Misc. Analyses
Stratified Analyses
By Sex
Total number of weight samples, by sex, for the 2016 cohort
weight.ls[[2]] %>%
janitor::tabyl(Gender) %>%
arrange(desc(n)) %>%
adorn_pct_formatting() %>%
tableStyle()| Gender | n | percent |
|---|---|---|
| M | 1097469 | 93.3% |
| F | 78526 | 6.7% |
Total number of people
weight.ls[[2]] %>%
distinct(PatientICN, Gender) %>%
tabyl(Gender) %>%
adorn_pct_formatting() %>%
tableStyle()| Gender | n | percent |
|---|---|---|
| F | 6200 | 6.3% |
| M | 92758 | 93.7% |
Association between number of weight samples collected, and Sex/Gender, for the 2016 cohort,
weight.ls[[2]] %>%
group_by(Gender) %>%
count(PatientICN) %>%
summarise(
mean = mean(n),
SD = sd(n),
median = median(n),
min = min(n),
max = max(n)
) %>%
tableStyle()| Gender | mean | SD | median | min | max |
|---|---|---|---|---|---|
| F | 12.67 | 12.61 | 9 | 1 | 277 |
| M | 11.83 | 19.05 | 8 | 1 | 1525 |
Men have, on average, slightly fewer weight values recorded than women in our sample.
Removing the major outlier,
weight.ls[[2]] %>%
group_by(Gender) %>%
count(PatientICN) %>%
filter(n < 1000) %>%
ggplot(aes(x = Gender, y = n)) %>%
add(geom_boxplot(fill = "maroon")) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(coord_flip()) %>%
add(labs(
x = "Sex",
y = "N Collected Weight Samples"
))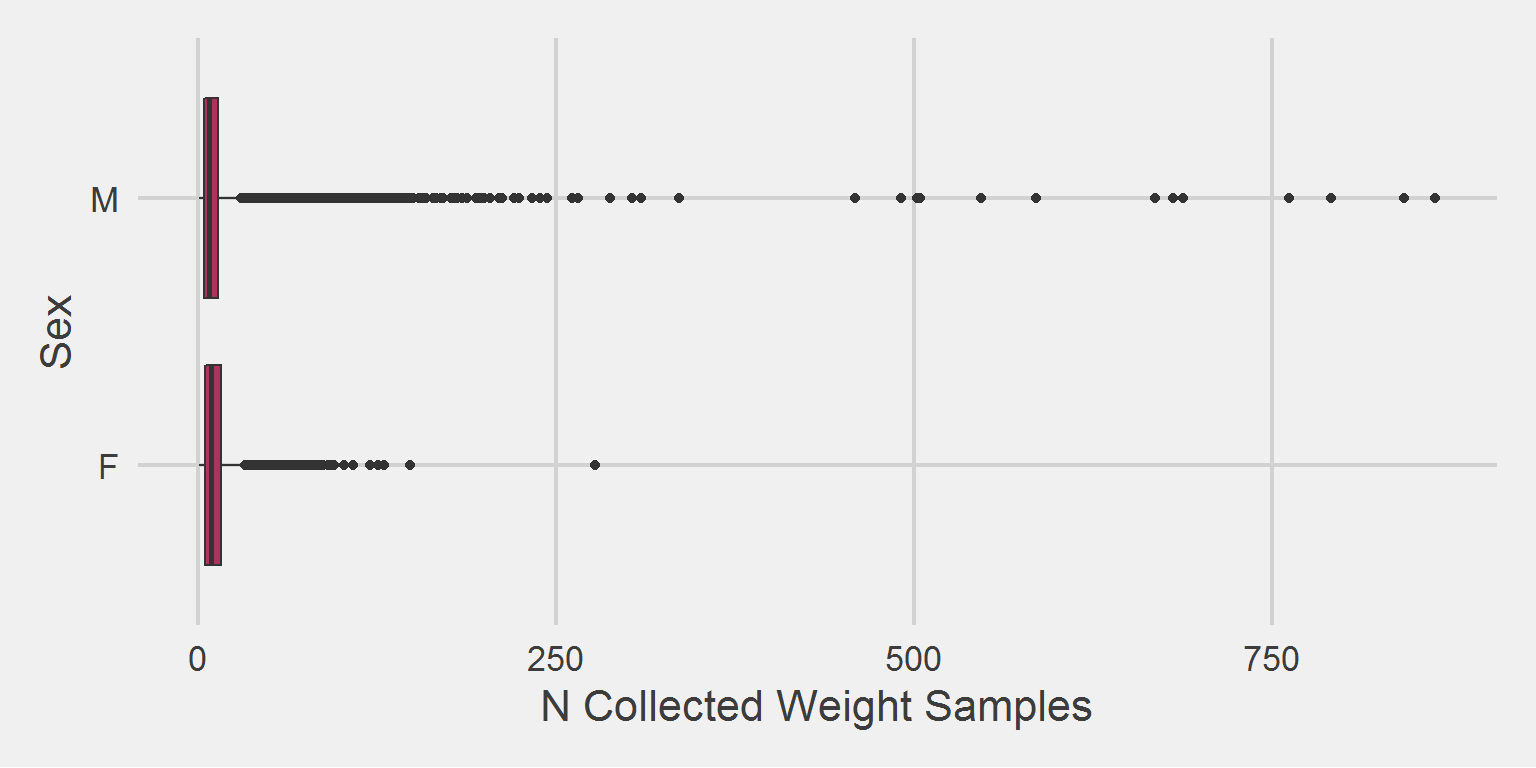
The distribution of outliers differs as well.
