I say this all the time but: learning dplyr + ggplot was one of the highest payoff things I've done in my career.
— Joshua G. Schraiber (@jgschraiber) January 22, 2019
Science and making scientific conclusions relies heavily on the analysis of data. This is in act what makes science science. We observe things, and/or do experiments and then come to some tentative conclusion about the world, This informs our theoretical understanding and allows or science to go on! Thus, while theory is important to science, data is king. So we need to be able to keep and analyze data.
Learning goals: By the end of this chapter you should be able to get data into R and explore it. Specifically, students will be well-positioned to analyze data by:
- Understanding the tidy data format.
- Using the mutate function in R to manipulate data.
- Using the summarize function in R to summarize the data
- (and doing so by groups with the group_by function.
- Combining these operations with the pipe %>% operator.
Tidy data
Figure 1: Watch this video about tidy data principles
Most often data are kept in a spreadsheet. If we’re luck data are tidy. 🤔 A tidy data structure usually makes statistical analysis and data visualization much easier.
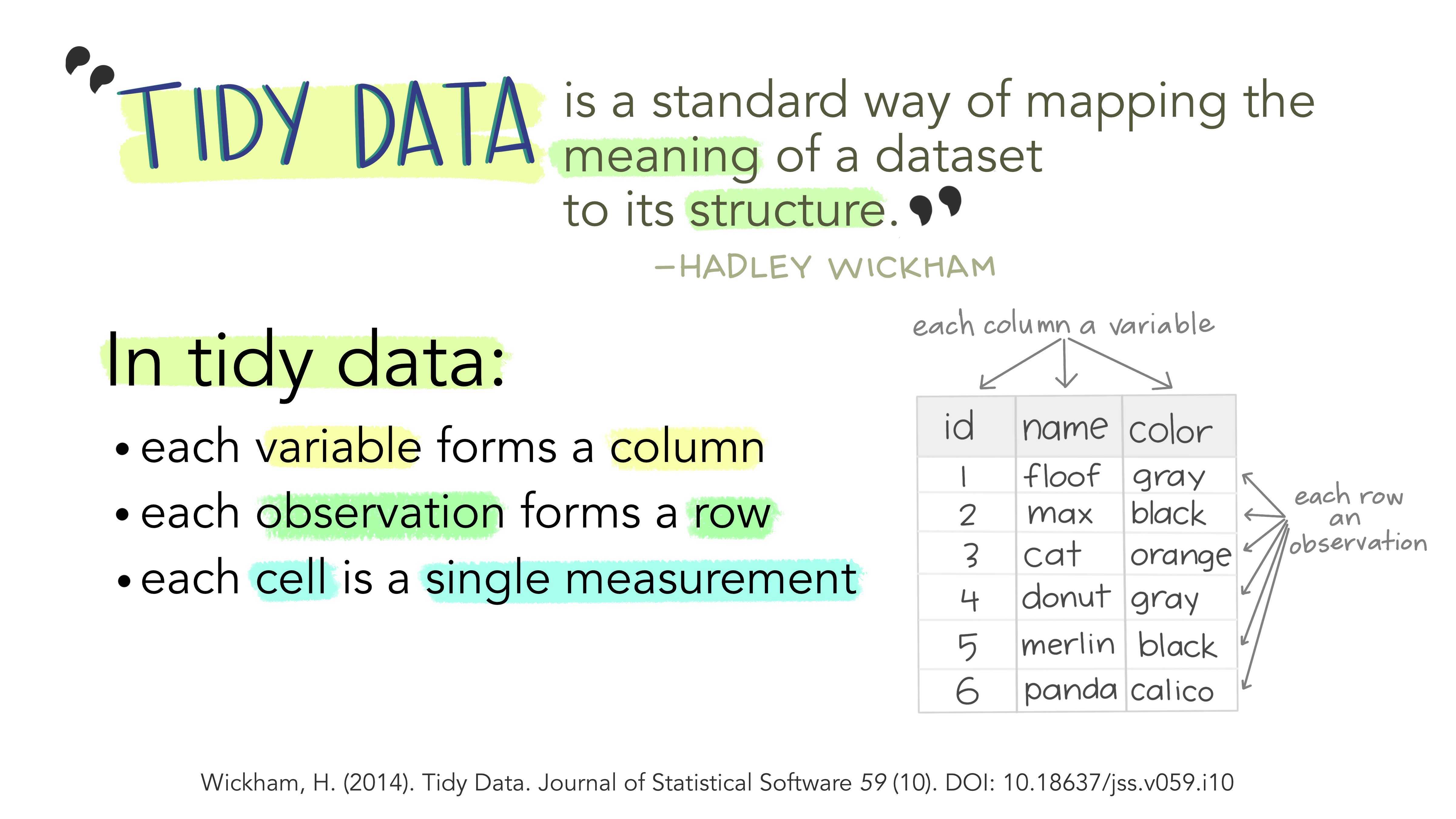
A tibble is the name for the primary structure that holds data in the tidyverse. Tibbles do not ensure that our data are tidy but they do make this easier. In a tidy tibble, each variable is a column (i.e. a vector), and all so while all entries in a column must be of the same class, each column in a tibble can have its own class. In a tidy tibble, rows unite values of various variables of a single entity (i.e. an individual at a time).
If you have spent time in base R you are likely familiar with matrices, arrays, and data frames - don’t even worry about these. A tibble is much like a data frame, but has numerous features that make them easier to deal with. If you care, see chapter 10 of R for Data Science (Grolemund and Wickham 2018) for more info. Anyways, in this course we will focus on vectors and tibbles, ignoring arrays and matrices, and avoiding lists for as long as possible.
Wrangling data in R

Lets get started with a fun data set that is commonly used for trying things out in R – some cute penguins. The penguins data are available in the palmerpenguins package.
install.packages("palmerpenguins")
library(palmerpenguins)Of course, the first thing we do when we have data is to look at it. so we use the gwalkr() function in the GWalkR package to bring up a simple GUI to explore our data.
