A.3 Solutions (03)
Here are the solutions of the exercises on essential dplyr commands of Chapter 3 (Section 3.5).
A.3.1 Exercise 1
Reshaping vs. reducing data
Discuss the main dplyr functions in terms of reshaping and reducing data (as introduced in Section 3.1.1).
Solution
Using the overview of essential dplyr functions (in Section 3.2), we can say:
arrange()reshapes data by sorting cases (rows);filter()andslice()reduce data by selecting cases (rows) by logical conditions or number;select()reduces data by selecting or reshapes data by reordering variables (columns);mutate()reduces and enhances data by computing new variables (columns) and adding them to the existing ones;summarise()reduces data by aggregating/collapsing multiple values of a variable (rows of a column) to a single one;
group_by()andungroup()reshape data (in a subtle way by) changing the unit of aggregation (in combination withmutate()andsummarise()). This is usually done as a preparatory step for reducing or enhancing data.
A.3.2 Exercise 2
Star and R wars
We start tackling the tidyverse by uncovering even more facts about the dplyr::starwars universe.
Answer the following questions by using pipes of basic dplyr commands (i.e., by arranging, filtering, selecting, grouping, counting, summarizing).
- Save the tibble
dplyr::starwarsasswand report its dimensions.
Known unknowns
How many missing (
NA) values doesswcontain?Which variable (column) has the most missing values?
Which individuals come from an unknown (missing)
homeworldbut have a knownbirth_yearor knownmass?
Solution
## Missing data: -----
# How many missing data points?
sum(is.na(sw)) # => 101 missing values.
#> [1] 105
# Which individuals come from an unknown (missing) homeworld
# but have a known birth_year or mass?
sw %>%
filter(is.na(homeworld), !is.na(mass) | !is.na(birth_year))
#> # A tibble: 3 × 14
#> name height mass hair_…¹ skin_…² eye_c…³ birth…⁴ sex gender homew…⁵
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 Yoda 66 17 white green brown 896 male mascu… <NA>
#> 2 IG-88 200 140 none metal red 15 none mascu… <NA>
#> 3 Qui-Gon Jinn 193 89 brown fair blue 92 male mascu… <NA>
#> # … with 4 more variables: species <chr>, films <list>, vehicles <list>,
#> # starships <list>, and abbreviated variable names ¹hair_color, ²skin_color,
#> # ³eye_color, ⁴birth_year, ⁵homeworld
# Which variable (column) has the most missing values?
colSums(is.na(sw)) # => birth_year has 44 missing values
#> name height mass hair_color skin_color eye_color birth_year
#> 0 6 28 5 0 0 44
#> sex gender homeworld species films vehicles starships
#> 4 4 10 4 0 0 0
colMeans(is.na(sw)) # (amounting to 50.1% of all cases).
#> name height mass hair_color skin_color eye_color birth_year
#> 0.00000000 0.06896552 0.32183908 0.05747126 0.00000000 0.00000000 0.50574713
#> sex gender homeworld species films vehicles starships
#> 0.04597701 0.04597701 0.11494253 0.04597701 0.00000000 0.00000000 0.00000000
## (x) Replace all missing values of `hair_color` (in the variable `sw$hair_color`) by "bald":
# sw$hair_color[is.na(sw$hair_color)] <- "bald"Gender issues
How many humans are contained in
swoverall and by gender?How many and which individuals in
sware neither male nor female?Of which species in
swexist at least 2 different gender values?
Solution
## (c) Gender issues: -----
# (+) How many humans are there of each gender?
sw %>%
filter(species == "Human") %>%
group_by(gender) %>%
count()
#> # A tibble: 2 × 2
#> # Groups: gender [2]
#> gender n
#> <chr> <int>
#> 1 feminine 9
#> 2 masculine 26
## Answer: 35 Humans in total: 9 females, 26 male.
# (+) How many and which individuals are neither male nor female?
sw %>%
filter(gender != "male", gender != "female")
#> # A tibble: 83 × 14
#> name height mass hair_…¹ skin_…² eye_c…³ birth…⁴ sex gender homew…⁵
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 Luke Skywa… 172 77 blond fair blue 19 male mascu… Tatooi…
#> 2 C-3PO 167 75 <NA> gold yellow 112 none mascu… Tatooi…
#> 3 R2-D2 96 32 <NA> white,… red 33 none mascu… Naboo
#> 4 Darth Vader 202 136 none white yellow 41.9 male mascu… Tatooi…
#> 5 Leia Organa 150 49 brown light brown 19 fema… femin… Aldera…
#> 6 Owen Lars 178 120 brown,… light blue 52 male mascu… Tatooi…
#> 7 Beru White… 165 75 brown light blue 47 fema… femin… Tatooi…
#> 8 R5-D4 97 32 <NA> white,… red NA none mascu… Tatooi…
#> 9 Biggs Dark… 183 84 black light brown 24 male mascu… Tatooi…
#> 10 Obi-Wan Ke… 182 77 auburn… fair blue-g… 57 male mascu… Stewjon
#> # … with 73 more rows, 4 more variables: species <chr>, films <list>,
#> # vehicles <list>, starships <list>, and abbreviated variable names
#> # ¹hair_color, ²skin_color, ³eye_color, ⁴birth_year, ⁵homeworld
# (+) Of which species are there at least 2 different gender values?
sw %>%
group_by(species, gender) %>%
count() %>% # table shows species by gender:
group_by(species) %>% # Which species appear more than once in this table?
count() %>%
filter(n > 1)
#> # A tibble: 4 × 2
#> # Groups: species [4]
#> species n
#> <chr> <int>
#> 1 Droid 2
#> 2 Human 2
#> 3 Kaminoan 2
#> 4 Twi'lek 2
# alternative (and shorter) solution:
sw %>%
group_by(species) %>%
summarise(n_gender_vals = n_distinct(gender)) %>%
filter(n_gender_vals >= 2)
#> # A tibble: 4 × 2
#> species n_gender_vals
#> <chr> <int>
#> 1 Droid 2
#> 2 Human 2
#> 3 Kaminoan 2
#> 4 Twi'lek 2Bonus task: R typically provides many ways to obtain a solution. Let’s gain an overview of the gender distribution in our
swdataset in three different ways:- Use a dplyr pipe to compute a summary table
tbthat counts the frequency of each gender insw.
- Use ggplot2 on the raw data of
