We want to (know what we can) learn about causation from observations: We know “correlation does not necessarily imply causation”, and that experiments are our best way to learn about causes. But we also understand that there is some use in observation, and we want to know how we can evaluate causal claims in observational studies.
What is a Cause?
Like much of statistics, understanding causation requires a healthy dose of imagination.
Imagine a series of parallel worlds: in one, a specific treatment is applied (e.g., drinking coffee, receiving a vaccine, raising taxes), and in another, it is not. By observing how an outcome of interest responds in each world, we can start to see whether the treatment influences that outcome. We say that a treatment causes the outcome if changing it would alter the outcome, on average. For quantitative treatments, we can envision multiple worlds where the treatment level varies by degree, observing how each variation impacts the outcome.
This concept of evaluating what would happen if we had changed a treatment is known as counterfactual thinking, and it is foundational to reasoning about causation.
Required reading:
- Calling bullshit Chapter 4. Causality. download here.
- How Tech Created a “Recipe for Loneliness” Brain C. Chen. BNew York Times. Nov 10, 2024.
DAGs, confounds, and experiments
THE curious associations with lung cancer found in relation to smoking habits do not, in the minds of some of us, lend themselves easily to the simple conclusion that the products of combustion reaching the surface of the bronchus induce, though after a long interval, the development of a cancer. If, for example, it were possible to infer that smoking cigarettes is a cause of this disease, it would equally be possible to infer on exactly similar grounds that inhaling cigarette smoke was a practice of considerable prophylactic value in preventing the disease, for the practice of inhaling is rarer among patients with cancer of the lung than with others.
Such results suggest that an error has been made,of an old kind, in arguing from correlation to causation, and that the possibility should be explored that the different smoking classes, non-smokers, cigarettesmokers, cigar smokers, pipe smokers, etc., have adopted their habits partly by reason of their personal temperaments and dispositions, and are not lightly to be assumed to be equivalent in their genotypic composition. Such differences in genetic make-up between these classes would naturally be associated with differences of disease incidence without the disease being causally connected with smoking. It would then seem not so paradoxical that the stronger fumes of pipes or cigars should be so much less associated with cancer than those of cigarettes, or that the practice of drawing cigarette smoke in bulk into the lung should have apparently a protective effect.
“Cancer and Smoking” Fisher (1958)

Figure 1: R.A. Fisher—a pipe enthusiast, notorious asshole, eugenicist, and widely regarded as the father of modern statistics and population genetics.
It is now well established that cigarette smoking causes cancer—but this wasn’t always the consensus. One of the most prominent statisticians in history, R.A. Fisher, famously argued that the observed relationship between smoking and cancer was spurious, insisting that smoking did not cause cancer. The extent to which his flawed logic was influenced by his own fondness for smoking, financial ties to the tobacco industry, or simple skepticism is unclear (Stolley (1991)). What is clear, however, is that Fisher raised a plausible argument—and that he was profoundly mistaken.
Randomized Controlled Experiments
Randomized controlled experiments are our strongest tool for learning about causation. By randomly assigning participants to different treatments, we effectively place them in these alternative realities we imagine and observe the outcomes of each treatment. In other words, we bring our hypothetical scenarios to life.
To distinguish between the claim that smoking causes cancer and Fisher’s claim that genetics is a confounder, we would ideally randomly assign some people to smoke and others not to smoke. However, for both ethical and logistical reasons, this is not feasible. So, we need alternative methods to approach this question, and that is our goal today! The directed acyclic graph is a visual approach that can get us started solving this problem.
DAGs
We represent causal relationships between variables by a Directed Acyclic Graph, or a DAG. Each variable is represented by a node (a point in the graph), and each causal relationship is shown as a directed arrow pointing from the cause to the effect. DAGs are directed because arrows point in the direction of causation. DAGs are Acyclic as arrows can’t loop back to a variable they originated from. This is because causation flows in only one direction (i.e. something cannot cause itself (See the Cosmological_argument from the philosophy of religion literature). DAGs help us map out complex relationships and clarify our assumptions about how variables are related. They can show us where potential biases or confounding might exist and guide us in deciding which variables we need to include or exclude in an analysis.
Above we introduced two simple causal models for the association between smoking and cancer
Smoking causes lug cancer: Figure 2A illustrates a straightforward model where smoking directly causes lung cancer, representing a front-door causal path from smoking to cancer. A front-door path is a direct causal pathway from one variable (smoking) to an outcome (lung cancer) without any intermediaries or confounders obstructing the causal effect. This does not imply that smoking is the only cause of lung cancer, nor that all smokers will develop lung cancer or that all non-smokers won’t. Rather, it suggests that if we could create a “clone” for each person, with one clone smoking and the other not, we would expect more lung cancer cases among the smoking clones due to this direct effect.
Genetics case smoking and lung cancer: Figure 2B represents Fisher’s argument that smoking doesn’t cause cancer directly but that both smoking and cancer result from a common cause—genetics. According to Fisher, the observed association between smoking and cancer is merely a statistical artifact of their shared genetic cause.