All weight in lbs. by sex, for the 2016 cohort only
weight.ls[[2]] %>%
group_by(Gender) %>%
summarise(
mean = mean(Weight),
SD = sd(Weight),
median = median(Weight),
min = min(Weight),
max = max(Weight)
) %>%
tableStyle()| Gender | mean | SD | median | min | max |
|---|---|---|---|---|---|
| F | 184.66 | 44.56 | 180.7 | 0 | 1479.9 |
| M | 209.48 | 48.45 | 204.0 | 0 | 1486.2 |
Both Males and Females have implausible maximum weight values.
weight.ls[[2]] %>%
ggplot(aes(x = Gender, y = Weight)) %>%
add(geom_boxplot(fill = "maroon")) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(coord_flip())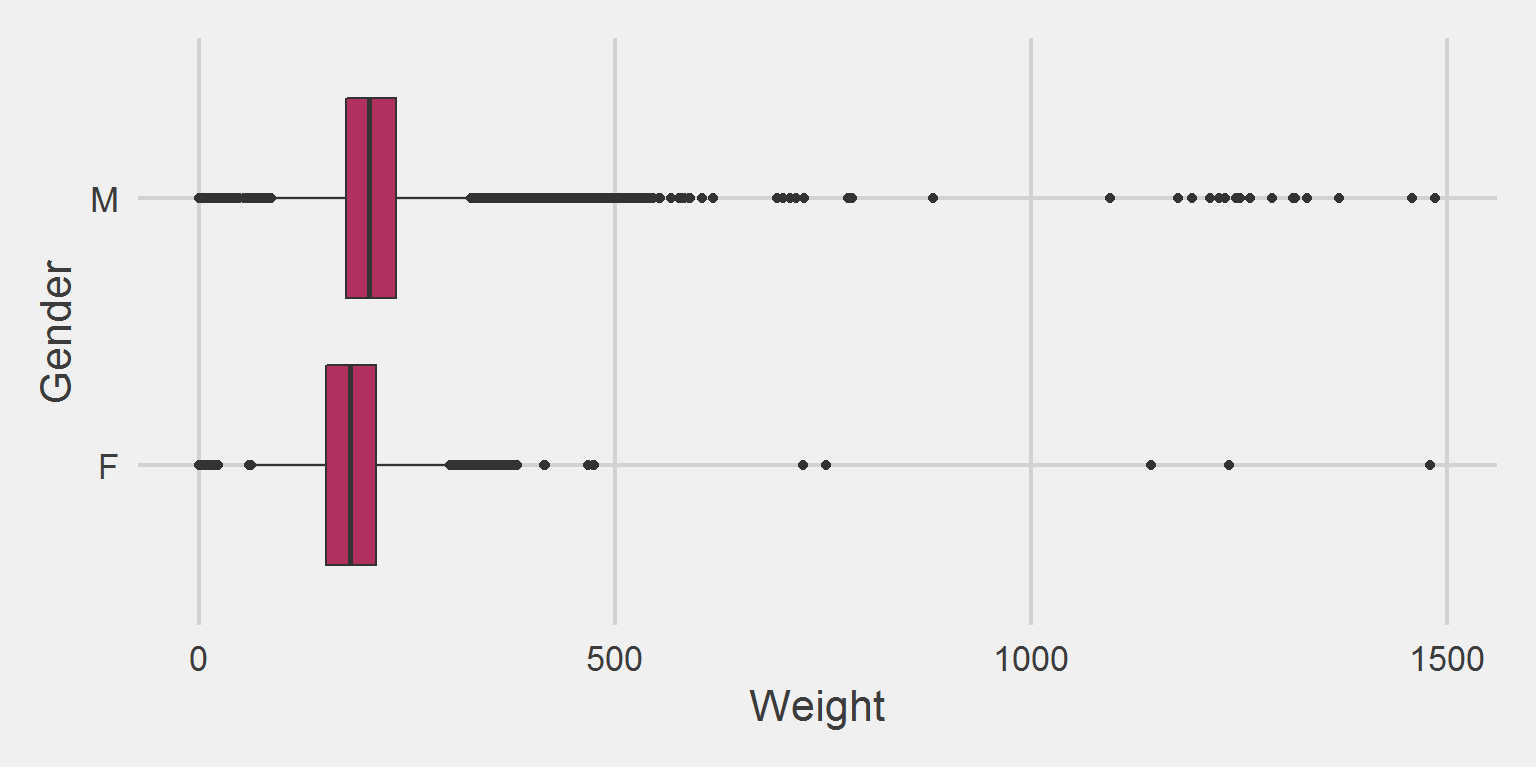
By Algorithm
raw <- weight.ls[[2]] %>%
select(PatientICN, Weight, Gender)
#---- apply Janney et al. 2016 ----#
Janney2016.df <- Janney2016.f(
DF = weight.ls[[2]],
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime",
startPoint = "VisitDateTime"
) %>%
select(PatientICN, Weight_OR, Gender) %>%
rename(Weight = Weight_OR) %>%
na.omit()
#---- apply Littman et al. 2012 ----#
Littman2012.df <- Littman2012.f(
DF = weight.ls[[2]],
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime"
) %>%
select(PatientICN, OutputMeasurement, Gender) %>%
rename(Weight = OutputMeasurement) %>%
na.omit()
#---- Coerce Maciejewski et al. 2016 to a workable format ----#
Maciejewski2016.df <- maciejewski %>%
filter(IO == "Output", SampleYear == "2016") %>%
mutate(PatientICN = as.character(PatientICN)) %>%
left_join(
raw %>%
distinct(PatientICN, Gender),
by = "PatientICN"
) %>%
select(PatientICN, Weight, Gender) %>%
na.omit()
#---- Apply Breland et al. 2017 ----#
Breland2017.df <- Breland2017.f(
DF = weight.ls[[2]],
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime"
) %>%
select(PatientICN, measures_aug_, Gender) %>%
rename(Weight = measures_aug_) %>%
na.omit()
#---- Apply Maguen et al. 2013 ----#
Maguen2013.df <- Maguen2013.f(
DF = weight.ls[[2]],
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime",
variables = c("AgeAtVisit", "Gender")
) %>%
select(PatientICN, Output, Gender) %>%
rename(Weight = Output) %>%
na.omit()
