mn_mass = mean(mass, na.rm = TRUE),
sd_mass = sd(mass, na.rm = TRUE),
# Counts of character variables:
n_names = n(),
n_species = n_distinct(species),
n_worlds = n_distinct(homeworld)
)#> # A tibble: 1 × 9
#> n_height mn_height sd_height n_mass mn_mass sd_mass n_names n_species n_worlds
#> <int> <dbl> <dbl> <int> <dbl> <dbl> <int> <int> <int>
#> 1 81 174. 34.8 59 97.3 169. 87 38 49While summarise() provides many different summary statistics by itself, it is even more useful in combination with group_by() (discussed next).
See ?dplyr::summarise for more help and additional examples.
Details
Note some details on summarise():
summarise()collapses multiple values into one value and returns a new tibble with as many columns as values computed.Each
summarise()step specifies a new variable name (without quotes), followed by=, and a function for computing the new variable from existing ones.Multiple
summarise()steps are separated by commas.Again, later variables may use earlier ones.
Again, variable names are unquoted.
See https://dplyr.tidyverse.org/reference/summarise.html for examples and useful functions in combination with
summarise().
Practice
Here are additional exercises to practice our dplyr prowess:
- Someone speculates that — on average — humans have longer names than droids, but droids are heavier than humans.
Can you compute some summaries (e.g., by combining
filter()withsummarise()commands) to check this?
Hint: The length of a character string s can be computed with nchar(s).
Solution
# Average name length and mass of humans:
sw %>%
filter(species == "Human") %>%
summarise(n_humans = n(),
# Name length:
mn_name_len = mean(nchar(name)),
sd_name_len = sd(nchar(name)),
# Descriptives of mass (from above):
n_mass = sum(!is.na(mass)),
mn_mass = mean(mass, na.rm = TRUE),
sd_mass = sd(mass, na.rm = TRUE))#> # A tibble: 1 × 6
#> n_humans mn_name_len sd_name_len n_mass mn_mass sd_mass
#> <int> <dbl> <dbl> <int> <dbl> <dbl>
#> 1 35 11.3 4.11 22 82.8 19.4# Average name length and mass of droids:
sw %>%
filter(species == "Droid") %>%
summarise(n_droids = n(),
# Name length:
mn_name_len = mean(nchar(name)),
sd_name_len = sd(nchar(name)),
# Descriptives of mass (from above):
n_mass = sum(!is.na(mass)),
mn_mass = mean(mass, na.rm = TRUE),
sd_mass = sd(mass, na.rm = TRUE))#> # A tibble: 1 × 6
#> n_droids mn_name_len sd_name_len n_mass mn_mass sd_mass
#> <int> <dbl> <dbl> <int> <dbl> <dbl>
#> 1 6 4.83 0.983 4 69.8 51.0Note:
It looks like the average name length is about twice as high for humans (but note that there are only six droids in the dataset).
The hypothesis about droids being heavier on average is wrong, as the mean differences point in the opposite direction (but both distributions contain missing values and show large variations).
- Apply all summary functions mentioned in
?dplyr::summariseto theswdata.
3.2.6 group_by() aggregates variables
Using group_by() does not change the data, but the unit of aggregation for other commands, which is particularly useful in combination with mutate() and summarise().