swto create a bar chart (A) that shows the same gender distribution.
- Use ggplot2 on the summary table
tbto create a bar chart (B) that shows the same gender distribution.
- Use a dplyr pipe to compute a summary table
Solution
- Computing the summary table
tbis straightforward:
# Summary table:
tb <- sw %>%
group_by(gender) %>%
count()
tb
#> # A tibble: 3 × 2
#> # Groups: gender [3]
#> gender n
#> <chr> <int>
#> 1 feminine 17
#> 2 masculine 66
#> 3 <NA> 4Creating the same gender distribution as two plots:
- A: from the raw data
sw
- B: from the summary table
tb
- A: from the raw data
# Define some colors:
my_cols <- unikn::usecol(c(Pinky, Seegruen, Seeblau, Grau))
# Plot A: From raw data
ggplot(sw, aes(x = gender)) +
geom_bar(aes(fill = gender)) +
# Set labels, colors, and theme:
labs(title = "Gender distribution of the sw Universe", tag = "A",
x = "Gender", y = "Frequency", caption = "Using raw data sw.") +
scale_fill_manual(name = "Gender:", values = my_cols, na.value = "black") +
ds4psy::theme_ds4psy()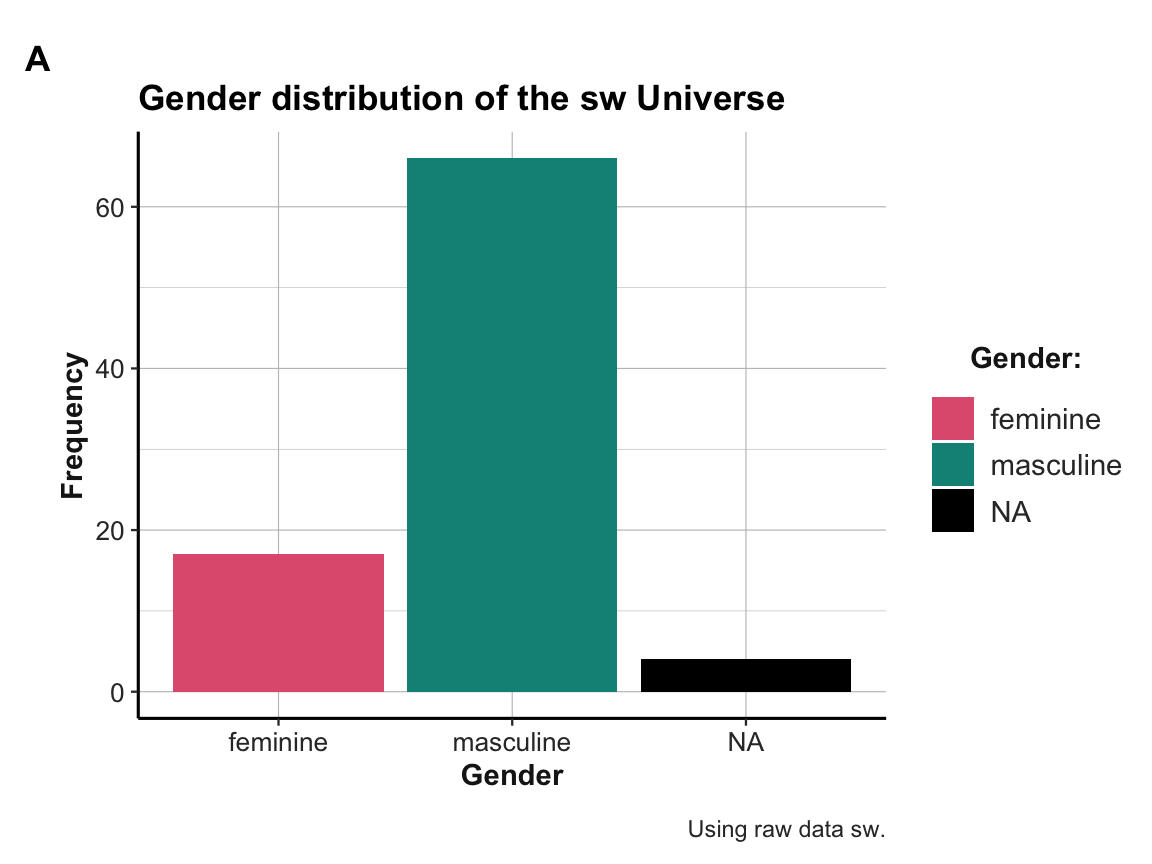
# Plot B: From summary table:
ggplot(tb, aes(x = gender, y = n)) +
geom_bar(aes(fill = gender), stat = "identity") +
# Set labels, colors, and theme:
labs(title = "Gender distribution of the sw Universe", tag = "B",
x = "Gender", y = "Frequency", caption = "Using summary data tb.") +
scale_fill_manual(name = "Gender:", values = my_cols, na.value = "black") +
ds4psy::theme_ds4psy()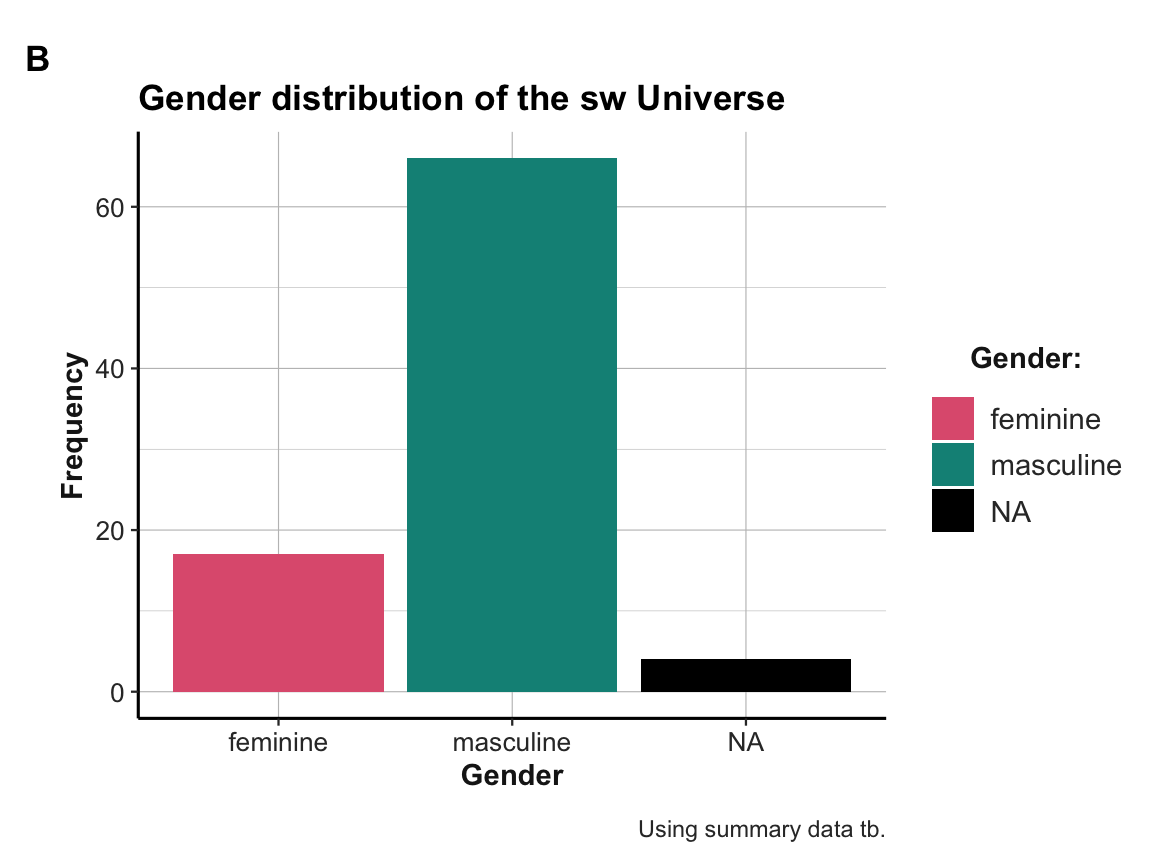
Note: Take a moment to reflect on the differences between both plots:
Both plots may look the same, but are internally quite different:
Plot A uses the raw data
swas its input and computes the frequency counts on the y-axis (as the default setting ofgeom_barisstat = "count"and the resulting counts are automatically mapped to the y-axis).Plot B uses the summary table
tbas its input and maps the existing values of the columnnon the y-axis. This requires setting the aesthetics toy = nand usingstat = "identity"forgeom_bar.)
Which of these ways we decide to use to plot summary results is a matter of preference, but has consequences:
On the one hand, using both a dplyr pipe and a ggplot2 command that re-computes the same counts (in Plot A) could help us to double-check our results. For instance, a common error is omitting the
na.valueargument when choosing your own colors withscale_fill_manual. This would drop the NA values from the plot — which we may never notice, if we had not computed the summary tabletbbefore.On the other hand, if we already have a summary table
tb, creating Plot B ensures that we are plotting exactly the contents of this table. Anything that changes the table will also change the plot.
Popular homes and heights
From which
homeworlddo the most indidividuals (rows) come from?What is the mean
heightof all individuals with orange eyes from the most popular homeworld?
Solution
## (d) Homeworld issues: -----
# (+) Popular homes: From which homeworld do the most indidividuals (rows) come from?
sw %>%
group_by(homeworld) %>%
count() %>%
arrange(desc(n))
#> # A tibble: 49 × 2
#> # Groups: homeworld [49]
#> homeworld n
#> <chr> <int>
#> 1 Naboo 11
#> 2 Tatooine 10
#> 3 <NA> 10
#> 4 Alderaan 3
#> 5 Coruscant 3
#> 6 Kamino 3
#> 7 Corellia 2
#> 8 Kashyyyk 2
#> 9 Mirial 2
#> 10 Ryloth 2
#> # … with 39 more rows
# => Naboo (with 11 individuals)
# (+) What is the mean height of all individuals with orange eyes from the most popular homeworld?
sw %>%
filter(homeworld == "Naboo", eye_color == "orange") %>%
summarise(n = n(),
mn_height = mean(height))
#> # A tibble: 1 × 2
#> n mn_height
#> <int> <dbl>
#> 1 3 209.
## Note:
sw %>%
filter(eye_color == "orange") # => 8 individuals
#> # A tibble: 8 × 14
#> name height mass hair_…¹ skin_…² eye_c…³ birth…⁴ sex gender homew…⁵
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 Jabba Desil… 175 1358 <NA> green-… orange 600 herm… mascu… Nal Hu…
#> 2 Ackbar 180 83 none brown … orange 41 male mascu… Mon Ca…
#> 3 Jar Jar Bin… 196 66 none orange orange 52 male mascu… Naboo
#> 4 Roos Tarpals 224 82 none grey orange NA male mascu… Naboo
#> 5 Rugor Nass 206 NA none green orange NA male mascu… Naboo
#> 6 Sebulba 112 40 none grey, … orange NA male mascu… Malast…
#> 7 Ben Quadina… 163 65 none grey, … orange NA male mascu… Tund
#> 8 Saesee Tiin 188 NA none pale orange NA male mascu… Iktotch
#> # … with 4 more variables: species <chr>, films <list>, vehicles <list>,
#> # starships <list>, and abbreviated variable names ¹hair_color, ²skin_color,
#> # ³eye_color, ⁴birth_year, ⁵homeworld
# (+) What is the mass and homeworld of the smallest droid?
sw %>%
filter(species == "Droid") %>%
arrange(height)
#> # A tibble: 6 × 14
#> name height mass hair_color skin_color eye_c…¹ birth…² sex gender homew…³
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
#> 1 R2-D2 96 32 <NA> white, bl… red 33 none mascu… Naboo
#> 2 R4-P17 96 NA none silver, r… red, b… NA none femin… <NA>
#> 3 R5-D4 97 32 <NA> white, red red NA none mascu… Tatooi…
#> 4 C-3PO 167 75 <NA> gold yellow 112 none mascu… Tatooi…
#> 5 IG-88 200 140 none metal red 15 none mascu… <NA>
#> 6 BB8 NA NA none none black NA none mascu… <NA>
#> # … with 4 more variables: species <chr>, films <list>, vehicles <list>,
#> # starships <list>, and abbreviated variable names ¹eye_color, ²birth_year,
#> # ³homeworldSize and mass issues
Compute the median, mean, and standard deviation of
heightfor all droids.Compute the average height and mass by species and save the result as
h_m.Sort
h_mto list the three species with the smallest individuals (in terms of meanheight).Sort
h_mto list the three species with the heaviest individuals (in terms of medianmass).