#---- Apply Goodrich 2016 ----#
Goodrich2016.df <- Goodrich2016.f(
DF = weight.ls[[2]],
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime",
startPoint = "VisitDateTime"
) %>%
select(PatientICN, output, Gender) %>%
rename(Weight = output) %>%
na.omit()
#---- Apply Chan & Raffa 2017 ----#
Chan2017.df <- Chan2017.f(
DF = weight.ls[[2]],
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime"
) %>%
select(PatientICN, measures_aug_, Gender) %>%
rename(Weight = measures_aug_) %>%
na.omit()
#---- Apply Jackson et al. 2015 ----#
Jackson2015.df <- Jackson2015.f(
DF = weight.ls[[2]],
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime",
startPoint = "VisitDateTime"
) %>%
left_join(
weight.ls[[2]] %>%
distinct(PatientICN, Gender),
by = "PatientICN"
) %>%
select(PatientICN, Gender, output) %>%
rename(Weight = output) %>%
na.omit()
#---- Apply Buta et al., 2018 ----#
Buta2018.df <- Buta2018.f(
DF = weight.ls[[2]],
id = "PatientICN",
measures = "BMI",
tmeasures = "WeightDateTime"
) %>%
select(PatientICN, Gender, Weight) %>%
na.omit()
#---- Apply Kazerooni & Lim, 2016 ----#
Kazerooni2016.df <- Kazerooni2016.f(
DF = weight.ls[[2]],
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime",
startPoint = "VisitDateTime"
) %>%
select(PatientICN, Gender, Weight) %>%
na.omit()
#---- Apply Noel et al., 2012 ----#
Noel2012.df <- Noel2012.f(
DF = weight.ls[[2]],
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime"
) %>%
distinct(PatientICN, Gender, FYQ, Qmedian) %>%
select(-FYQ) %>%
rename(Weight = Qmedian) %>%
na.omit()
#---- Apply Rosenberger et al., 2011 ----#
Rosenberger2011.df <- Rosenberger2011.f(
DF = weight.ls[[2]],
id = "PatientICN",
tmeasures = "WeightDateTime",
startPoint = "VisitDateTime",
pad = 1
) %>%
select(PatientICN, Gender, Weight) %>%
na.omit()
# stack original (raw), janney 2016, littman 2012, maciejewski 2016,
# Breland 2017, Maguen 2013, Goodrich 2016, Chan & Raffa 2017,
# Jackson 2015, Buta 2018, Kazerooni & Lim 2016, Noel 2012,
# and Rosenberger 2011 processed weights
algos.fac <- c("Raw Weights",
"Janney 2016",
"Littman 2012",
"Maciejewski 2016",