When used by itself, group_by() returns the same tibble in a grouped form. For instance, the following commands will group sws by species:
# Grouping does not change the data, but lists its groups:
group_by(sws, species) # => 38 groups of species#> # A tibble: 87 × 8
#> # Groups: species [38]
#> name height mass birth_year sex gender homeworld species
#> <chr> <int> <dbl> <dbl> <chr> <chr> <chr> <chr>
#> 1 Luke Skywalker 172 77 19 male masculine Tatooine Human
#> 2 C-3PO 167 75 112 none masculine Tatooine Droid
#> 3 R2-D2 96 32 33 none masculine Naboo Droid
#> 4 Darth Vader 202 136 41.9 male masculine Tatooine Human
#> 5 Leia Organa 150 49 19 female feminine Alderaan Human
#> 6 Owen Lars 178 120 52 male masculine Tatooine Human
#> 7 Beru Whitesun lars 165 75 47 female feminine Tatooine Human
#> 8 R5-D4 97 32 NA none masculine Tatooine Droid
#> 9 Biggs Darklighter 183 84 24 male masculine Tatooine Human
#> 10 Obi-Wan Kenobi 182 77 57 male masculine Stewjon Human
#> # … with 77 more rows#> # A tibble: 87 × 8
#> # Groups: species [38]
#> name height mass birth_year sex gender homeworld species
#> <chr> <int> <dbl> <dbl> <chr> <chr> <chr> <chr>
#> 1 Luke Skywalker 172 77 19 male masculine Tatooine Human
#> 2 C-3PO 167 75 112 none masculine Tatooine Droid
#> 3 R2-D2 96 32 33 none masculine Naboo Droid
#> 4 Darth Vader 202 136 41.9 male masculine Tatooine Human
#> 5 Leia Organa 150 49 19 female feminine Alderaan Human
#> 6 Owen Lars 178 120 52 male masculine Tatooine Human
#> 7 Beru Whitesun lars 165 75 47 female feminine Tatooine Human
#> 8 R5-D4 97 32 NA none masculine Tatooine Droid
#> 9 Biggs Darklighter 183 84 24 male masculine Tatooine Human
#> 10 Obi-Wan Kenobi 182 77 57 male masculine Stewjon Human
#> # … with 77 more rowsThis seems rather mundane, but becomes very powerful when combining the group_by() statement with a subsequent mutate() or summarise() command.
Grouped mutates
When combining group_by() with a subsequent mutate(), the scope of the variables computed by mutate() is the group defined by group_by().
For instance, the following pipe counts the number of individuals of each species and computes their mean height within each species:
#> # A tibble: 87 × 10
#> # Groups: species [38]
#> name height mass birth…¹ sex gender homew…² species n_ind…³ mn_he…⁴
#> <chr> <int> <dbl> <dbl> <chr> <chr> <chr> <chr> <int> <dbl>
#> 1 Luke Skywa… 172 77 19 male mascu… Tatooi… Human 35 177.
#> 2 C-3PO 167 75 112 none mascu… Tatooi… Droid 6 131.
#> 3 R2-D2 96 32 33 none mascu… Naboo Droid 6 131.
#> 4 Darth Vader 202 136 41.9 male mascu… Tatooi… Human 35 177.
#> 5 Leia Organa 150 49 19 fema… femin… Aldera… Human 35 177.
#> 6 Owen Lars 178 120 52 male mascu… Tatooi… Human 35 177.
#> 7 Beru White… 165 75 47 fema… femin… Tatooi… Human 35 177.
#> 8 R5-D4 97 32 NA none mascu… Tatooi… Droid 6 131.
#> 9 Biggs Dark… 183 84 24 male mascu… Tatooi… Human 35 177.
#> 10 Obi-Wan Ke… 182 77 57 male mascu… Stewjon Human 35 177.