Solution
## Size and mass issues (group summaries): -----
# (+) Compute the median, mean, and standard deviation of `height` for all droids.
sw %>%
filter(species == "Droid") %>%
summarise(n = n(),
not_NA_h = sum(!is.na(height)),
md_height = median(height, na.rm = TRUE),
mn_height = mean(height, na.rm = TRUE),
sd_height = sd(height, na.rm = TRUE))
#> # A tibble: 1 × 5
#> n not_NA_h md_height mn_height sd_height
#> <int> <int> <int> <dbl> <dbl>
#> 1 6 5 97 131. 49.1
# (+) Compute the average height and mass by species and save the result as `h_m`:
h_m <- sw %>%
group_by(species) %>%
summarise(n = n(),
not_NA_h = sum(!is.na(height)),
mn_height = mean(height, na.rm = TRUE),
not_NA_m = sum(!is.na(mass)),
md_mass = median(mass, na.rm = TRUE)
)
h_m
#> # A tibble: 38 × 6
#> species n not_NA_h mn_height not_NA_m md_mass
#> <chr> <int> <int> <dbl> <int> <dbl>
#> 1 Aleena 1 1 79 1 15
#> 2 Besalisk 1 1 198 1 102
#> 3 Cerean 1 1 198 1 82
#> 4 Chagrian 1 1 196 0 NA
#> 5 Clawdite 1 1 168 1 55
#> 6 Droid 6 5 131. 4 53.5
#> 7 Dug 1 1 112 1 40
#> 8 Ewok 1 1 88 1 20
#> 9 Geonosian 1 1 183 1 80
#> 10 Gungan 3 3 209. 2 74
#> # … with 28 more rows
# (+) Use `h_m` to list the 3 species with the smallest individuals (in terms of mean height)?
h_m %>% arrange(mn_height) %>% slice(1:3)
#> # A tibble: 3 × 6
#> species n not_NA_h mn_height not_NA_m md_mass
#> <chr> <int> <int> <dbl> <int> <dbl>
#> 1 Yoda's species 1 1 66 1 17
#> 2 Aleena 1 1 79 1 15
#> 3 Ewok 1 1 88 1 20
# (+) Use `h_m` to list the 3 species with the heaviest individuals (in terms of median mass)?
h_m %>% arrange(desc(md_mass)) %>% slice(1:3)
#> # A tibble: 3 × 6
#> species n not_NA_h mn_height not_NA_m md_mass
#> <chr> <int> <int> <dbl> <int> <dbl>
#> 1 Hutt 1 1 175 1 1358
#> 2 Kaleesh 1 1 216 1 159
#> 3 Wookiee 2 2 231 2 124Bonus tasks
The following bonus tasks are more difficult, but can be solved with a single dplyr pipe:
How many individuals come from the three most frequent (known) species?
Which individuals are more than 20% lighter (in terms of mass) than the average mass of individuals of their own homeworld?
Solution
# How many individuals come from the 3 most frequent (known) species?
sw %>%
group_by(species) %>%
count %>%
arrange(desc(n)) %>%
filter(n > 1)
#> # A tibble: 9 × 2
#> # Groups: species [9]
#> species n
#> <chr> <int>
#> 1 Human 35
#> 2 Droid 6
#> 3 <NA> 4
#> 4 Gungan 3
#> 5 Kaminoan 2
#> 6 Mirialan 2
#> 7 Twi'lek 2
#> 8 Wookiee 2
#> 9 Zabrak 2
# Which individuals are more than 20% lighter (in terms of mass)
# than the average mass of individuals of their own homeworld?
sw %>%
select(name, homeworld, mass) %>%
group_by(homeworld) %>%
mutate(n_notNA_mass = sum(!is.na(mass)),
mn_mass = mean(mass, na.rm = TRUE),
lighter = mass < (mn_mass - (.20 * mn_mass))
) %>%
filter(lighter == TRUE)
#> # A tibble: 5 × 6
#> # Groups: homeworld [4]
#> name homeworld mass n_notNA_mass mn_mass lighter
#> <chr> <chr> <dbl> <int> <dbl> <lgl>
#> 1 R2-D2 Naboo 32 6 64.2 TRUE
#> 2 Leia Organa Alderaan 49 2 64 TRUE
#> 3 R5-D4 Tatooine 32 8 85.4 TRUE
#> 4 Yoda <NA> 17 3 82 TRUE
#> 5 Padmé Amidala Naboo 45 6 64.2 TRUEA.3.3 Exercise 3
Sleeping mammals
The dataset ggplot2::msleep contains a mammals sleep dataset (see ?msleep for details and the definition of variables).
- Save the data as
spand check the dimensions, variable types, and number of missing values in the dataset.
Solution
# Check:
dim(sp) # 83 x 11 variables
#> [1] 83 11
glimpse(sp) # 5 <chr> and 6 <dbl>
#> Rows: 83
#> Columns: 11
#> $ name <chr> "Cheetah", "Owl monkey", "Mountain beaver", "Greater shor…
#> $ genus <chr> "Acinonyx", "Aotus", "Aplodontia", "Blarina", "Bos", "Bra…
#> $ vore <chr> "carni", "omni", "herbi", "omni", "herbi", "herbi", "carn…
#> $ order <chr> "Carnivora", "Primates", "Rodentia", "Soricomorpha", "Art…
#> $ conservation <chr> "lc", NA, "nt", "lc", "domesticated", NA, "vu", NA, "dome…
#> $ sleep_total <dbl> 12.1, 17.0, 14.4, 14.9, 4.0, 14.4, 8.7, 7.0, 10.1, 3.0, 5…
#> $ sleep_rem <dbl> NA, 1.8, 2.4, 2.3, 0.7, 2.2, 1.4, NA, 2.9, NA, 0.6, 0.8, …
#> $ sleep_cycle <dbl> NA, NA, NA, 0.1333333, 0.6666667, 0.7666667, 0.3833333, N…
#> $ awake <dbl> 11.9, 7.0, 9.6, 9.1, 20.0, 9.6, 15.3, 17.0, 13.9, 21.0, 1…
#> $ brainwt <dbl> NA, 0.01550, NA, 0.00029, 0.42300, NA, NA, NA, 0.07000, 0…
#> $ bodywt <dbl> 50.000, 0.480, 1.350, 0.019, 600.000, 3.850, 20.490, 0.04…
sum(is.na(sp)) # 136 missing values
#> [1] 136Arranging and filtering data
Use the dplyr-verbs arrange(), group_by(), and filter() to answer the following questions by creating ordered subsets of the data:
Arrange the rows (alphabetically) by
vore,order, andname, and report thegenusof the top three mammals.What is the most common type of
vorein the data? How many omnivores are there?What is the most common
orderin the dataset? Are there more exemplars of theorder“Carnivora” or “Primates”?Which two mammals of the order “Primates” have the longest and shortest
sleep_totaltimes?
Solution
# Arranging rows:
sp1 <- sp %>%
arrange(vore, order, name)
sp1$genus[1:3] # => top 3: "Vulpes" "Phoca" "Acinonyx"
#> [1] "Vulpes" "Phoca" "Acinonyx"
# Counting common vores:
# (a) short solution:
sp %>%
count(vore) %>%
arrange(desc(n))
#> # A tibble: 5 × 2
#> vore n
#> <chr> <int>
#> 1 herbi 32
#> 2 omni 20
#> 3 carni 19
#> 4 <NA> 7
#> 5 insecti 5
# (a) is the same as (b):
sp %>%
group_by(vore) %>%
tally() %>%
arrange(desc(n))
#> # A tibble: 5 × 2
#> vore n
#> <chr> <int>
#> 1 herbi 32
#> 2 omni 20
#> 3 carni 19
#> 4 <NA> 7
#> 5 insecti 5
# (b) is the same as (c):
sp %>%
group_by(vore) %>%
summarise(n = n()) %>%
arrange(desc(n))
#> # A tibble: 5 × 2
#> vore n
#> <chr> <int>
#> 1 herbi 32
#> 2 omni 20
#> 3 carni 19
#> 4 <NA> 7
#> 5 insecti 5
# => 32 herbivores, 20 omnivores
# Counting common orders:
sp %>%
count(order) %>%
arrange(desc(n))
#> # A tibble: 19 × 2
#> order n
#> <chr> <int>
#> 1 Rodentia 22
#> 2 Carnivora 12
#> 3 Primates 12
#> 4 Artiodactyla 6
#> 5 Soricomorpha 5
#> 6 Cetacea 3
#> 7 Hyracoidea 3
#> 8 Perissodactyla 3
#> 9 Chiroptera 2
#> 10 Cingulata 2
#> 11 Didelphimorphia 2
#> 12 Diprotodontia 2
#> 13 Erinaceomorpha 2
#> 14 Proboscidea 2
#> 15 Afrosoricida 1
#> 16 Lagomorpha 1
#> 17 Monotremata 1
#> 18 Pilosa 1
#> 19 Scandentia 1
# OR:
sp %>%
group_by(order) %>%
count() %>%
arrange(desc(n))
#> # A tibble: 19 × 2
#> # Groups: order [19]
#> order n
#> <chr> <int>
#> 1 Rodentia 22
#> 2 Carnivora 12
#> 3 Primates 12
#> 4 Artiodactyla 6
#> 5 Soricomorpha 5
#> 6 Cetacea 3
#> 7 Hyracoidea 3
#> 8 Perissodactyla 3
#> 9 Chiroptera 2
#> 10 Cingulata 2
#> 11 Didelphimorphia 2
#> 12 Diprotodontia 2
#> 13 Erinaceomorpha 2
#> 14 Proboscidea 2
#> 15 Afrosoricida 1
#> 16 Lagomorpha 1
#> 17 Monotremata 1
#> 18 Pilosa 1
#> 19 Scandentia 1
# => 22 Rodentia, 12 Carnivora = 12 Primates
# Primates with longest and shortest sleep_total times:
sp %>%
filter(order == "Primates") %>%
arrange(desc(sleep_total))
#> # A tibble: 12 × 11
#> name genus vore order conse…¹ sleep…² sleep…³ sleep…⁴ awake brainwt bodywt
#> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 Owl m… Aotus omni Prim… <NA> 17 1.8 NA 7 0.0155 0.48
#> 2 Slow … Nyct… carni Prim… <NA> 11 NA NA 13 0.0125 1.4
#> 3 Potto Pero… omni Prim… lc 11 NA NA 13 NA 1.1
#> 4 Patas… Eryt… omni Prim… lc 10.9 1.1 NA 13.1 0.115 10
#> 5 Macaq… Maca… omni Prim… <NA> 10.1 1.2 0.75 13.9 0.179 6.8
#> 6 Grivet Cerc… omni Prim… lc 10 0.7 NA 14 NA 4.75
#> 7 Galago Gala… omni Prim… <NA> 9.8 1.1 0.55 14.2 0.005 0.2
#> 8 Chimp… Pan omni Prim… <NA> 9.7 1.4 1.42 14.3 0.44 52.2
#> 9 Squir… Saim… omni Prim… <NA> 9.6 1.4 NA 14.4 0.02 0.743
#> 10 Mongo… Lemur herbi Prim… vu 9.5 0.9 NA 14.5 NA 1.67
#> 11 Baboon Papio omni Prim… <NA> 9.4 1 0.667 14.6 0.18 25.2
#> 12 Human Homo omni Prim… <NA> 8 1.9 1.5 16 1.32 62
#> # … with abbreviated variable names ¹conservation, ²sleep_total, ³sleep_rem,
#> # ⁴sleep_cycle
# => max: Owl monkey: 17 hours, min: Human 8 hours.Computing new variables
Solve the following tasks by combining the dplyr commands mutate, group_by, and summarise:
Compute a variable
sleep_awake_sumthat adds thesleep_totaltime and theawaketime of each mammal. What result do you expect and get?Which animals have the smallest and largest brain to body ratio (in terms of weight)? How many mammals have a larger ratio than humans?