"Breland 2017",
"Maguen 2013",
"Goodrich 2016",
"Chan 2017",
"Jackson 2015",
"Buta 2018",
"Kazerooni 2016",
"Noel 2012",
"Rosenberger 2011")
DF <- bind_rows(
list(`Raw Weights` = raw,
`Janney 2016` = Janney2016.df,
`Littman 2012` = Littman2012.df,
`Maciejewski 2016` = Maciejewski2016.df,
`Breland 2017` = Breland2017.df,
`Maguen 2013` = Maguen2013.df,
`Goodrich 2016` = Goodrich2016.df,
`Chan 2017` = Chan2017.df,
`Jackson 2015` = Jackson2015.df,
`Buta 2018` = Buta2018.df,
`Kazerooni 2016` = Kazerooni2016.df,
`Noel 2012` = Noel2012.df,
`Rosenberger 2011` = Rosenberger2011.df),
.id = "Algorithm"
)
rm(
Buta2018.df,
Kazerooni2016.df,
Noel2012.df,
Rosenberger2011.df,
Breland2017.df,
Chan2017.df,
Goodrich2016.df,
Jackson2015.df,
Janney2016.df,
Littman2012.df,
Maciejewski2016.df,
Maguen2013.df,
raw
)
DF$Algorithm <- factor(DF$Algorithm, levels = algos.fac, labels = algos.fac)
DF$Type <- case_when(
DF$Algorithm == "Raw Weights" ~ 1,
DF$Algorithm %in% algos.fac[c(2, 7, 9, 11:13)] ~ 2,
DF$Algorithm %in% algos.fac[c(3:6, 8, 10)] ~ 3
)
DF$Type <- factor(DF$Type, 1:3, c("Raw", "Time-Periods", "Time-Series"))Total weight measurements, by algorithm and Sex.
DF %>%
tabyl(Algorithm, Gender) %>%
adorn_percentages("col") %>%
adorn_pct_formatting() %>%
adorn_ns() %>%
tableStyle()| Algorithm | F | M |
|---|---|---|
| Raw Weights | 12.3% (78526) | 12.2% (1097469) |
| Janney 2016 | 1.9% (12268) | 2.1% (187562) |
| Littman 2012 | 12.1% (77105) | 12.1% (1084556) |
| Maciejewski 2016 | 12.1% (77240) | 11.9% (1067596) |
| Breland 2017 | 12.3% (78462) | 12.2% (1096715) |
| Maguen 2013 | 10.9% (69824) | 10.8% (967469) |
| Goodrich 2016 | 1.9% (12268) | 2.1% (187518) |
| Chan 2017 | 12.2% (78150) | 12.1% (1091964) |
| Jackson 2015 | 2.4% (15479) | 2.6% (236022) |
| Buta 2018 | 11.9% (76284) | 11.7% (1055712) |
| Kazerooni 2016 | 0.8% (5094) | 0.7% (66867) |
| Noel 2012 | 6.9% (44332) | 7.1% (638676) |
| Rosenberger 2011 | 2.2% (14116) | 2.4% (213099) |
We can test the overall differences in distributions by algorithm and sex, though for the table above, it is wholly unnecessary.