#> # … with 77 more rows, and abbreviated variable names ¹birth_year, ²homeworld,
#> # ³n_individuals, ⁴mn_heightAs before, the new variables (here: n_individuals and mn_height) are added to the tibble, but now their values are computed relative to the group_by() variable (here: species) as the unit of aggregation. Interestingly, this implies that there exists no such thing as “the mean” of a variable, as any mean is always relative to some unit of aggregation. By changing the unit of aggregation, we can compute many different means for the same variable. For instance, we can compute the mean height of individuals overall, by species, by gender, etc.:
sws %>%
mutate(mn_height_1 = mean(height, na.rm = TRUE)) %>% # aggregates over ALL cases
group_by(species) %>%
mutate(mn_height_2 = mean(height, na.rm = TRUE)) %>% # aggregates over current group (species)
group_by(gender) %>%
mutate(mn_height_3 = mean(height, na.rm = TRUE)) %>% # aggregates over current group (gender)
group_by(name) %>%
mutate(mn_height_4 = mean(height, na.rm = TRUE)) %>% # aggregates over current group (name)
select(name, height, mn_height_1:mn_height_4)#> # A tibble: 87 × 6
#> # Groups: name [87]
#> name height mn_height_1 mn_height_2 mn_height_3 mn_height_4
#> <chr> <int> <dbl> <dbl> <dbl> <dbl>
#> 1 Luke Skywalker 172 174. 177. 177. 172
#> 2 C-3PO 167 174. 131. 177. 167
#> 3 R2-D2 96 174. 131. 177. 96
#> 4 Darth Vader 202 174. 177. 177. 202
#> 5 Leia Organa 150 174. 177. 165. 150
#> 6 Owen Lars 178 174. 177. 177. 178
#> 7 Beru Whitesun lars 165 174. 177. 165. 165
#> 8 R5-D4 97 174. 131. 177. 97
#> 9 Biggs Darklighter 183 174. 177. 177. 183
#> 10 Obi-Wan Kenobi 182 174. 177. 177. 182
#> # … with 77 more rowsGrouped summaries
Our summarise() commands above yielded some summary of one or several variables as one line of output. When combining group_by() with a subsequent summarise(), we obtain the corresponding summary for each group:
sws %>%
group_by(species) %>%
summarise(n_individuals = n(),
mn_height = mean(height, na.rm = TRUE),
mn_mass = mean(mass, na.rm = TRUE)
) %>%
arrange(desc(mn_height))#> # A tibble: 38 × 4
#> species n_individuals mn_height mn_mass
#> <chr> <int> <dbl> <dbl>
#> 1 Quermian 1 264 NaN
#> 2 Wookiee 2 231 124
#> 3 Kaminoan 2 221 88
#> 4 Kaleesh 1 216 159
#> 5 Gungan 3 209. 74
#> 6 Pau'an 1 206 80
#> 7 Besalisk 1 198 102
#> 8 Cerean 1 198 82
#> 9 Chagrian 1 196 NaN
#> 10 Nautolan 1 196 87
#> # … with 28 more rowsNote that the group_by() followed by summarise() returns a new tibble, with 38 rows (= groups of species) and
- 1 column of the group variable (here
species) and
- 3 columns of the 3 newly summarised variables.
Here, we also arranged this output tibble by descending means of height.
3.2.6.1 Grouping by multiple variables
Using group_by() with multiple variables yields a tibble containing the combination of all variable levels.
For instance, how many combinations of hair_color and eye_color exist, we could count them as follows:
#> # A tibble: 87 × 14
#> # Groups: hair_color, eye_color [35]
#> name height mass hair_…¹ skin_…² eye_c…³ birth…⁴ sex gender homew…⁵
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 Luke Skywa… 172 77 blond fair blue 19 male mascu… Tatooi…
#> 2 C-3PO 167 75 <NA> gold yellow 112 none mascu… Tatooi…
#> 3 R2-D2 96 32 <NA> white,… red 33 none mascu… Naboo
#> 4 Darth Vader 202 136 none white yellow 41.9 male mascu… Tatooi…
#> 5 Leia Organa 150 49 brown light brown 19 fema… femin… Aldera…
#> 6 Owen Lars 178 120 brown,… light blue 52 male mascu… Tatooi…
#> 7 Beru White… 165 75 brown light blue 47 fema… femin… Tatooi…
#> 8 R5-D4 97 32 <NA> white,… red NA none mascu… Tatooi…
#> 9 Biggs Dark… 183 84 black light brown 24 male mascu… Tatooi…
#> 10 Obi-Wan Ke… 182 77 auburn… fair blue-g… 57 male mascu… Stewjon