What is the minimum, average (mean), and maximum sleep cycle length for each
vore? (Hint: First group the data bygroup_by, then usesummariseon thesleep_cyclevariable, but also count the number ofNAvalues for eachvore. When computing grouped summaries,NAvalues can be removed byna.rm = TRUE.)Replace your
summariseverb in the previous task bymutate. What do you get as a result? (Hint: The last two tasks illustrate the difference betweenmutateand groupedmutatecommands.)
Solution
# Computing `sleep_awake_sum`:
sp2 <- sp %>%
mutate(sleep_awake_sum = sleep_total + awake)
sp2$sleep_awake_sum # => all 24 hours
#> [1] 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00
#> [13] 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00
#> [25] 24.00 24.00 24.00 24.00 24.00 24.00 24.05 24.00 24.00 24.00 24.00 24.00
#> [37] 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00
#> [49] 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.05
#> [61] 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00 24.00
#> [73] 24.00 24.00 24.00
#> [ reached getOption("max.print") -- omitted 8 entries ]
# Computing brain to body ratios:
sp3 <- sp %>%
mutate(brain_to_body_wt = brainwt / bodywt) %>%
arrange(brain_to_body_wt)
sp3$name[1] # => smallest ratio: Cow
#> [1] "Cow"
sp3 %>%
arrange(desc(brain_to_body_wt))
#> # A tibble: 83 × 12
#> name genus vore order conse…¹ sleep…² sleep…³ sleep…⁴ awake brainwt bodywt
#> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 Thirt… Sper… herbi Rode… lc 13.8 3.4 0.217 10.2 0.004 0.101
#> 2 Owl m… Aotus omni Prim… <NA> 17 1.8 NA 7 0.0155 0.48
#> 3 Lesse… Cryp… omni Sori… lc 9.1 1.4 0.15 14.9 0.00014 0.005
#> 4 Squir… Saim… omni Prim… <NA> 9.6 1.4 NA 14.4 0.02 0.743
#> 5 Macaq… Maca… omni Prim… <NA> 10.1 1.2 0.75 13.9 0.179 6.8
#> 6 Littl… Myot… inse… Chir… <NA> 19.9 2 0.2 4.1 0.00025 0.01
#> 7 Galago Gala… omni Prim… <NA> 9.8 1.1 0.55 14.2 0.005 0.2
#> 8 Mole … Spal… <NA> Rode… <NA> 10.6 2.4 NA 13.4 0.003 0.122
#> 9 Tree … Tupa… omni Scan… <NA> 8.9 2.6 0.233 15.1 0.0025 0.104
#> 10 Human Homo omni Prim… <NA> 8 1.9 1.5 16 1.32 62
#> # … with 73 more rows, 1 more variable: brain_to_body_wt <dbl>, and abbreviated
#> # variable names ¹conservation, ²sleep_total, ³sleep_rem, ⁴sleep_cycle
# => largest ratio: Thirteen-lined ground squirrel,
# 9 mammals have larger brain_to_body_wt than Human.
# Computing sleep cycle length for each `vore`:
sp %>%
group_by(vore) %>%
summarise(n = n(),
n_NA = sum(is.na(sleep_cycle)),
non_NA = sum(!is.na(sleep_cycle)),
min_cyc = min(sleep_cycle, na.rm = TRUE),
mn_cyc = mean(sleep_cycle, na.rm = TRUE),
max_cyc = max(sleep_cycle, na.rm = TRUE)
)
#> # A tibble: 5 × 7
#> vore n n_NA non_NA min_cyc mn_cyc max_cyc
#> <chr> <int> <int> <int> <dbl> <dbl> <dbl>
#> 1 carni 19 14 5 0.333 0.373 0.417
#> 2 herbi 32 20 12 0.117 0.418 1
#> 3 insecti 5 2 3 0.117 0.161 0.2
#> 4 omni 20 9 11 0.133 0.592 1.5
#> 5 <NA> 7 6 1 0.183 0.183 0.183
# Replacing summarise by mutate:
sp4 <- sp %>%
group_by(vore) %>%
mutate(n = n(),
n_NA = sum(is.na(sleep_cycle)),
non_NA = sum(!is.na(sleep_cycle)),
min_cyc = min(sleep_cycle, na.rm = TRUE),
mn_cyc = mean(sleep_cycle, na.rm = TRUE),
max_cyc = max(sleep_cycle, na.rm = TRUE)
)
# => A tibble that contains grouped summaries as 6 new variables:
sp4 %>%
select(vore, name, sleep_cycle, n:max_cyc) %>%
arrange(vore)
#> # A tibble: 83 × 9
#> # Groups: vore [5]
#> vore name sleep…¹ n n_NA non_NA min_cyc mn_cyc max_cyc
#> <chr> <chr> <dbl> <int> <int> <int> <dbl> <dbl> <dbl>
#> 1 carni Cheetah NA 19 14 5 0.333 0.373 0.417
#> 2 carni Northern fur seal 0.383 19 14 5 0.333 0.373 0.417
#> 3 carni Dog 0.333 19 14 5 0.333 0.373 0.417
#> 4 carni Long-nosed armadillo 0.383 19 14 5 0.333 0.373 0.417
#> 5 carni Domestic cat 0.417 19 14 5 0.333 0.373 0.417
#> 6 carni Pilot whale NA 19 14 5 0.333 0.373 0.417
#> 7 carni Gray seal NA 19 14 5 0.333 0.373 0.417
#> 8 carni Thick-tailed opposum NA 19 14 5 0.333 0.373 0.417
#> 9 carni Slow loris NA 19 14 5 0.333 0.373 0.417
#> 10 carni Northern grasshopper… NA 19 14 5 0.333 0.373 0.417
#> # … with 73 more rows, and abbreviated variable name ¹sleep_cycleA.3.4 Exercise 4
Outliers
This exercise examines different possibilities for defining outliers and uses the outliers dataset of the ds4psy package (also available as out.csv at http://rpository.com/ds4psy/data/out.csv) to illustate and compare them.
With respect to your insights into dplyr, this exercise helps disentangling mutate from grouped mutate commands.
Data on outliers
Use the outliers data (from the ds4psy package) or use the following read_csv() command to load the data into an R object entitled outliers:
# Data:
outliers <- ds4psy::outliers # from ds4psy package
## Alternatively, load csv data from online source (as comma-separated file):
# outliers_2 <- readr::read_csv("http://rpository.com/ds4psy/data/out.csv") # from online source
## Verify equality:
# all.equal(ds4psy::outliers, outliers_2)
## Alternatively from a local data file:
# outliers <- read_csv("out.csv") # from current directoryNot all outliers are alike
An outlier can be defined as an individual whose value in some variable deviates by more than a given criterion (e.g., two standard deviations) from the mean of the variable. However, this definition is incomplete unless it also specifies the reference group over which the means and deviations are computed. In the following, we explore the implications of different reference groups.
Basic tasks
Save the data into a tibble
outliersand report its number of observations and variables, and their types.How many missing data values are there in
outliers?What is the gender (or
sex) distribution in this sample?Create a plot that shows the distribution of
heightvalues for each gender.