# Let's use the gwalkr function in the GWalkR package to look at these data (ignore the code below)
visConfig <- '[{"config":{"defaultAggregated":false,"geoms":["auto"],"coordSystem":"generic","limit":-1},"encodings":{"dimensions":[{"fid":"c3BlY2llcw==","name":"species","basename":"species","semanticType":"nominal","analyticType":"dimension"},{"fid":"aXNsYW5k","name":"island","basename":"island","semanticType":"nominal","analyticType":"dimension"},{"fid":"c2V4","name":"sex","basename":"sex","semanticType":"nominal","analyticType":"dimension"},{"fid":"eWVhcg==","name":"year","basename":"year","semanticType":"nominal","analyticType":"dimension"},{"fid":"gw_mea_key_fid","name":"Measure names","analyticType":"dimension","semanticType":"nominal"}],"measures":[{"fid":"YmlsbF9sZW5ndGhfbW0=","name":"bill_length_mm","basename":"bill_length_mm","analyticType":"measure","semanticType":"quantitative","aggName":"sum"},{"fid":"YmlsbF9kZXB0aF9tbQ==","name":"bill_depth_mm","basename":"bill_depth_mm","analyticType":"measure","semanticType":"quantitative","aggName":"sum"},{"fid":"ZmxpcHBlcl9sZW5ndGhfbW0=","name":"flipper_length_mm","basename":"flipper_length_mm","analyticType":"measure","semanticType":"quantitative","aggName":"sum"},{"fid":"Ym9keV9tYXNzX2c=","name":"body_mass_g","basename":"body_mass_g","analyticType":"measure","semanticType":"quantitative","aggName":"sum"},{"fid":"gw_count_fid","name":"Row count","analyticType":"measure","semanticType":"quantitative","aggName":"sum","computed":true,"expression":{"op":"one","params":[],"as":"gw_count_fid"}},{"fid":"gw_mea_val_fid","name":"Measure values","analyticType":"measure","semanticType":"quantitative","aggName":"sum"}],"rows":[{"fid":"ZmxpcHBlcl9sZW5ndGhfbW0=","name":"flipper_length_mm","basename":"flipper_length_mm","analyticType":"measure","semanticType":"quantitative","aggName":"sum"}],"columns":[{"fid":"Ym9keV9tYXNzX2c=","name":"body_mass_g","basename":"body_mass_g","analyticType":"measure","semanticType":"quantitative","aggName":"sum"}],"color":[],"opacity":[],"size":[],"shape":[],"radius":[],"theta":[],"longitude":[],"latitude":[],"geoId":[],"details":[],"filters":[],"text":[]},"layout":{"showActions":true,"showTableSummary":false,"stack":"stack","interactiveScale":false,"zeroScale":true,"size":{"mode":"auto","width":1520,"height":150},"format":{},"geoKey":"name","resolve":{"x":false,"y":false,"color":false,"opacity":false,"shape":false,"size":false}},"visId":"gw_t6SM","name":"Chart 1"}]';gwalkr(penguins, visConfig=visConfig)Figure 2: Ineractive visualization of the palmer penguin data set
As we look at the penguins data set below we can get a sense of the utility of a tibble. We can see, not only the first few values of the data set, but also the class of each variable (chr for character, fct for factor, dbl for double – a continuous class of data). Recall that we can get a sense of a dataset with the glimpse() function in dplyr, and scroll through it with View(). Note to make things easier to look at, I only show species, sex, body_mass_g, and bill_length_mm.