These are not the only plausible causal models for an association between smoking and cancer. I present three other possibilities in Figure 2.
Genetics influences smoking, and smoking, in turn, causes cancer. Figure 2C introduces a “pipe” model. In this model, genetics influences smoking, and smoking, in turn, causes cancer. Statistically, this model is challenging to evaluate because altering genetic factors in an experiment wouldn’t directly “cause” cancer. Simply including genetics as a variable in a linear model could obscure smoking’s effect. One approach is to “match” on genetics and then compare cancer outcomes. For example, a 2017 study examined lung cancer rates in monozygotic twins where only one smoked, finding higher cancer rates in smoking twins (Hjelmborg et al. 2017).
Genetics and smoking contribute to cancer. Figure 2D introduces a colider – Here, both genetics and smoking contribute to cancer, “colliding” in their effects on this outcome. There are two paths from smoking to cancer: 1) the front-door causal path—where smoking causes cancer directly, and 2) the back-door non-causal path, where the connection between smoking and cancer is confounded by genetics. This model challenges us to appropriately separate and attribute causes.
A more complex and realistic model including the effects of the environment on cancer and smoking is presented in 2E. Noe that in this model genes do not cause the environment and the environment does not cause genes.
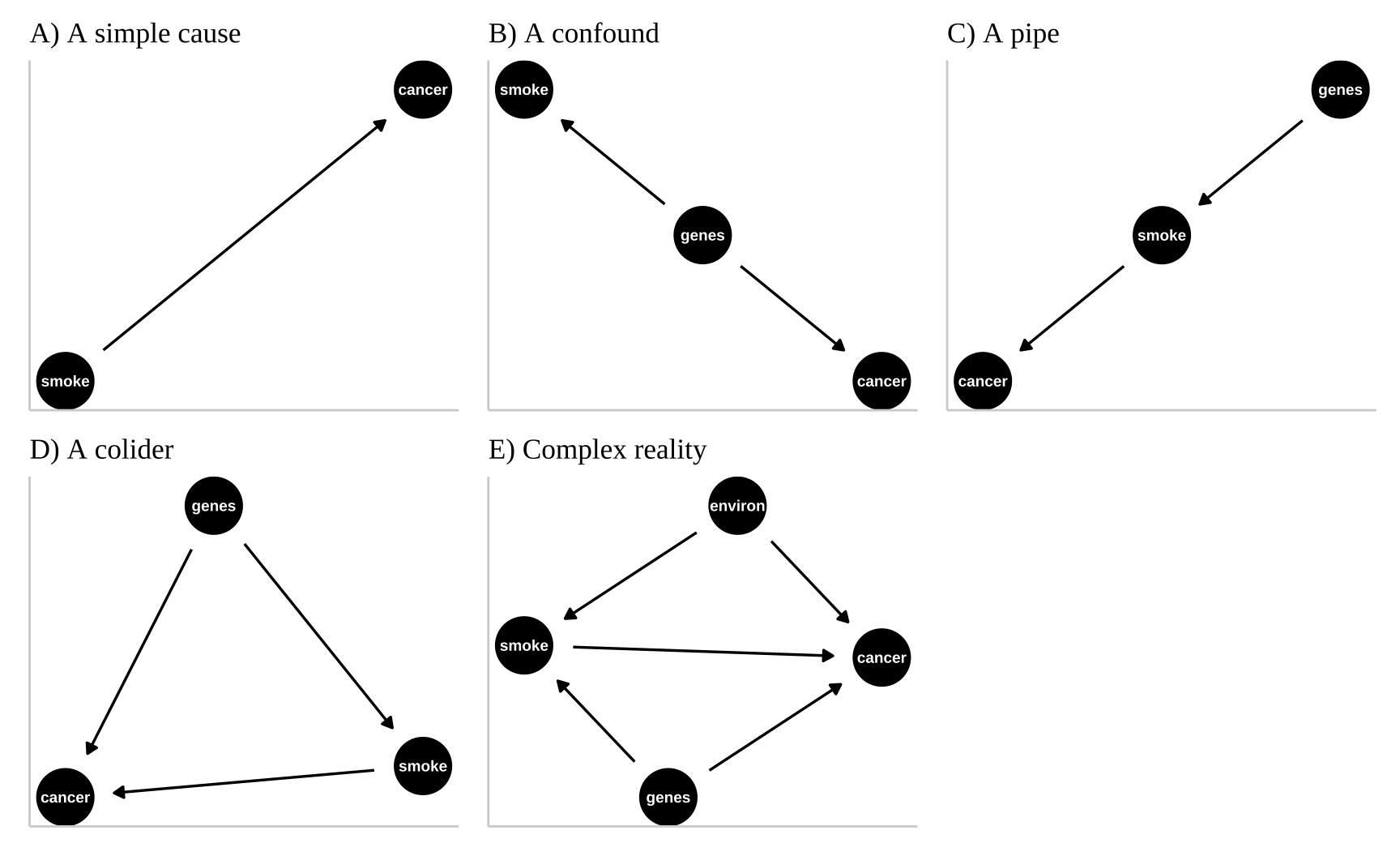
Figure 2: We could represent this causal claim with the simplest causal graph we can imagine. This is our first formal introduction to a Directed Acyclic Graph (herefater DAG). This is Directed because WE are pointing a causal arrow from smoking to cancer. It is acyclic because causality in these models only flows in one direction, and its a graph because we are looking at it. These DAGs are the backbone of causal thinking because they allow us to lay our causal models out there for the world to see. Here we will largely use DAGs to consider potential causal paths, but these can be used for mathematical and statistical analyses.