Solution
# Load and inspect data:
# outliers <- read_csv("out.csv") # read in csv-file
# outliers <- as_tibble(data) # if data is not already a tibble
dim(outliers) # => 1000 observations (rows) x 3 variables (columns)
#> [1] 1000 3
# Missing data points:
sum(is.na(outliers)) # => 18 missing values
#> [1] 18
# Gender distribution:
outliers %>%
group_by(sex) %>%
count()
#> # A tibble: 2 × 2
#> # Groups: sex [2]
#> sex n
#> <chr> <int>
#> 1 female 507
#> 2 male 493
# => 50.7% females, 49.3% males.
# Distributions of `height` as density plot:
ggplot(outliers, aes(x = height)) +
geom_density(fill = "gold", alpha = 2/3) +
geom_density(aes(fill = sex), alpha = 1/3) +
labs(title = "Distribution of heights overall and by gender",
fill = "Gender") +
# scale_fill_brewer(palette = "Set1") + # using Brewer palette
scale_fill_manual(name = "Gender:", values = unikn::usecol(c(Pinky, Seeblau))) + # unikn colors
theme_bw()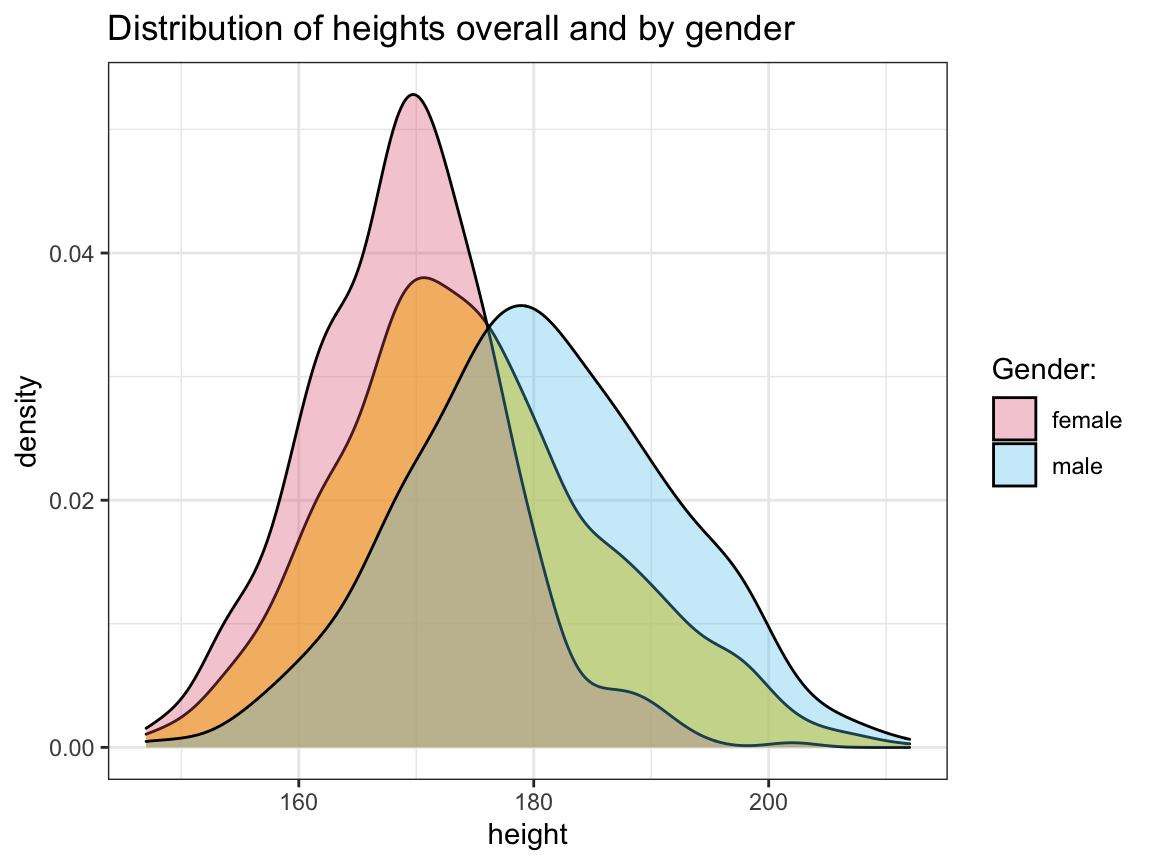
# Note: To avoid the warning about removing 18 cases with NA-values,
# we could first filter out those cases:
# non_NA_data <- filter(outliers, !is.na(height))
# Alternative solution as 2 histograms:
ggplot(outliers) +
facet_wrap(~sex) +
geom_histogram(aes(x = height, fill = sex), binwidth = 5, color = "grey10") +
labs(title = "Distribution of heights by gender",
x = "Height", y = "Frequency") +
# scale_fill_brewer(name = "Gender:", palette = "Set1") + # using Brewer palette
scale_fill_manual(name = "Gender:", values = unikn::usecol(c(Pinky, Seeblau))) + # unikn colors
theme_bw()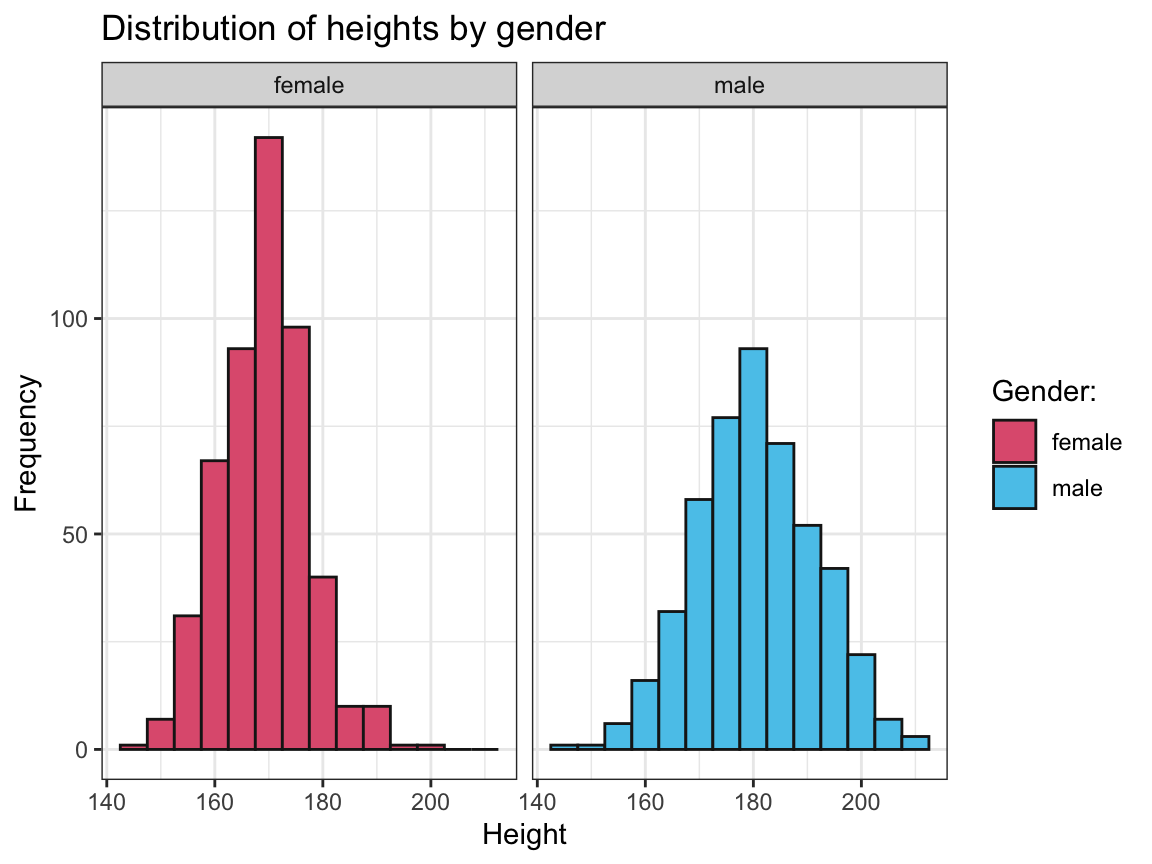
Defining different outliers
Compute 2 new variables that signal and distinguish between 2 types of outliers in terms of height:
outliers relative to the
heightof the overall sample (i.e., individuals withheightvalues deviating more than 2 SD from the overall mean ofheight);outliers relative to the
heightof some subgroup’s mean and SD. Here, a suitable subgroup to consider is every person’s gender (i.e., individuals withheightvalues deviating more than 2 SD from the meanheightof their own gender).
Hints: As both variable signal whether or not someone is an outlier they should be defined as logicals (being either TRUE or FALSE) and added as new columns to data (via appropriate mutate commands). While the 1st variable can be computed based on the mean and SD of the overall sample, the 2nd variable can be computed after grouping outliers by gender and then computing and using the corresponding mean and SD values. The absolute difference between 2 numeric values x and y is provided by abs(x - y).
Relative outliers
Now use the 2 new outlier variables to define (or filter) 2 subsets of the data that contain 2 subgroups of people:
out_1: Individuals (females and males) withheightvalues that are outliers relative to both the entire sample and the sample of their own gender. How many such individuals are inoutliers?out_2: Individuals (females and males) withheightvalues that are not outliers relative to the entire population, but are outliers relative to their own gender. How many such individuals are inoutliers?