penguins# A tibble: 344 × 4
species sex body_mass_g bill_length_mm
<fct> <fct> <int> <dbl>
1 Adelie male 3750 39.1
2 Adelie female 3800 39.5
3 Adelie female 3250 40.3
4 Adelie <NA> NA NA
5 Adelie female 3450 36.7
6 Adelie male 3650 39.3
7 Adelie female 3625 38.9
8 Adelie male 4675 39.2
9 Adelie <NA> 3475 34.1
10 Adelie <NA> 4250 42
# ℹ 334 more rowsWhile glimpse() and View() are among the very handy tidyverse functions, the real utility of tidyverse is that it gives us a unified way to deal with data. Usually when we get data we want to handle/clean it, summarize it, visualize the results and develop a statistical model from it. This is where tidyverse really shines!
We first focus on handling data with the dplyr package. Today we’ll talk about using it to mutate, and summarize() our data.
Figure 3: Watch this video about using dplyr to deal with data in R
Modify data with mutate
Say we are curious about the ratio of body mass to wing length in these penguins. We can add this column by with the mutate function in the dplyr package. The easiest way to do this is to:
- Start with our data,
penguins,
- Pipe
%>%it forward and
- Make a new column with
mutate. We can name this new column whatever we want. This adds a column (see data and Figure 4 below).
# A tibble: 344 × 5
species sex body_mass_g bill_length_mm wt_bill_ratio
<fct> <fct> <int> <dbl> <dbl>
1 Adelie male 3750 39.1 95.9
2 Adelie female 3800 39.5 96.2
3 Adelie female 3250 40.3 80.6
4 Adelie <NA> NA NA NA
5 Adelie female 3450 36.7 94.0
6 Adelie male 3650 39.3 92.9
7 Adelie female 3625 38.9 93.2
8 Adelie male 4675 39.2 119.
9 Adelie <NA> 3475 34.1 102.
10 Adelie <NA> 4250 42 101.
# ℹ 334 more rowsmutate() cartoon:
mutate() adds (or overwrites) columns from data we have in the tibble (Figure 4).
 can add columns by combining existing ones. Taken from David Keyes' Blog https://rfortherestofus.com/2024/07/dplyr-functions-animated.](images/mutate.gif)
Figure 4: An illustration of how mutate can add columns by combining existing ones. Taken from David Keyes’ Blog https://rfortherestofus.com/2024/07/dplyr-functions-animated.
summarize data
Having the entire raw data at our fingertips is useful, but we often want to describe our data. Common descriptions include the mean (R function, mean()), maximum (R function, max()), and minimum (R function, min()) values of a variable (see the next chapter for more info). We can reduce large tibbles to simple summaries with the summarize() function.