When Correlation Is (Not) Good Enough
As we explore causation, we might wonder when it’s essential to understand causes.
We don’t need to understand causation to make predictions under the status quo. If our goal is simply to make accurate predictions, we can build a strong multiple regression model and use it to predict outcomes without needing to know the underlying causes. For example, if I want to buy good corn, I can go to a farm stand that reliably sells tasty corn without needing to know if its quality comes from the soil, sunlight, or the farmers playing Taylor Swift each morning to “excite” the corn. Similarly, if I were selling insurance, I would only need to predict who is likely to develop lung cancer—not necessarily understand the cause.
We need to understand causation when we want to intervene (or make causal claims). If I want to grow my own tasty corn, I’d need to know what specifically makes the farm’s corn delicious. It wouldn’t be useful to fertilize my soil if all that’s needed is a daily dose of Taylor Swift’s music. Likewise, if I were offering public health advice, I would need to know that smoking causes cancer to credibly recommend that people quit smoking to reduce their risk of lung cancer.
Multiple Regression and Causal Inference
So far, we’ve discussed how to construct and interpret causal models. This process is incredibly valuable: creating a causal model clarifies our assumptions and reasoning.
But how can we use these models in statistical analysis? As it turns out, they can be quite useful! To illustrate this, I’ll simulate data under different causal models and apply various linear regressions to the simulated data to examine the outcomes.
I’m demonstrating these concepts through simulation because, with simulated data, we know the true underlying model. This allows us to clearly see what happens when models are misspecified.
Set Up
In evolutionary studies, fitness is the central metric of interest. Although it is challenging to measure and define directly, we can often assess traits related to it, such as the number of offspring an organism produces. For this example, let’s assume this measure is sufficiently representative of fitness.
Now, suppose we are studying a species of fish and want to determine if being larger (in terms of length) increases fitness, measured as the number of eggs produced. To make things more interesting, let’s assume that fish inhabit environments whose quality we can measure. For this example, we’ll assume that all measurements are accurate, unbiased, and follow a normal distribution.
Causal Model 1: The Confound
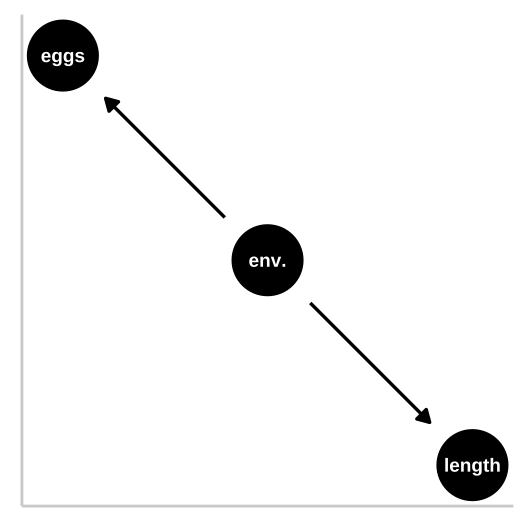
Let’s begin with a simple confound—where a high-quality environment leads to larger fish and increased fitness, but size itself has no direct effect on fitness. Here’s how we’ll set up the simulation:
- One hundred fish (
n=100) - Each fish grows in environments of varying quality (normally distributed, with a mean, \(\mu = 50\), and standard deviation, \(\sigma = 5\)).
- Length is influenced by environmental quality (predicted length for each fish matches environmental quality, with a standard deviation of two).
- The number of eggs is also influenced by environmental quality (predicted egg count for each fish is half the environmental quality, with a standard deviation of six, rounded to the nearest integer because fish cannot lay a fractional number of eggs).
In this model, fish length has no causal impact on the number of eggs laid.
We use the rnorm() function to simulate data from a normal distribution as follows:
We know that fish length does not cause fish to lay more eggs—since we did not model it this way. Nonetheless, a plot and a statistical test show a strong association between length and egg count if we exclude environmental quality from the model.