tmp <- DF %>%
tabyl(Algorithm, Gender) %>%
adorn_percentages("col") %>%
pivot_longer(!Algorithm, names_to = "Sex", values_to = "Prop")
test <- mgcv::gam(
Prop ~ Sex + s(Algorithm, bs = 're'),
data = tmp,
family = binomial,
method = 'REML'
)
broom::tidy(test)# A tibble: 1 x 5
term edf ref.df statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 s(Algorithm) 0.00000209 12 0.000000145 1.00No difference by Algorithm or Sex
library(mgcViz)
plot(sm(getViz(test), 1)) %>%
add(l_fitLine(color = "red")) %>%
add(l_ciLine(mul = 5, color = "blue", linetype = 2)) %>%
add(l_points())
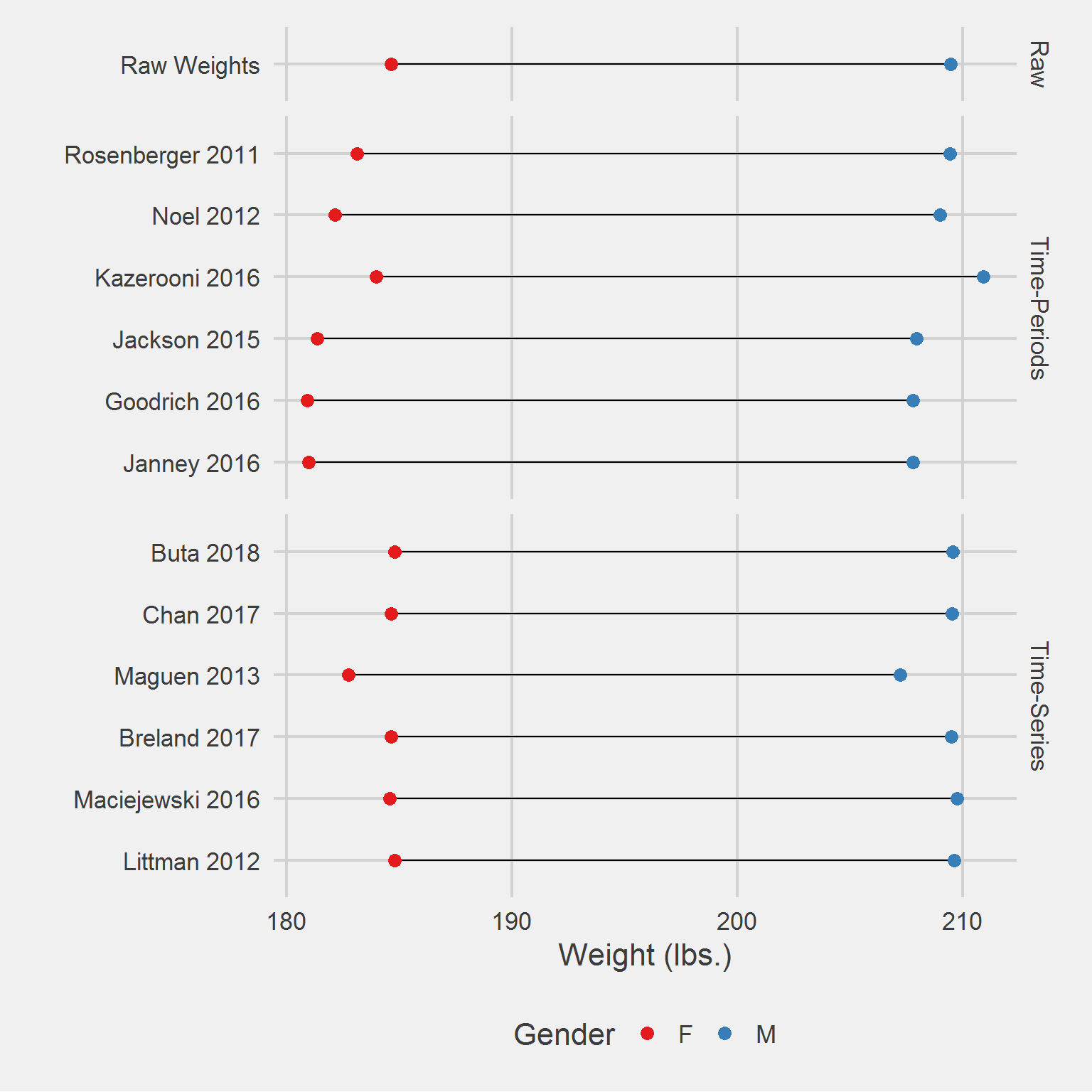
Distribution of weight measurements, by algorithm,
tmp <- DF %>%
filter(!is.na(Weight)) %>%
group_by(Type, Algorithm, Gender) %>%
dplyr::summarise(
weight_n = n(),
mean = mean(Weight),
SD = sd(Weight),
median = median(Weight),
IQR = IQR(Weight),
min = min(Weight),
max = max(Weight)
)
tableStyle(tmp)| Type | Algorithm | Gender | weight_n | mean | SD | median | IQR | min | max |
|---|---|---|---|---|---|---|---|---|---|
| Raw | Raw Weights | F | 78526 | 184.66 | 44.56 | 180.70 | 59.80 | 0.00 | 1479.90 |
| Raw | Raw Weights | M | 1097469 | 209.48 | 48.45 | 204.00 | 60.00 | 0.00 | 1486.20 |
| Time-Periods | Janney 2016 | F | 12268 | 181.00 | 42.37 | 177.00 | 56.90 | 78.40 | 473.99 |
| Time-Periods | Janney 2016 | M | 187562 | 207.80 | 45.17 | 202.55 | 56.20 | 80.00 | 546.00 |
| Time-Periods | Goodrich 2016 | F | 12268 | 180.93 | 42.25 | 177.00 | 56.90 | 80.25 | 382.00 |
| Time-Periods | Goodrich 2016 | M | 187518 | 207.78 | 45.13 | 202.50 | 56.20 | 80.00 | 500.00 |
| Time-Periods | Jackson 2015 | F | 15479 | 181.36 | 42.24 | 177.00 | 57.63 | 75.63 | 375.00 |
| Time-Periods | Jackson 2015 | M | 236022 | 207.94 | 45.24 | 202.70 | 56.19 | 78.00 | 553.00 |
| Time-Periods | Kazerooni 2016 | F | 5094 | 183.98 | 43.26 | 180.20 | 59.00 | 75.62 | 467.38 |
| Time-Periods | Kazerooni 2016 | M | 66867 | 210.90 | 47.76 | 205.70 | 59.80 | 0.00 | 1233.70 |
| Time-Periods | Noel 2012 | F | 44332 | 182.17 | 42.61 | 178.00 | 57.71 | 72.90 | 467.38 |