Solution
## Defining different outliers: -----
# Included in data_out (below), but also possible to do separately:
# Compute the number, means and SD of height values in 2 ways:
# 1. overall:
outliers %>%
summarise(n = n(),
n_not_NA = sum(!is.na(height)),
mn_height = mean(height, na.rm = TRUE),
sd_height = sd(height, na.rm = TRUE))
#> # A tibble: 1 × 4
#> n n_not_NA mn_height sd_height
#> <int> <int> <dbl> <dbl>
#> 1 1000 982 175. 11.3
# 2. by gender:
outliers %>%
group_by(sex) %>%
summarise(n = n(),
n_not_NA = sum(!is.na(height)),
mn_height = mean(height, na.rm = TRUE),
sd_height = sd(height, na.rm = TRUE))
#> # A tibble: 2 × 5
#> sex n n_not_NA mn_height sd_height
#> <chr> <int> <int> <dbl> <dbl>
#> 1 female 507 501 169. 8.19
#> 2 male 493 481 181. 11.0
# Detecting and marking outliers (by logical variables):
# Compute the means, SDs, and corresponding outliers in 2 ways:
crit <- 2 # criterion value for detecting outliers (in SD units)
data_out <- outliers %>%
# 1. Compute means, SD, and outliers for overall sample:
mutate(mn_height = mean(height, na.rm = TRUE),
sd_height = sd(height, na.rm = TRUE),
out_height = abs(height - mn_height) > (crit * sd_height)) %>%
group_by(sex) %>%
# 2. Compute same metrics for subgroups (by sex):
mutate(mn_sex_height = mean(height, na.rm = TRUE),
sd_sex_height = sd(height, na.rm = TRUE),
out_sex_height = abs(height - mn_sex_height) > (crit * sd_sex_height))
knitr::kable(head(data_out))| id | sex | height | mn_height | sd_height | out_height | mn_sex_height | sd_sex_height | out_sex_height |
|---|---|---|---|---|---|---|---|---|
| nr.1 | female | 164 | 174.7006 | 11.26015 | FALSE | 169.0679 | 8.193619 | FALSE |
| nr.2 | male | 178 | 174.7006 | 11.26015 | FALSE | 180.5676 | 11.026677 | FALSE |
| nr.3 | female | 177 | 174.7006 | 11.26015 | FALSE | 169.0679 | 8.193619 | FALSE |
| nr.4 | male | 188 | 174.7006 | 11.26015 | FALSE | 180.5676 | 11.026677 | FALSE |
| nr.5 | male | 193 | 174.7006 | 11.26015 | FALSE | 180.5676 | 11.026677 | FALSE |
| nr.6 | female | 168 | 174.7006 | 11.26015 | FALSE | 169.0679 | 8.193619 | FALSE |
## Relative outliers: -----
# Filter specific combinations of outliers:
# 1. Outliers relative to the entire population AND to their own gender:
out_1 <- data_out %>%
filter(out_height & out_sex_height) %>%
arrange(sex, height)
nrow(out_1) # => 21 individuals.
#> [1] 21
# 2. Outliers relative to their own gender, but NOT relative to the entire population:
out_2 <- data_out %>%
filter(!out_height & out_sex_height) %>%
arrange(sex, height)
nrow(out_2) # => 24 individuals.
#> [1] 24Bonus plots
Visualize the raw values and distributions of
heightfor both types of outliers (out_1andout_2) in 2 separate plots.Interpret both plots by describing the
heightandsexcombination of the individuals shown in each plot.
Solution
# Visualization and interpretation of both types of outliers:
# 1. Showing out_1:
ggplot(out_1, aes(x = sex, y = height)) +
geom_violin(aes(fill = sex)) +
geom_jitter(size = 4, alpha = 2/3) +
# scale_fill_manual(values = c("firebrick", "steelblue3")) +
scale_fill_manual(name = "Gender:", values = unikn::usecol(c(Pinky, Seeblau))) + # unikn colors
labs(title = "Outliers relative to both overall sample and gender",
x = "Gender", y = "Height (in cm)",
fill = "Gender:") +
theme_bw()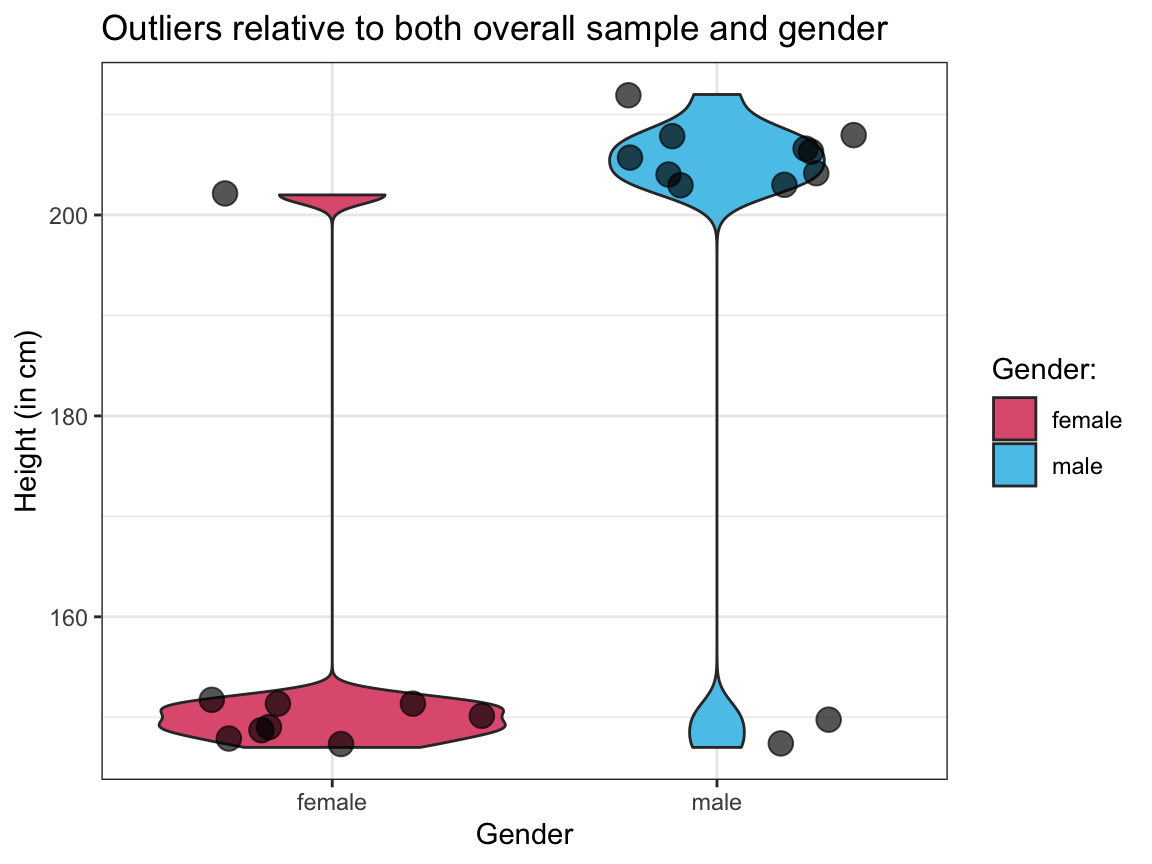
# Interpretation:
# `out_1` contains mostly short women (except for 1 tall woman)
# and mostly tall men (except for 2 short men).
# 2. Showing out_2:
ggplot(out_2, aes(x = sex, y = height)) +
geom_violin(aes(fill = sex)) +
geom_jitter(size = 4, alpha = 2/3) +
# scale_fill_manual(values = c("firebrick", "steelblue3")) +
scale_fill_manual(name = "Gender:", values = unikn::usecol(c(Pinky, Seeblau))) + # unikn colors
labs(title = "Outliers relative to gender but not overall sample",
x = "Gender", y = "Height (in cm)",
fill = "Gender:") +
theme_bw()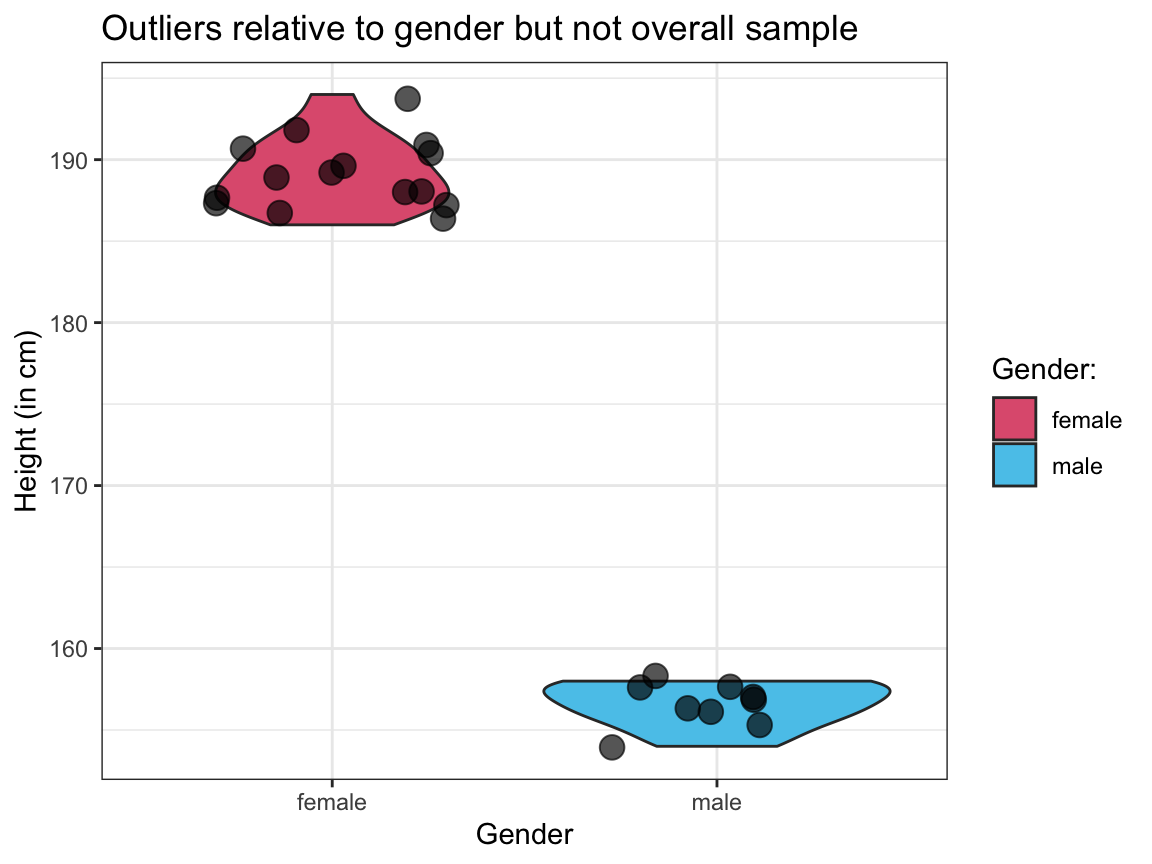
A.3.5 Exercise 5
Revisiting positive psychology
In previous exercises, we used the p_info data — available as posPsy_p_info in the ds4psy package or as http://rpository.com/ds4psy/data/posPsy_participants.csv — from a study on the effectiveness of web-based positive psychology interventions (Woodworth et al., 2018).
