3.1 Determining the Primary Structures of Proteins
3.1.1 Enzymatic cleavage
One can determine the primary structure of proteins via enzymatic cleavage:
Figure 3.2: Enzymes Used in Cleaving Polypeptides
Figure 3.2 displays a list of enzymes used in determining the primary structure of proteins (and the cleavage points of these enzymes).
3.1.2 Short peptide sequencing
Figure 3.3: Sequencing Short Peptides
Short peptides can also be sequenced as shown in figure 3.3. Note that “FDNB” (i.e., 1-Fluoro-2,4-dinitrobenzene) is also known as Sanger’s reagent. FDNB reacts with the free amine group in amino acids to produce dinitrophenyl-amino acids.
3.1.3 Mass spectrometry
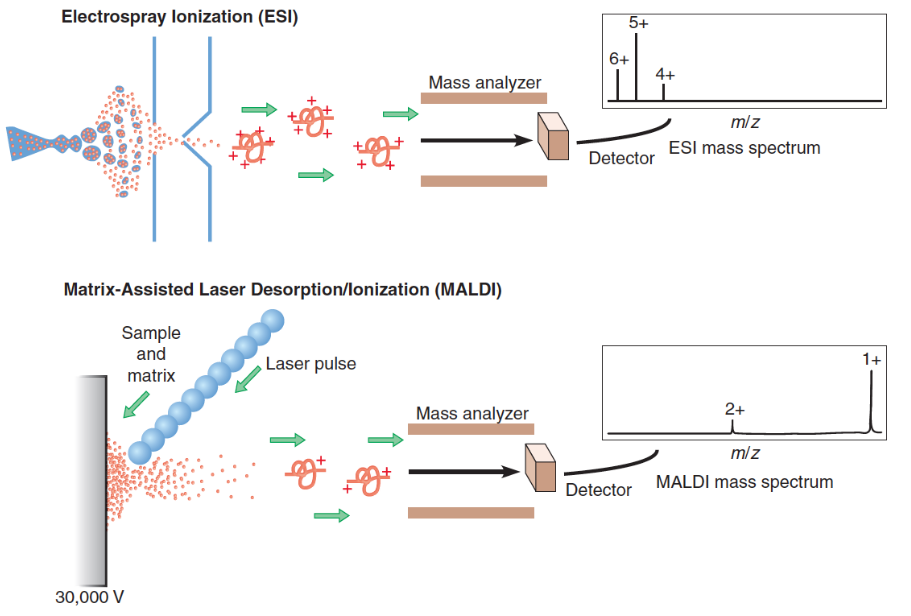
Figure 3.4: Mass Spectrometry to Determine Protein Primary Structures
While there are two kinds of mass spectrometry (shown in figure 3.4) performed to determine the primary structure of a protein, the molecules in either kind of mass spectrometry are first induced to pass through an electric or a magnetic field.
The path taken a molecule is a function of its mass-to-charge ratio (i.e., m/z ratio). This property can then be used to deduce the mass of molecules to a single Dalton precision!
3.1.3.1 MALDI-based protein identification
Figure 3.5: MALDI in Use for Determining Protein Properties
The above graphic (i.e., figure 3.5) aims to give an idea of what is possible in terms of identifying a protein from a database comparison (by measuring peptides from digestion).