As we see in the code below and Figure 5 below and summarize() reduces the number of columns in our tibble. As with, mutate() we set the name of the new summary column to whatever we wish.
na.rm = TRUE tells R to ignore missing values when calculating summaries.
# A tibble: 1 × 3
mean_wt_bill_ratio min_wt_bill_ratio max_wt_bill_ratio
<dbl> <dbl> <dbl>
1 95.8 57.6 132.Note: The tibble this generates has as many rows as the classes of things we’re summarizing (in this case, eight, representing all combinations of sex (including unknown), and species we have).
summarize() cartoon:
summarize() reduces our big data set down to summaries of variables (Figure 5).
 provides data summaries of columns. Taken from David Keyes' Blog https://rfortherestofus.com/2024/07/dplyr-functions-animated.](images/summarize.gif)
Figure 5: An illustration of how summarize provides data summaries of columns. Taken from David Keyes’ Blog https://rfortherestofus.com/2024/07/dplyr-functions-animated.
Combine summarize with group_by() to summarize by groups.
We often want to compare values of variables for different types of individuals in our sample. For example, we might want to compare the mean and ranges of wt_bill_ratio for different species and sexes of penguins. We can do this by grouping by group_by() categories before summarizing (be sure to ungroup() when you’re done!)
# A tibble: 8 × 5
sex species mean_wt_bill_ratio min_wt_bill_ratio max_wt_bill_ratio
<fct> <fct> <dbl> <dbl> <dbl>
1 fema… Adelie 90.6 75.1 108.
2 fema… Chinst… 75.9 57.6 90.2
3 fema… Gentoo 103. 91.4 117.
4 male Adelie 100. 80.9 127.
5 male Chinst… 77.1 63.1 92.3
6 male Gentoo 111. 95.8 132.
7 <NA> Adelie 93.5 79.3 102.
8 <NA> Gentoo 101. 92.1 110. Figure 6 shows two different visulizations of the relationship between penguin body mass and bill length by sex and species (generated from Figure 2). This shows that different choices in our exploratory data visualizations can reveal different patterns, so rememebr to mess around a bit with this.