More specifically, we used this data in
Exercise 6 of Chapter 1 and
Exercise 5 of Chapter 2
to explore the participant information and create some corresponding plots.
(See Section B.1 of Appendix B for background information on this data.)
Answer the same questions as in those exercises by verifying your earlier base R results and ggplot2 graphs by pipes of dplyr functions. Do your graphs and quantitative results support the same conclusions?
From Exercise 6 of Chapter 1
Questions from Exercise 6 of Chapter 1:
Examine the participant information in p_info by describing each of its variables:
How many individuals are contained in the dataset?
What percentage of them is female (i.e., has a
sexvalue of 1)?How many participants were in one of the 3 treatment groups (i.e., have an
interventionvalue of 1, 2, or 3)?What is the participants’ mean education level? What percentage has a university degree (i.e., an
educvalue of at least 4)?What is the age range (
mintomax) of participants? What is the average (mean and median) age?Describe the range of
incomelevels present in this sample of participants. What percentage of participants self-identifies as having a below-average income (i.e., anincomevalue of 1)?
Solution
# 1. How many individuals are contained in the dataset?
N <- nrow(p_info) # OR
N # 295
#> [1] 295
# Note:
p_info %>% count()
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 295
# would yield a tibble (with only 1 element: 295).
# 2. What percentage of them is female (i.e., has a `sex` value of 1)?
p_info %>%
group_by(sex) %>%
summarise(n_sex = n(), # number
pc_sex = n_sex/N * 100 # percentage
)
#> # A tibble: 2 × 3
#> sex n_sex pc_sex
#> <dbl> <int> <dbl>
#> 1 1 251 85.1
#> 2 2 44 14.9
# 3. How many participants were in one of the 3 treatment groups (i.e., have an `intervention` value of 1, 2, or 3)?
p_info %>%
filter(intervention < 4) %>%
count()
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 222
# => 222 individuals
# OR:
t_iv <- p_info %>%
group_by(intervention) %>%
summarise(n_iv = n(), # number
pc_iv = n_iv/N * 100 # percentage
)
t_iv
#> # A tibble: 4 × 3
#> intervention n_iv pc_iv
#> <dbl> <int> <dbl>
#> 1 1 72 24.4
#> 2 2 76 25.8
#> 3 3 74 25.1
#> 4 4 73 24.7
sum(t_iv$n_iv[1:3])
#> [1] 222
# => 222 in elements 1:3 of vector n_iv
# 4. What is the participants' mean education level?
p_info %>%
summarise(mn_edu = mean(educ))
#> # A tibble: 1 × 1
#> mn_edu
#> <dbl>
#> 1 3.98
# What percentage has a university degree (i.e., an `educ` value of at least 4)?
p_info %>%
group_by(educ) %>%
summarise(n_edu = n(), # number
pc_edu = n_edu/N * 100 # percentage
)
#> # A tibble: 5 × 3
#> educ n_edu pc_edu
#> <dbl> <int> <dbl>
#> 1 1 14 4.75
#> 2 2 21 7.12
#> 3 3 39 13.2
#> 4 4 104 35.3
#> 5 5 117 39.7
# 5. What is the age range (`min` to `max`) of participants?
# What is the average (mean and median) age?
p_info %>%
summarise(n = n(),
min_age = min(age),
mn_age = mean(age),
max_age = max(age))
#> # A tibble: 1 × 4
#> n min_age mn_age max_age
#> <int> <dbl> <dbl> <dbl>
#> 1 295 18 43.8 83
# 6. Describe the range of `income` levels present in this sample of participants.
# What percentage of participants self-identifies as a below-average income
# (i.e., an `income` value of 1)?
p_info %>%
group_by(income) %>%
summarise(n_income = n(),
pc_income = n_income/N * 100)
#> # A tibble: 3 × 3
#> income n_income pc_income
#> <dbl> <int> <dbl>
#> 1 1 73 24.7
#> 2 2 136 46.1
#> 3 3 86 29.2From Exercise 5 of Chapter 2
Questions from Exercise 5 of Chapter 2:
Use the p_info data to create some plots that descripte the sample of participants:
- A histogram that shows the distribution of participant
agein 3 ways:- overall,
- separately for each
sex, and - separately for each
intervention.
Solution
When using dplyr instead of ggplot2, we can replace the information contained in the histogram by a table of descriptives:
# Age distribution (as a table):
age_overall <- p_info %>%
summarise(n = n(), # number
min_age = min(age), # minimum
mn_age = mean(age), # mean
md_age = median(age), # median
sd_age = sd(age), # standard deviation
max_age = max(age) # maximum
)
age_overall
#> # A tibble: 1 × 6
#> n min_age mn_age md_age sd_age max_age
#> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 295 18 43.8 44 12.4 83
# Age distribution by sex (as a table):
age_by_sex <- p_info %>%
group_by(sex) %>%
summarise(n = n(), # number
min_age = min(age), # minimum
mn_age = mean(age), # mean
md_age = median(age), # median
sd_age = sd(age), # standard deviation
max_age = max(age) # maximum
)
age_by_sex
#> # A tibble: 2 × 7
#> sex n min_age mn_age md_age sd_age max_age
#> <dbl> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 251 19 43.9 44 12.1 83
#> 2 2 44 18 43.0 44 14.1 71
# Age distribution by intervention (as a table):
age_by_iv <- p_info %>%
group_by(intervention) %>%
summarise(n = n(), # number
min_age = min(age), # minimum
mn_age = mean(age), # mean
md_age = median(age), # median
sd_age = sd(age), # standard deviation
max_age = max(age) # maximum
)
age_by_iv
#> # A tibble: 4 × 7
#> intervention n min_age mn_age md_age sd_age max_age
#> <dbl> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 72 22 44.6 45 12.1 71
#> 2 2 76 18 45.4 45.5 12.5 83
#> 3 3 74 19 43.3 44 12.2 71
#> 4 4 73 19 41.7 40 12.8 75- A bar plot that
- shows how many participants took part in each
intervention; or - shows how many participants of each
sextook part in eachintervention.
- shows how many participants took part in each
Solution
Using dplyr instead of ggplot2:
# Number of participants per intervention:
# (was contained in age_by_iv above):
age_by_iv %>%
select(intervention, n)
#> # A tibble: 4 × 2
#> intervention n
#> <dbl> <int>
#> 1 1 72
#> 2 2 76
#> 3 3 74
#> 4 4 73
# N and percentage by intervention and sex:
p_info %>%
group_by(intervention, sex) %>%
summarise(n_iv_sex = n(), # number
pc_iv_sex = n_iv_sex/N * 100 # percentage
)
#> # A tibble: 8 × 4
#> # Groups: intervention [4]
#> intervention sex n_iv_sex pc_iv_sex
#> <dbl> <dbl> <int> <dbl>
#> 1 1 1 62 21.0
#> 2 1 2 10 3.39
#> 3 2 1 66 22.4
#> 4 2 2 10 3.39
#> 5 3 1 62 21.0
#> 6 3 2 12 4.07
#> 7 4 1 61 20.7
#> 8 4 2 12 4.07A.3.6 Exercise 6
Surviving the Titanic
The Titanic data in datasets contains basic information on the Age, Class, Sex, and Survival status for the people on board of the fatal maiden voyage of the Titanic. This data is saved as a 4-dimensional array resulting from cross-tabulating 2201 observations on four variables, but can easily be transformed into a tibble titanic by calling titanic <- as_tibble(datasets::Titanic).
# ?datasets::Titanic # see documentation
# dim(Titanic) # see also dim(FFTrees::titanic)
titanic <- tibble::as_tibble(datasets::Titanic)
knitr::kable(head(titanic), caption = "The head of the Titanic dataset (as a tibble).")| Class | Sex | Age | Survived | n |
|---|---|---|---|---|
| 1st | Male | Child | No | 0 |
| 2nd | Male | Child | No | 0 |
| 3rd | Male | Child | No | 35 |
| Crew | Male | Child | No | 0 |
| 1st | Female | Child | No | 0 |
| 2nd | Female | Child | No | 0 |
Use dplyr pipes to answer each of the following questions by a summary table that counts the sum of particular groups of survivors.