#> # … with 77 more rows, 4 more variables: species <chr>, films <list>,
#> # vehicles <list>, starships <list>, and abbreviated variable names
#> # ¹hair_color, ²skin_color, ³eye_color, ⁴birth_year, ⁵homeworldA common application of using group_by() with multiple varialbes is to count the number of cases (here: individuals) in each sub-group:
# Counting the frequency of cases in groups:
sw %>%
group_by(hair_color, eye_color) %>%
count() %>%
arrange(desc(n)) #> # A tibble: 35 × 3
#> # Groups: hair_color, eye_color [35]
#> hair_color eye_color n
#> <chr> <chr> <int>
#> 1 black brown 9
#> 2 brown brown 9
#> 3 none black 9
#> 4 brown blue 7
#> 5 none orange 7
#> 6 none yellow 6
#> 7 blond blue 3
#> 8 none blue 3
#> 9 none red 3
#> 10 black blue 2
#> # … with 25 more rows# The same using summarise and n():
sw %>%
group_by(hair_color, eye_color) %>%
summarise(n = n()) %>%
arrange(desc(n)) #> # A tibble: 35 × 3
#> # Groups: hair_color [13]
#> hair_color eye_color n
#> <chr> <chr> <int>
#> 1 black brown 9
#> 2 brown brown 9
#> 3 none black 9
#> 4 brown blue 7
#> 5 none orange 7
#> 6 none yellow 6
#> 7 blond blue 3
#> 8 none blue 3
#> 9 none red 3
#> 10 black blue 2
#> # … with 25 more rowsSee ?dplyr::group_by for more help and additional examples.
Details
Note some details on group_by():
group_by()changes the unit of aggregation for other commands (especiallymutate()andsummarise()).Again, variable names are unquoted.
When using
group_by()with multiple variables, they are separated by commas.Using
group_by()withmutate()results in a tibble that has the same number of cases (rows) as the original tibble. By contrast, usinggroup_by()withsummarise()results in a new tibble with all combinations of variable levels as its cases (rows).
Practice
Here are some exercises that combine multiple dplyr commands:
- In the last practice section above, we used two combinations of
filter()andsummarise()to check the hypotheses that — on average — humans have longer names than droids, but droids are heavier than humans. Now that we learned aboutgroup_by(), try to perform this check in one pipe.
Solution
Here is one of many possible ways of computing the average name length and mass for the two desired species:
# Average name length and mass of humans vs. droids:
sw %>%
filter(species == "Human" | species == "Droid") %>%
group_by(species) %>%
summarise(n_cases = n(),
# Name length:
mn_name_len = mean(nchar(name)),
sd_name_len = sd(nchar(name)),
# Descriptives of mass (from above):
n_mass = sum(!is.na(mass)),
mn_mass = mean(mass, na.rm = TRUE),
sd_mass = sd(mass, na.rm = TRUE))- Yoda says: “Taller creatures heavier are than smaller ones.”
Test his hypothesis for theswdataset in one pipe by
- selecting only the relevant variables
name,height, andmass, - computing variables for the median height and a logical variable
is_tallthat isTRUEif and only if an individual is taller than the median height, - grouping the data by
is_tall; - counting the cases and computing the mean mass for tall vs. non-tall individuals.
Solution
sw %>%
select(name, height, mass) %>%
mutate(md_height = median(height, na.rm = TRUE),
is_tall = height > md_height) %>%
group_by(is_tall) %>%
summarise(n = n(),
mn_mass = mean(mass, na.rm = TRUE),
sd_mass = sd(mass, na.rm = TRUE))#> # A tibble: 3 × 4
#> is_tall n mn_mass sd_mass
#> <lgl> <int> <dbl> <dbl>
#> 1 FALSE 43 103. 234.
#> 2 TRUE 38 91.1 25.7
#> 3 NA 6 NaN NAInterpretation
- Yoda seems wrong: The three tall creatures (with a
heightabove the median) have an average mass value of 91.1 kg, whereas the 43 smaller creatures (with aheightbelow or equal to the median) have an average mass value of 103 kg.
You just smoked a median-split analysis in a pipe — congratulations!