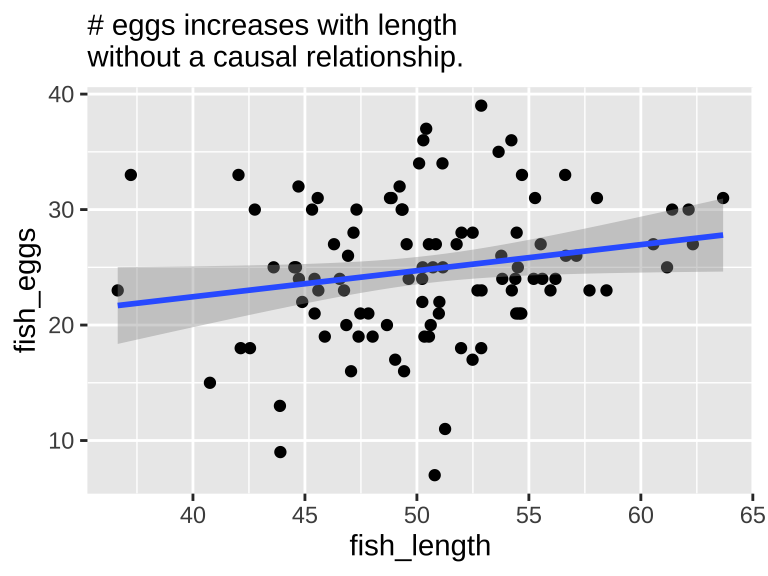
Our statistical analysis will not show cause
We can build a simple linear model predicting the number of fish eggs as a function of fish length. We can see that the prediction is good, and makes sense – egg number reliably increases (0.039) with fish length (slope = 0.226, df = (1,98), F = 4.01, p-value = 0.048). But we know this is not a causal relationship (because we didn’t have this cause in our simulation).
| Model with confound only | ||||
| lm(fish_eggs~ fish_length, data = confounded_fish) | ||||
| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| fish_length | 0.226 | 0.113 | 2.003 | 0.048 |
A Model Including the True Cause
Let’s say we didn’t have a clear idea of which trait causally increased the number of eggs a fish laid. In this example, it’s plausible (although not necessarily true) to believe that both environmental quality and fish length influence the number of eggs laid. So, let’s build a model that includes both variables, including the confounding variable, environmental quality. Then, we’ll find our estimates and use the appropriate Type II sums of squares to evaluate significance. Finally, we’ll use the emtrends() and confint() functions from the emmeans package to estimate uncertainty in our slope estimates.
library(emmeans)
fish_lm_w_confound <- lm(fish_eggs ~ env_quality + fish_length, confounded_fish)
anova_table <- Anova(fish_lm_w_confound, type = "II")
env_qual_est <- emtrends(fish_lm_w_confound, var = "env_quality") %>% confint()
fish_length_est <- emtrends(fish_lm_w_confound, var = "fish_length") %>% confint()| ANOVA Table: Model with Cause & Confound | ||||
| Model: lm(fish_eggs ~ env_quality + fish_length, data = confounded_fish) | ||||
| term | sumsq | df | F_value | P_value |
|---|---|---|---|---|
| env_quality | 53.971 | 1 | 1.536 | 0.218 |
| fish_length | 6.575 | 1 | 0.187 | 0.666 |
| Residuals | 3408.435 | 97 | NA | NA |
| Slope Estimates and Uncertainty: Model with Cause & Confound | |||||
| Model: lm(fish_eggs ~ env_quality + fish_length, data = confounded_fish) | |||||
| variable | slope | SE | df | lower.CL | upper.CL |
|---|---|---|---|---|---|
| env_quality | 0.403 | 0.325 | 97 | -0.242 | 1.048 |
| fish_length | -0.135 | 0.312 | 97 | -0.754 | 0.484 |
The Good News is that now that we’ve added the true cause, we no longer find that fish_length is statistically associated with the number of eggs laid.
The Bad News is that, because we still have the wrong variable in our model, we lose the ability to correctly identify that environmental quality strongly predicts the number of eggs laid.
What to do?
First let’s look at all the relationships in our data.
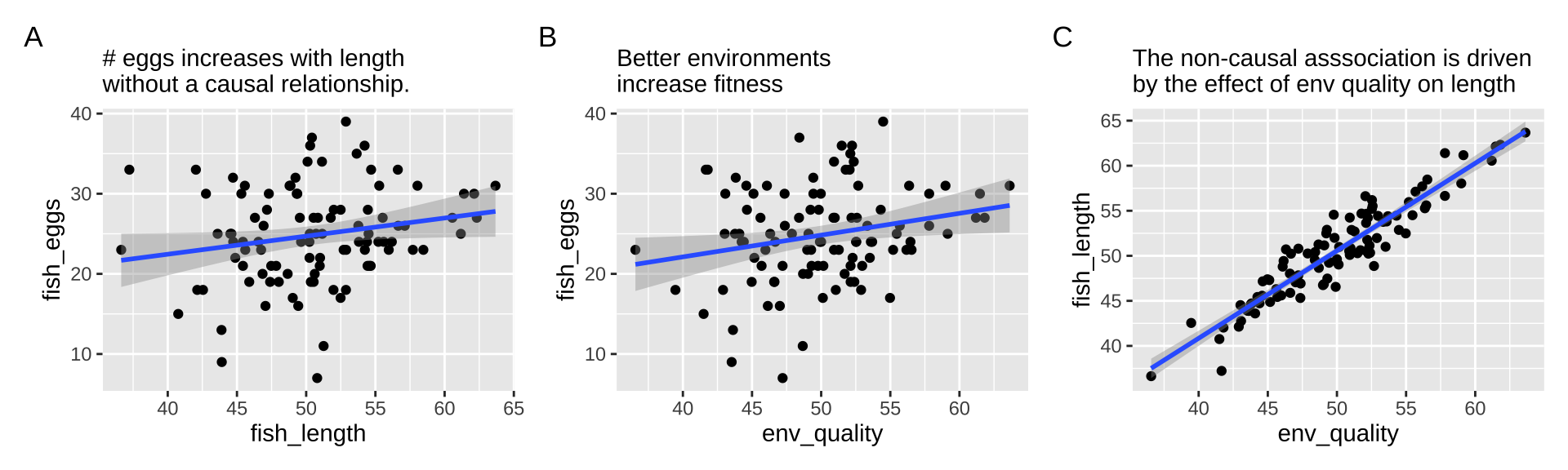
The right thing to do in this case is to just build a model with the environmental quality.