- Determine the number of survivors by
Sex: Were female passengers more likely to survive than male passengers?
# Survivors by Sex:
# (a) simple solution:
titanic %>%
group_by(Sex, Survived) %>%
summarise(Number = sum(n))
#> # A tibble: 4 × 3
#> # Groups: Sex [2]
#> Sex Survived Number
#> <chr> <chr> <dbl>
#> 1 Female No 126
#> 2 Female Yes 344
#> 3 Male No 1364
#> 4 Male Yes 367
# (b) complex solution (computing percentages overall and per subgroup):
t_51 <- titanic %>%
group_by(Sex, Survived) %>%
summarise(nr_all = sum(n),
pc_all = round(nr_all/sum(titanic$n) * 100, 1)) %>%
group_by(Sex) %>%
mutate(nr_sex = sum(nr_all),
pc_sex = round(nr_all/nr_sex * 100, 1))
knitr::kable(t_51, caption = "Number of Titanic survivors by `Sex`.")| Sex | Survived | nr_all | pc_all | nr_sex | pc_sex |
|---|---|---|---|---|---|
| Female | No | 126 | 5.7 | 470 | 26.8 |
| Female | Yes | 344 | 15.6 | 470 | 73.2 |
| Male | No | 1364 | 62.0 | 1731 | 78.8 |
| Male | Yes | 367 | 16.7 | 1731 | 21.2 |
# Corresponding Chi-square test:
tab_2x2 <- xtabs(n ~ Survived + Sex, titanic)
tab_2x2
#> Sex
#> Survived Female Male
#> No 126 1364
#> Yes 344 367
csq_tst <- chisq.test(tab_2x2, correct = FALSE)
csq_tst
#>
#> Pearson's Chi-squared test
#>
#> data: tab_2x2
#> X-squared = 456.87, df = 1, p-value < 2.2e-16
csq_tst$expected
#> Sex
#> Survived Female Male
#> No 318.1736 1171.8264
#> Yes 151.8264 559.1736
csq_tst$observed
#> Sex
#> Survived Female Male
#> No 126 1364
#> Yes 344 367Answer: Yes, the variables Sex and Survived are related.
Whereas females were about three times as likely to survive than to die, males were about four times as likely to die than to survive.
- Determine the number of survivors by
Age: Were children more likely to survive than adults?
# Survivors by Age:
# (a) simple solution:
titanic %>%
group_by(Age, Survived) %>%
summarise(Number = sum(n))
#> # A tibble: 4 × 3
#> # Groups: Age [2]
#> Age Survived Number
#> <chr> <chr> <dbl>
#> 1 Adult No 1438
#> 2 Adult Yes 654
#> 3 Child No 52
#> 4 Child Yes 57
# (b) complex solution (computing percentages overall and per subgroup):
t_52 <- titanic %>%
group_by(Age, Survived) %>%
summarise(nr_age_sur = sum(n),
pc_age_sur = round(nr_age_sur/sum(titanic$n) * 100, 1)) %>%
group_by(Age) %>%
mutate(nr_age = sum(nr_age_sur),
pc_age = round(nr_age_sur/nr_age * 100, 1))
knitr::kable(t_52, caption = "Number of Titanic survivors by `Age`.")| Age | Survived | nr_age_sur | pc_age_sur | nr_age | pc_age |
|---|---|---|---|---|---|
| Adult | No | 1438 | 65.3 | 2092 | 68.7 |
| Adult | Yes | 654 | 29.7 | 2092 | 31.3 |
| Child | No | 52 | 2.4 | 109 | 47.7 |
| Child | Yes | 57 | 2.6 | 109 | 52.3 |
# Corresponding Chi-square test:
tab_2x2 <- xtabs(n ~ Survived + Age, titanic)
tab_2x2
#> Age
#> Survived Adult Child
#> No 1438 52
#> Yes 654 57
csq_tst <- chisq.test(tab_2x2, correct = FALSE)
csq_tst
#>
#> Pearson's Chi-squared test
#>
#> data: tab_2x2
#> X-squared = 20.956, df = 1, p-value = 4.701e-06
csq_tst$expected
#> Age
#> Survived Adult Child
#> No 1416.2108 73.78919
#> Yes 675.7892 35.21081
csq_tst$observed
#> Age
#> Survived Adult Child
#> No 1438 52
#> Yes 654 57Answer: Yes, the variables Age and Survived are related. Whereas adults were more than twice as likely to die than to survive, the chances of survival for children were almost 50%.
- Consider the number of survivors as a function of both
SexandAge. Does the pattern observed in 1. hold equally for children and adults?
# Survivors by Age and Sex:
t_53 <- titanic %>%
group_by(Age, Sex, Survived) %>%
summarise(nr_age_sex_sur = sum(n),
pc_age_sex_sur = round(nr_age_sex_sur/sum(titanic$n) * 100, 1)) %>%
group_by(Age, Sex) %>%
mutate(nr_age_sex = sum(nr_age_sex_sur),
pc_age_sex = round(nr_age_sex_sur/nr_age_sex * 100, 1))
knitr::kable(t_53, caption = "Number of Titanic survivors by `Age` and `Sex`.")| Age | Sex | Survived | nr_age_sex_sur | pc_age_sex_sur | nr_age_sex | pc_age_sex |
|---|---|---|---|---|---|---|
| Adult | Female | No | 109 | 5.0 | 425 | 25.6 |
| Adult | Female | Yes | 316 | 14.4 | 425 | 74.4 |
| Adult | Male | No | 1329 | 60.4 | 1667 | 79.7 |
| Adult | Male | Yes | 338 | 15.4 | 1667 | 20.3 |
| Child | Female | No | 17 | 0.8 | 45 | 37.8 |
| Child | Female | Yes | 28 | 1.3 | 45 | 62.2 |
| Child | Male | No | 35 | 1.6 | 64 | 54.7 |
| Child | Male | Yes | 29 | 1.3 | 64 | 45.3 |
Answer: Female adults have a higher chance of survival than male adults (74.4% vs. 20.3%) and female children have a higher chance of survival than male children (62.2% vs. 45.3%, respectively). However, the relative advantage for being female rather than male is much higher for adults (as the survival chances of female adults are 54.1% percentage points higher than for male adults) than for children (as the survival chance of female children are only 16.9% percentage points higher than for male children). Thus, being female was more of an advantage for adults than for children (and the survival rate of female adults was higher than the survival rate for children of either sex).
Statistically, this suggests an interaction between the factors Age and Sex on the chances of survival, though note that there were only 109 children overall (i.e., 4.95% of the passenger population).
- The documentation of the
Titanicdata suggests that the policy women and children first was “not entirely successful in saving the women and children in the third class”. Verify this by creating corresponding contingency tables (i.e., counts of survivors).
# Survivors by Sex for each class:
t_54a <- titanic %>%
# filter(Class == "3rd") %>%
group_by(Class, Sex, Survived) %>%
summarise(count = sum(n),
pc = round(count/sum(titanic$n) * 100, 2))
knitr::kable(t_54a, caption = "Number of Titanic survivors by `Class` and `Sex`.")| Class | Sex | Survived | count | pc |
|---|---|---|---|---|
| 1st | Female | No | 4 | 0.18 |
| 1st | Female | Yes | 141 | 6.41 |
| 1st | Male | No | 118 | 5.36 |
| 1st | Male | Yes | 62 | 2.82 |
| 2nd | Female | No | 13 | 0.59 |
| 2nd | Female | Yes | 93 | 4.23 |
| 2nd | Male | No | 154 | 7.00 |
| 2nd | Male | Yes | 25 | 1.14 |
| 3rd | Female | No | 106 | 4.82 |
| 3rd | Female | Yes | 90 | 4.09 |
| 3rd | Male | No | 422 | 19.17 |
| 3rd | Male | Yes | 88 | 4.00 |
| Crew | Female | No | 3 | 0.14 |
| Crew | Female | Yes | 20 | 0.91 |
| Crew | Male | No | 670 | 30.44 |
| Crew | Male | Yes | 192 | 8.72 |
# Survivors by Age for each class:
t_54b <- titanic %>%
# filter(Class == "3rd") %>%
group_by(Class, Age, Survived) %>%
summarise(count = sum(n),
pc = round(count/sum(titanic$n) * 100, 2))
knitr::kable(t_54b, caption = "Number of Titanic survivors by `Class` and `Age`.")| Class | Age | Survived | count | pc |
|---|---|---|---|---|
| 1st | Adult | No | 122 | 5.54 |
| 1st | Adult | Yes | 197 | 8.95 |
| 1st | Child | No | 0 | 0.00 |
| 1st | Child | Yes | 6 | 0.27 |
| 2nd | Adult | No | 167 | 7.59 |
| 2nd | Adult | Yes | 94 | 4.27 |
| 2nd | Child | No | 0 | 0.00 |
| 2nd | Child | Yes | 24 | 1.09 |
| 3rd | Adult | No | 476 | 21.63 |
| 3rd | Adult | Yes | 151 | 6.86 |
| 3rd | Child | No | 52 | 2.36 |
| 3rd | Child | Yes | 27 | 1.23 |
| Crew | Adult | No | 673 | 30.58 |
| Crew | Adult | Yes | 212 | 9.63 |
| Crew | Child | No | 0 | 0.00 |
| Crew | Child | Yes | 0 | 0.00 |
Answer: Both regularities (i.e., much higher survival chance of females and for children) are lower for 3rd class passengers (despite still showing the same trends).
This concludes our exercises on dplyr — but the topic of data transformation will stay relevant throughout this book.