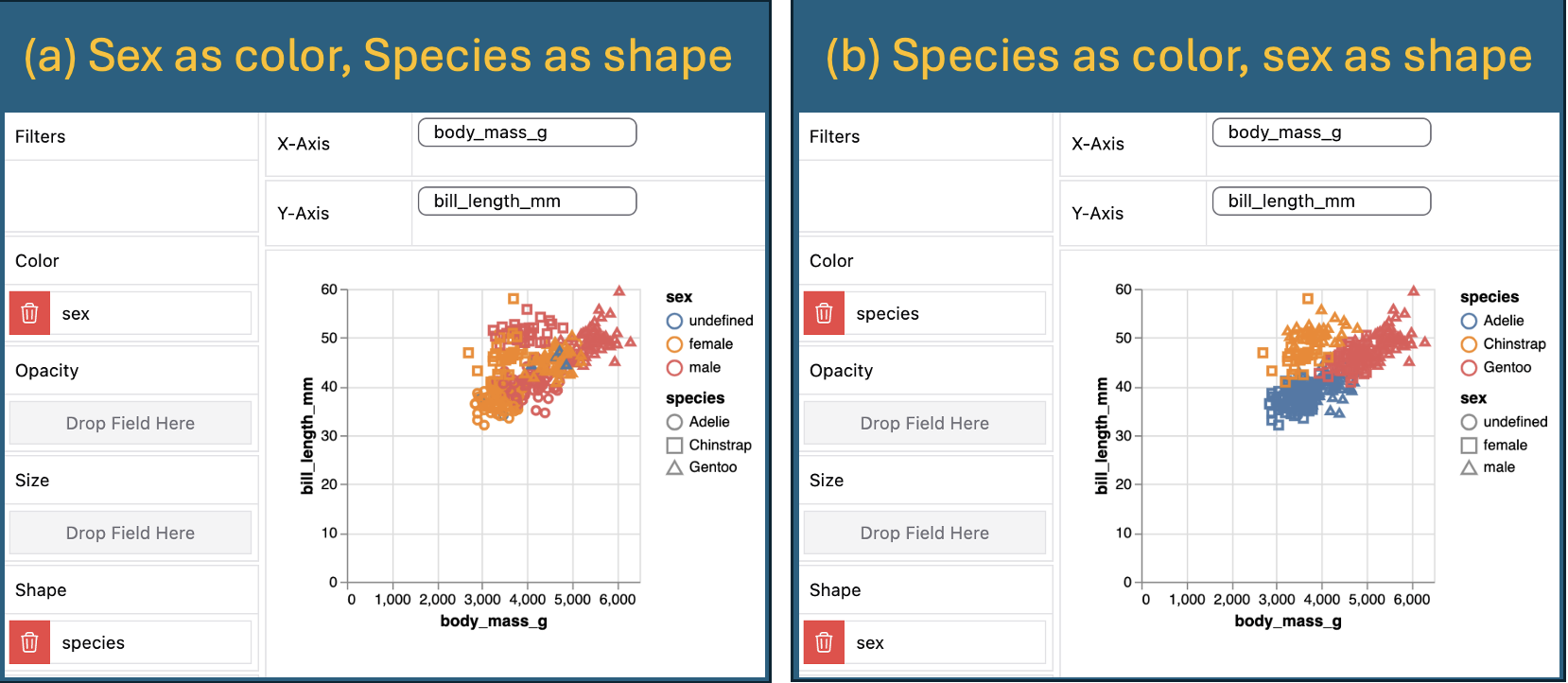
Figure 6: Two views of the relationship between penguin body mass and bill length by sex and species.
Change variable type with mutate()
 investigating Floyd Mayweather's boxing greatness.](https://fivethirtyeight.com/wp-content/uploads/2017/08/roederbutler-undefeated-1.png)
Figure 7: Data from an article investigating Floyd Mayweather’s boxing greatness.
The authors of an article at fivethirtyeight.com ( link) looked at how long different boxers have gone undefeated. Figure 7 shows that Floyd Mayweather has gone a long time without a loss.
Let’s check out some of these data in R. I provide a simple summary (link) of the number of consecutive wins each boxer (name) had to start their career.
undefeated <- read_csv("https://github.com/ybrandvain/datasets/raw/master/undefeated.csv")
undefeated# A tibble: 92 × 4
name url date wins
<chr> <chr> <date> <chr>
1 Juro Fukuda http://boxrec.com/boxer/195508 1943-11-26 27
2 Hilaire Pratesi http://boxrec.com/boxer/56069 1955-06-27 26
3 Rocky Marciano http://boxrec.com/boxer/9032 1955-09-21 49
4 Joe Devlin http://boxrec.com/boxer/69735 1960-04-29 1 5
5 Dick Tobin http://boxrec.com/boxer/118985 1960-06-07 12
6 Ray Schlamp http://boxrec.com/boxer/32768 1964-06-11 15
7 Laszlo Papp http://boxrec.com/boxer/34798 1964-10-09 27
8 Alex Mack http://boxrec.com/boxer/93050 1969-11-25 13
9 Agustin Senin http://boxrec.com/boxer/79869 1972-09-27 42
10 Roberto Ayala http://boxrec.com/boxer/94505 1973-12-18 17
# ℹ 82 more rowsLooking at these data we see a problem. *wins should be a number as it described the number of consecutive wins to start a career, but it is a character <chr> instead.** We can see what went wrong here (although in other cases, we may need to use the View() function). Here Joe Devlin is marked as have 1 5 consecutive wins to start his career. 1 5 is obviously not a number. Is it one, five or fifteen? We have two options to fix this.
- We could treat it as missing data, or
- Find out the truth and fix it.