| Model with cause only | ||||
| lm(fish_eggs~ env_quality, data = confounded_fish) | ||||
| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| env_quality | 0.272 | 0.117 | 2.33 | 0.022 |
Causal model 2: The pipe
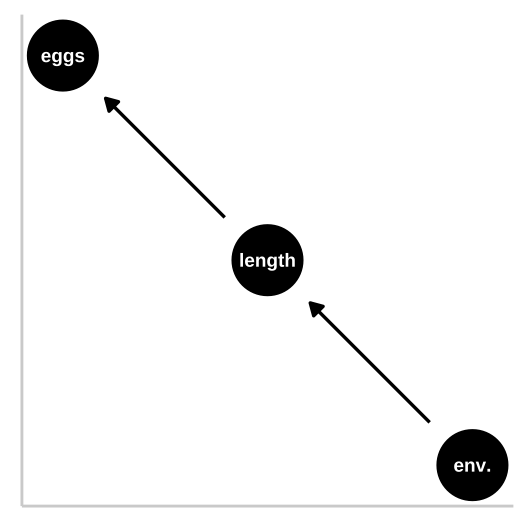
So now let’s look at a pipe in which the environment causes fish length and fish length causes fitness, but environment itself has has no impact on fitness. First let’s simulate:
- One hundred fish (
n = 100) - Environmental quality for each fish (normally distributed with mean \(\mu = 50\) and standard deviation \(\sigma = 5\))
- Fish length based on environmental quality (predicted length = environmental quality with standard deviation 2)
- Egg count based on fish length (predicted egg count = half the fish length, standard deviation 5, rounded to the nearest integer)
In this scenario, we know that environmental quality does not directly cause fish to lay more eggs, as the model does not include a direct link from quality to egg count. However, both a plot and a statistical test reveal a strong association between environmental quality and egg count when fish length is not included in the model.
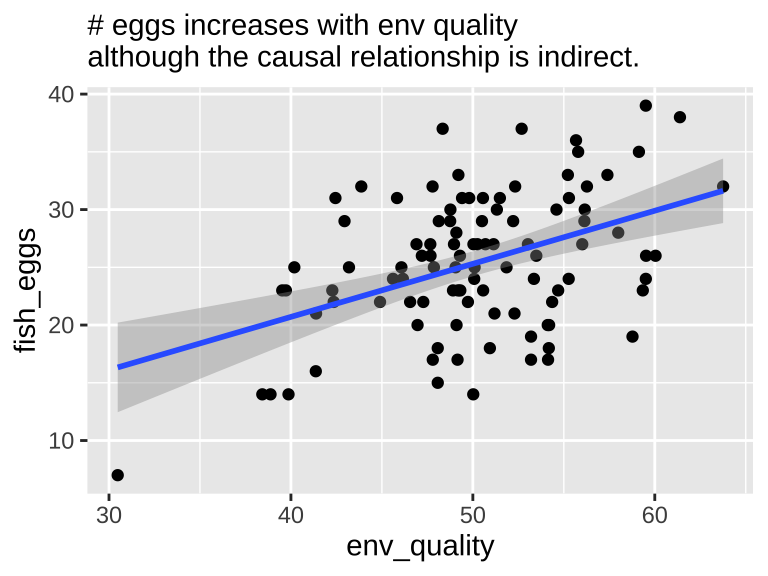
Our Statistical Analysis Cannot Prove Causation
We can fit a simple linear model predicting egg count based solely on environmental quality. This model will show a positive and reliable association (slope = 0.4596; \(r^2 =\) 0.191) —- egg count appears to increase with environmental quality. Of course, this is not a causal relationship, as we did not model environmental quality as a direct cause of egg production.
| Model with indirect cause only | ||||
| lm(fish_eggs ~ env_quality, pipe_fish) | ||||
| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| env_quality | 0.46 | 0.095 | 4.816 | 0 |
Adding the immediate cause into our model
So, let’s build a model including both the immediate cause, fish length, as well as the indirect cause, environmental quality. We see pretty strange behavior here - when both variables are in the statistical model, neither is significantly associated with the number of eggs laid.