However, before getting too excited, we should try to understand why our results came out in this way.
To explain Yoda’s mistake, let’s look at a scatterplot that plots mass as a function of height (and colors the points by the value of our is_tall variable):
sw_tall <- sw %>%
select(name, height, mass) %>%
mutate(md_height = median(height, na.rm = TRUE),
is_tall = height > md_height)
# All individuals:
ggplot(sw_tall, aes(x = height, y = mass)) +
geom_point(aes(color = is_tall), size = 2) +
geom_text(aes(label = name), hjust = -.2, angle = 45, size = 2, alpha = 2/3) +
coord_cartesian(ylim = c(0, 1700)) +
scale_color_manual(values = c("firebrick", "steelblue", "gold")) +
labs(title = "Individual's mass by height",
x = "Height (in cm)", y = "Mass (in kg)") +
theme_classic()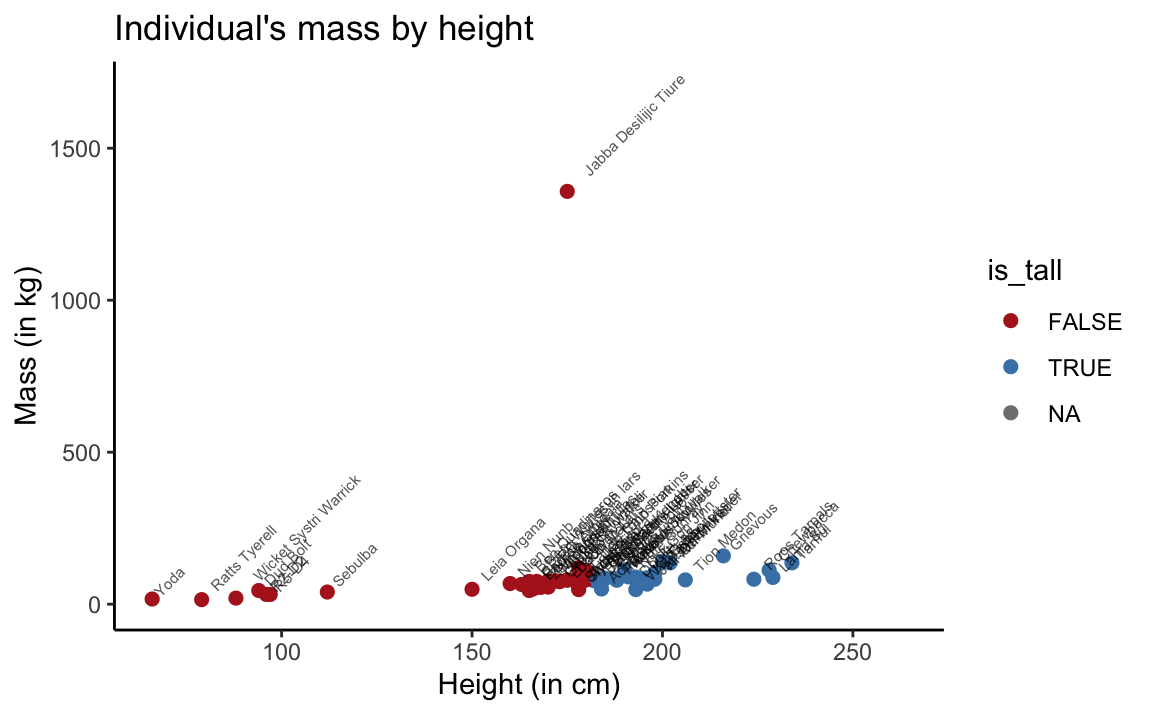
Figure 3.4: Scatterplot of mass by height in the full sw dataset.
Note that we used coord_cartesian to restrict the range of y values shown to ylim = c(0, 1700).
The scatterplot shows that the sw data contains a blatant outlier: Jabba Desilijic Tiure, the crime lord aka. ‘Jabba the Hutt’, has a mass of 1358 kg despite his below-average height of 175 cm.