| Time-Periods | Noel 2012 | M | 638676 | 208.99 | 45.86 | 203.60 | 57.30 | 70.00 | 588.90 |
| Time-Periods | Rosenberger 2011 | F | 14116 | 183.13 | 43.41 | 179.00 | 57.90 | 0.00 | 1144.00 |
| Time-Periods | Rosenberger 2011 | M | 213099 | 209.44 | 45.95 | 204.00 | 57.00 | 0.00 | 1314.50 |
| Time-Series | Littman 2012 | F | 77105 | 184.79 | 43.71 | 180.90 | 59.60 | 77.38 | 413.80 |
| Time-Series | Littman 2012 | M | 1084556 | 209.62 | 48.02 | 204.00 | 59.73 | 75.20 | 546.00 |
| Time-Series | Maciejewski 2016 | F | 77240 | 184.57 | 43.72 | 180.60 | 59.88 | 62.00 | 413.80 |
| Time-Series | Maciejewski 2016 | M | 1067596 | 209.76 | 47.99 | 204.00 | 59.70 | 64.00 | 546.00 |
| Time-Series | Breland 2017 | F | 78462 | 184.66 | 43.79 | 180.78 | 59.80 | 75.00 | 473.99 |
| Time-Series | Breland 2017 | M | 1096715 | 209.51 | 48.12 | 204.00 | 59.90 | 75.20 | 694.46 |
| Time-Series | Maguen 2013 | F | 69824 | 182.76 | 42.81 | 179.00 | 58.50 | 70.30 | 382.00 |
| Time-Series | Maguen 2013 | M | 967469 | 207.23 | 46.18 | 202.00 | 57.90 | 74.00 | 541.10 |
| Time-Series | Chan 2017 | F | 78150 | 184.66 | 43.82 | 180.80 | 59.80 | 59.20 | 725.32 |
| Time-Series | Chan 2017 | M | 1091964 | 209.52 | 48.14 | 204.00 | 59.90 | 54.00 | 727.50 |
| Time-Series | Buta 2018 | F | 76284 | 184.81 | 43.81 | 181.00 | 59.95 | 60.00 | 415.79 |
| Time-Series | Buta 2018 | M | 1055712 | 209.58 | 48.17 | 204.00 | 60.00 | 66.50 | 540.00 |
tmp %>%
ggplot(aes(mean, Algorithm)) %>%
add(geom_line(aes(group = Algorithm))) %>%
add(geom_point(aes(color = Gender), size = 3)) %>%
add(facet_grid(rows = vars(Type), scales = "free", space = "free")) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(scale_color_brewer(palette = "Set1")) %>%
add(labs(
x = "Weight (lbs.)",
y = ""
))
Test/model if there is a difference by algorithm choice
test_mean <- mgcv::gam(
mean ~ Gender + s(Algorithm, bs = 're'),
data = tmp,
family = gaussian,
method = 'REML'
)
broom::tidy(test_mean)# A tibble: 1 x 5
term edf ref.df statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 s(Algorithm) 9.96 12 4.87 0.000569This would suggest that there are some differences by Algorithm on the mean; mean weight is expected to differ by sex.