| ANOVA Table: Model with Indirect & Direct Cause | ||||
| Model: lm(fish_eggs ~ env_quality + fish_length, data = pipe_fish) | ||||
| term | sumsq | df | F_value | P_value |
|---|---|---|---|---|
| fish_length | 85.841 | 1 | 2.936 | 0.090 |
| env_quality | 0.472 | 1 | 0.016 | 0.899 |
| Residuals | 2835.860 | 97 | NA | NA |
| Slope Estimates and Uncertainty: Model with Indirect & Direct Cause | |||||
| Model: lm(fish_eggs ~ env_quality + fish_length, data = pipe_fish) | |||||
| variable | slope | SE | df | lower.CL | upper.CL |
|---|---|---|---|---|---|
| fish_length | 0.478 | 0.279 | 97 | -0.076 | 1.033 |
| env_quality | -0.039 | 0.306 | 97 | -0.646 | 0.568 |
What to do?
First let’s look at all the relationships in our data
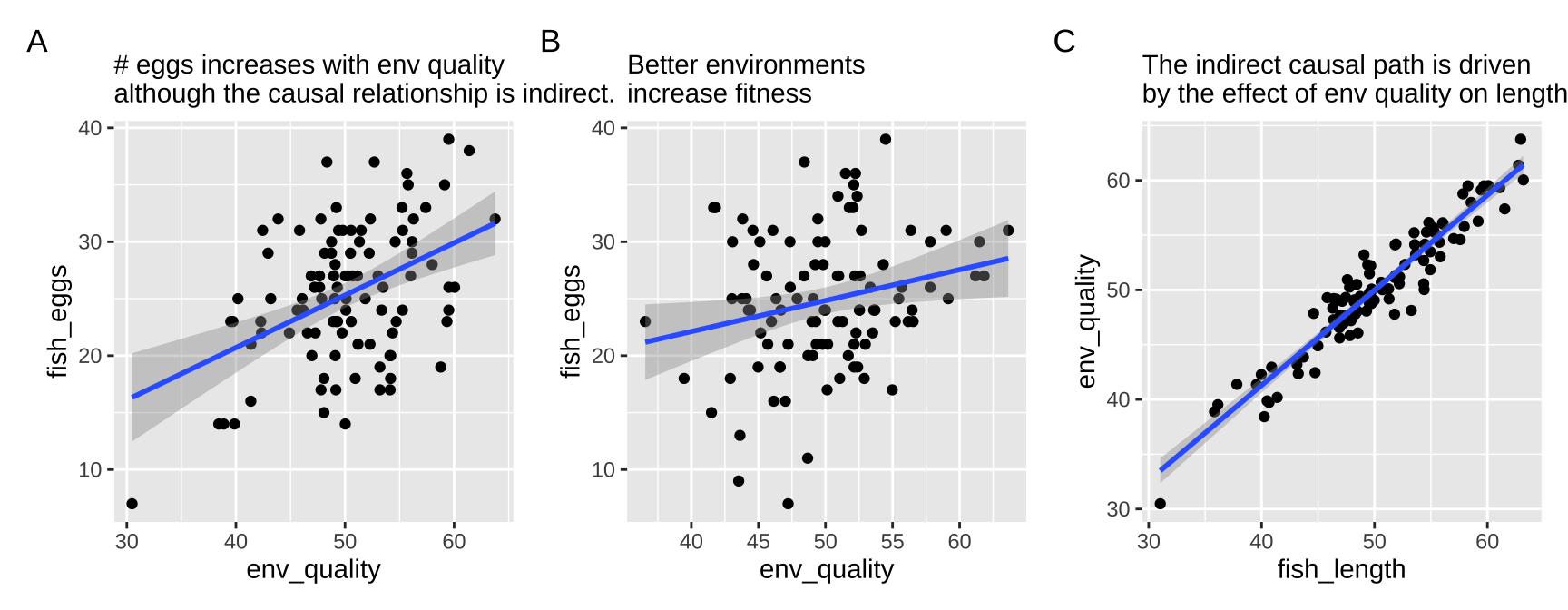
The right thing to do in this case is to just build a model with the fish length.
| Model with direct cause only | ||||
| lm(fish_eggs~ fish_length, data = pipe_fish) | ||||
| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| fish_length | 0.445 | 0.086 | 5.181 | 0 |
Causal model 3: Collider
A collider is something caused by two variables. Say in our example both fish length and environmental quality caused fish to lay more eggs. Here egg number is a colider. Lets simulate it!