Let’s do the first option first, because it’s easiest, and then the second option (because it’s correct)
A simple mutate() to change class
The easiest option is to take our data and simply change wins to a number by using the as.numeric() function inside of a mutate. Here we can overwrite wins, as follows:
undefeated %>%
mutate(wins = as.numeric(wins))# A tibble: 92 × 4
name url date wins
<chr> <chr> <date> <dbl>
1 Juro Fukuda http://boxrec.com/boxer/195508 1943-11-26 27
2 Hilaire Pratesi http://boxrec.com/boxer/56069 1955-06-27 26
3 Rocky Marciano http://boxrec.com/boxer/9032 1955-09-21 49
4 Joe Devlin http://boxrec.com/boxer/69735 1960-04-29 NA
5 Dick Tobin http://boxrec.com/boxer/118985 1960-06-07 12
6 Ray Schlamp http://boxrec.com/boxer/32768 1964-06-11 15
7 Laszlo Papp http://boxrec.com/boxer/34798 1964-10-09 27
8 Alex Mack http://boxrec.com/boxer/93050 1969-11-25 13
9 Agustin Senin http://boxrec.com/boxer/79869 1972-09-27 42
10 Roberto Ayala http://boxrec.com/boxer/94505 1973-12-18 17
# ℹ 82 more rowsYou can see that this solved the problem – wins is now a <dbl>. But in doing so our entry, 1 5 turned into an NA. This is a bummer.
The actual value is likely either one, five, or fifteen, and it would be best to know the truth and make it right. Consulting the Consulting the original dataset, we see that the actual answer is fifteen. So let’s try to fix this!
Use Large Language Models to help you code
The problem was, this was hard for me. I knew I wanted to combine the case_when() function with mutate, but I kept getting errors. So I turned to ChatGPT for help. Importantly, I could clearly state my problem, so I gave ChatGPT this prompt
I am coding in R using the tidyverse and have the following problem. In my tibble (called,
undefeated), there is a column called “wins”, this should be a numeric vector<dbl>, but it is read as a character<chr>. Upon some inspection, i found that one value, which should have been 15, was actually “1 5”. How do I combine mutate and case_when, to convert “1 5” into 15, and make sure the new “wins” column is numeric?
ChatGPT responded
You can use mutate along with case_when and some string manipulation functions to clean the "1 5" value and ensure that the "wins" column is numeric. Here’s how you can do it:
undefeated <- undefeated %>%
mutate(
wins = case_when(
wins == "1 5" ~ "15", # Convert "1 5" to "15"
TRUE ~ wins # Keep other values as they are
),
wins = as.numeric(wins) # Convert the entire column to numeric
)Chat GPT went on to explain:
This code does the following:
- Uses
mutateto modify thewinscolumn.
case_whenchecks if the value inwinsis"1 5", and if so, replaces it with"15".
- The
TRUE ~ winsline keeps all other values in the column unchanged.
- After correcting
"1 5", the entirewinscolumn is converted to numeric usingas.numeric.
This will ensure that the wins column is correctly transformed into a numeric vector.
Let’s use the glimpse() function to check it out and see if it worked
glimpse(undefeated)Rows: 92
Columns: 4
$ name <chr> "Juro Fukuda", "Hilaire Pratesi", "Rocky Marciano", "Jo…
$ url <chr> "http://boxrec.com/boxer/195508", "http://boxrec.com/bo…
$ date <date> 1943-11-26, 1955-06-27, 1955-09-21, 1960-04-29, 1960-0…
$ wins <dbl> 27, 26, 49, 15, 12, 15, 27, 13, 42, 17, 9, 21, 15, 14, …THIS TOTALLY WORKED!!! So if you have some coding know how, you can use ChatGPT or other LLMs to help you when you run into tough problems.
Quiz
Figure 8: The accompanying quiz link
Key functions
Many require the dplyr package. I type dplyr:: before these, as it is a(n optional) way to let R know that a function comes from the dplyr package.
dplyr::mutate(): Add a new column, usually as some function of existing columns.
dplyr::summarize(): Condense a data set into summaries of variables.
dplyr::group_by(): Tell tidyverse that you want to perform the next operation on a groups (noted by the variable wegroup_by). Undo this with [ungroup()] when you are done.
as.numeric(): Change the class of a vector to numeric.