Only considering creatures with a mass up to 170 kg suggests that Yoda’s hypothesis is perfectly valid when this outlier is excluded:
# Only showing mass values from 0 to 180:
ggplot(sw_tall, aes(x = height, y = mass)) +
geom_abline(aes(intercept = lm(mass ~ height)$coefficients[1], slope = lm(mass ~ height)$coefficients[2]),
linetype = 2, col = "orange") +
# stat_ellipse(aes(color = is_tall), alpha = .5) +
geom_point(aes(color = is_tall), size = 2) +
geom_text(aes(label = name), hjust = -.2, angle = 45, size = 2, alpha = 2/3) +
coord_cartesian(ylim = c(0, 170)) +
scale_color_manual(values = c("firebrick", "steelblue", "gold")) +
labs(title = "Individual's mass by height without outlier",
x = "Height (in cm)", y = "Mass (in kg)") +
theme_classic()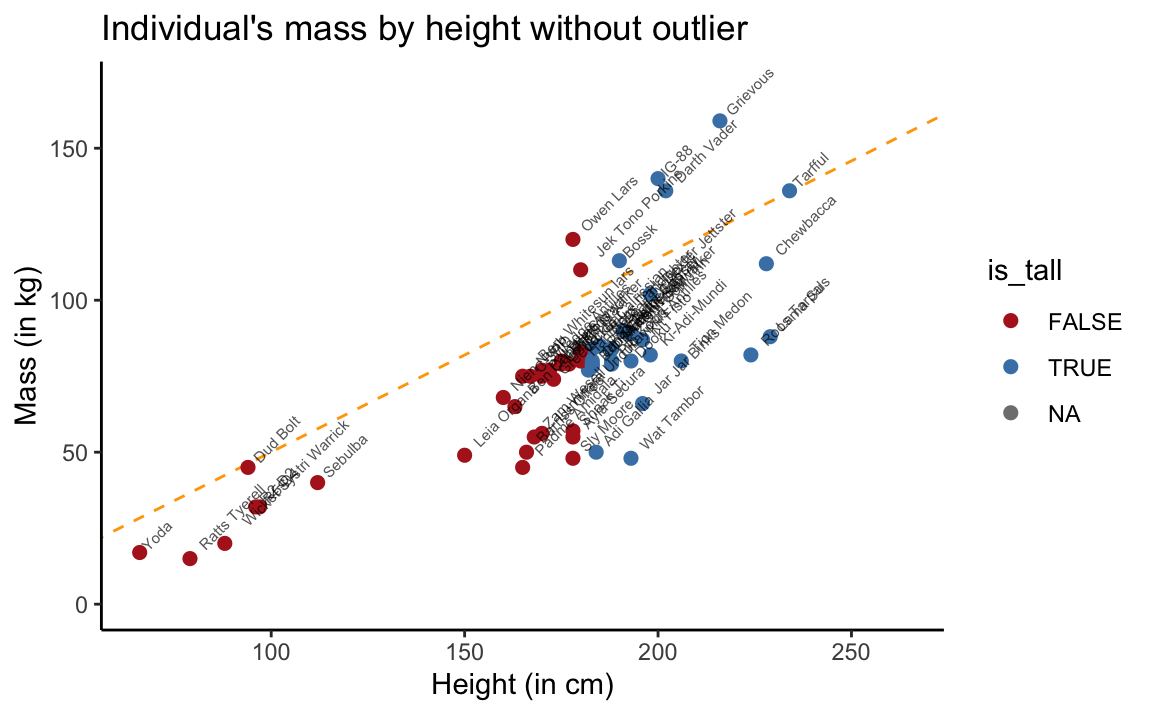
Figure 3.5: Scatterplot of mass by height without Jabba the Hutt.
This is yet another instance of the lesson taught by Anscombe’s quartet (in Section 2.1): We should never interpret the results of some statistical calculation without properly inspecting the underlying data.