- One thousand fish
- Environmental quality for each fish (normally distributed with mean \(\mu = 50\) and standard deviation \(\sigma = 5\))
- Fish length is independent of quality (ormally distributed with mean \(\mu = 50\) and standard deviation \(\sigma = 5\))
- Egg count based on fish length and environmental quality (predicted egg count = (Fish length + Environmental quality) /4, standard deviation 8, rounded to the nearest integer)
n_fish <-1000
collider_fish <- tibble(
env_quality = rnorm(n = n_fish, mean = 50, sd = 8), # Simulating environment
fish_length = rnorm(n = n_fish, mean = 50, sd = 8),
fish_eggs = rnorm(n = n_fish, mean = (env_quality+fish_length) / 4, sd = 2) %>% round()
) %>%
mutate(fish_eggs = ifelse(fish_eggs <0,0,fish_eggs))Let’s check out our data:
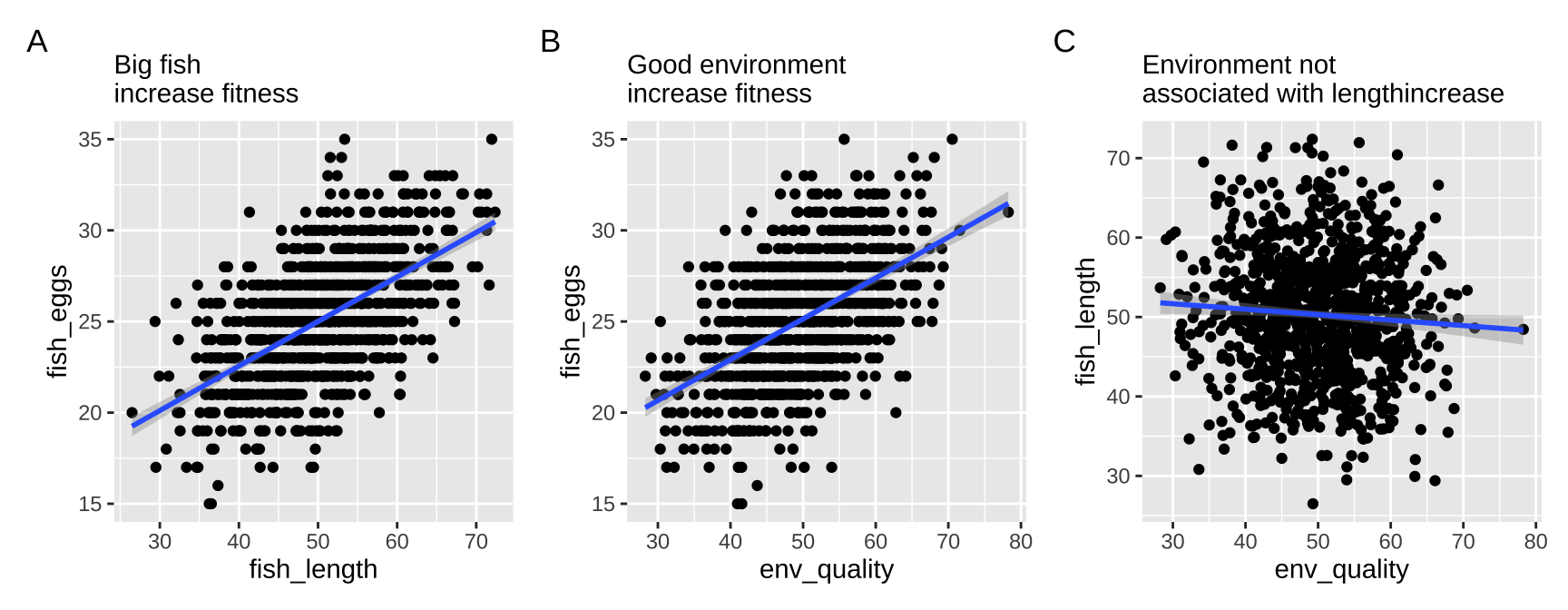
We see everyhting that is expected, and our stats back this up.
library(emmeans)
fish_lm_w_collider <- lm(fish_eggs~ fish_length + env_quality, collider_fish)
anova_table <- Anova(fish_lm_w_collider, type = "II")
fish_length_est <- emtrends(fish_lm_w_collider, var = "fish_length") %>% confint()
env_qual_est <- emtrends(fish_lm_w_collider, var = "env_quality") %>% confint()| ANOVA Table: Modeling a collider | ||||
| Model: lm(fish_eggs ~ env_quality + fish_length, data = collider_fish) | ||||
| term | sumsq | df | F_value | P_value |
|---|---|---|---|---|
| fish_length | 4213.963 | 1 | 1057.773 | 0 |
| env_quality | 3608.377 | 1 | 905.761 | 0 |
| Residuals | 3971.856 | 997 | NA | NA |
| Slope Estimates and Uncertainty: Modeling a collider | |||||
| Model: lm(fish_eggs ~ env_quality + fish_length, data = collider_fish) | |||||
| variable | slope | SE | df | lower.CL | upper.CL |
|---|---|---|---|---|---|
| fish_length | 0.261 | 0.008 | 997 | 0.245 | 0.277 |
| env_quality | 0.242 | 0.008 | 997 | 0.226 | 0.258 |
Here the correct model is to include both causes. exliding one will likely lead to unbiased estimates of the effect of the other, but it will reduce power and increase uncertainty.
Here’s an edited version:
Beware of Collider Bias
Suppose we’re interested in fish that lay a large number of eggs, and we want to explore factors that might help them lay even more. If we focus only on fish that laid more than the average number of eggs (e.g., 30), we might observe an unexpected pattern:
ggplot(collider_fish %>% filter(fish_eggs >= 30),
aes(x = env_quality, y = fish_length)) +
geom_point() +
geom_smooth(method = "lm", se = FALSE)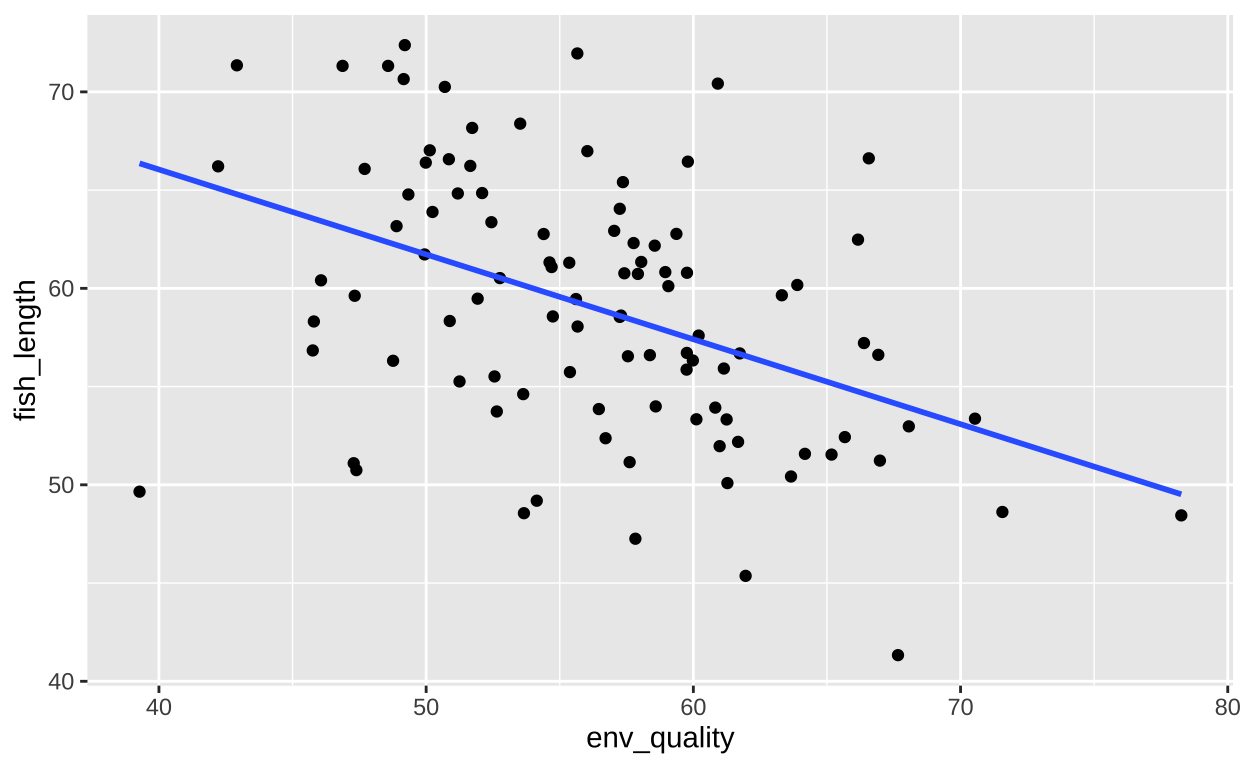
Even though our predictors are uncorrelated, we see a significant negative relationship between them. This correlation is artificially induced because we’ve conditioned on a high outcome. This phenomenon, known as collider bias, should be carefully avoided.
| Model with direct cause only | ||||
| lm(fish_length ~ env_quality, data = collider_fish %>% filter(fish_eggs>=30)) | ||||
| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| env_quality | -0.432 | 0.088 | -4.892 | 0 |
What to do?
The examples above show the complexity in deciphering causes without experiments. But they also show us the light about how we can infer causation, because causal diagrams can point to testable hypotheses. If we cannot do experiments, causal diagrams offer us a glimpse into how we can infer causation. Perhaps the best way to do this is by matching – if we can match subjects that are identical for all causal paths except the one we are testing, we can then test for a statistical association, ad make a causal claim we can believe in.
The field of causal inference is developing rapidly. If you want to hear more, the popular book, The Book of Why (Pearl and Mackenzie 2018) is a good place to start. The field of causal inference offers many exciting techniques for deciphering cause. For now we will just say that a good biological intution and carefull thinking about your model and causation is a great starting place.
Quiz
Be sure to complete the quiz, based on
- This reading
- Calling bullshit Chapter 4. Causality. download here.
- How Tech Created a “Recipe for Loneliness” Brian C. Chen. BNew York Times. Nov 10, 2024.
Figure 3: Quiz on causal inference here