test_sd <- mgcv::gam(
SD ~ Gender + s(Algorithm, bs = 're'),
data = tmp,
family = gaussian,
method = 'REML'
)
broom::tidy(test_sd)# A tibble: 1 x 5
term edf ref.df statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 s(Algorithm) 10.5 12 7.03 0.0000380And there certainly is some difference in standard deviation by algorithm
Number of Patients/Veterans Retained, by algorithm, by Sex,
tmp <- DF %>%
filter(!is.na(Weight)) %>%
group_by(Type, Algorithm, Gender) %>%
distinct(PatientICN) %>%
count()
tableStyle(tmp)| Type | Algorithm | Gender | n |
|---|---|---|---|
| Raw | Raw Weights | F | 6200 |
| Raw | Raw Weights | M | 92758 |
| Time-Periods | Janney 2016 | F | 5933 |
| Time-Periods | Janney 2016 | M | 89809 |
| Time-Periods | Goodrich 2016 | F | 5935 |
| Time-Periods | Goodrich 2016 | M | 89811 |
| Time-Periods | Jackson 2015 | F | 6007 |
| Time-Periods | Jackson 2015 | M | 90552 |
| Time-Periods | Kazerooni 2016 | F | 1698 |
| Time-Periods | Kazerooni 2016 | M | 22289 |
| Time-Periods | Noel 2012 | F | 6200 |
| Time-Periods | Noel 2012 | M | 92758 |
| Time-Periods | Rosenberger 2011 | F | 3929 |
| Time-Periods | Rosenberger 2011 | M | 59476 |
| Time-Series | Littman 2012 | F | 6026 |
| Time-Series | Littman 2012 | M | 90104 |
| Time-Series | Maciejewski 2016 | F | 6200 |
| Time-Series | Maciejewski 2016 | M | 92758 |
| Time-Series | Breland 2017 | F | 6200 |
| Time-Series | Breland 2017 | M | 92758 |
| Time-Series | Maguen 2013 | F | 6164 |
| Time-Series | Maguen 2013 | M | 92188 |
| Time-Series | Chan 2017 | F | 6027 |
| Time-Series | Chan 2017 | M | 90105 |
| Time-Series | Buta 2018 | F | 5749 |
| Time-Series | Buta 2018 | M | 84410 |
tmp %>%
ggplot(aes(x = Algorithm, y = n)) %>%
add(geom_point(aes(fill = Gender), size = 4, pch = 21)) %>%
add(facet_grid(
cols = vars(Gender),
rows = vars(Type),
scales = "free",
space = "free_y"
)) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(scale_fill_brewer(palette = "Set1")) %>%
add(coord_flip()) %>%
add(labs(
x = "",
y = "N Patients Retained"
))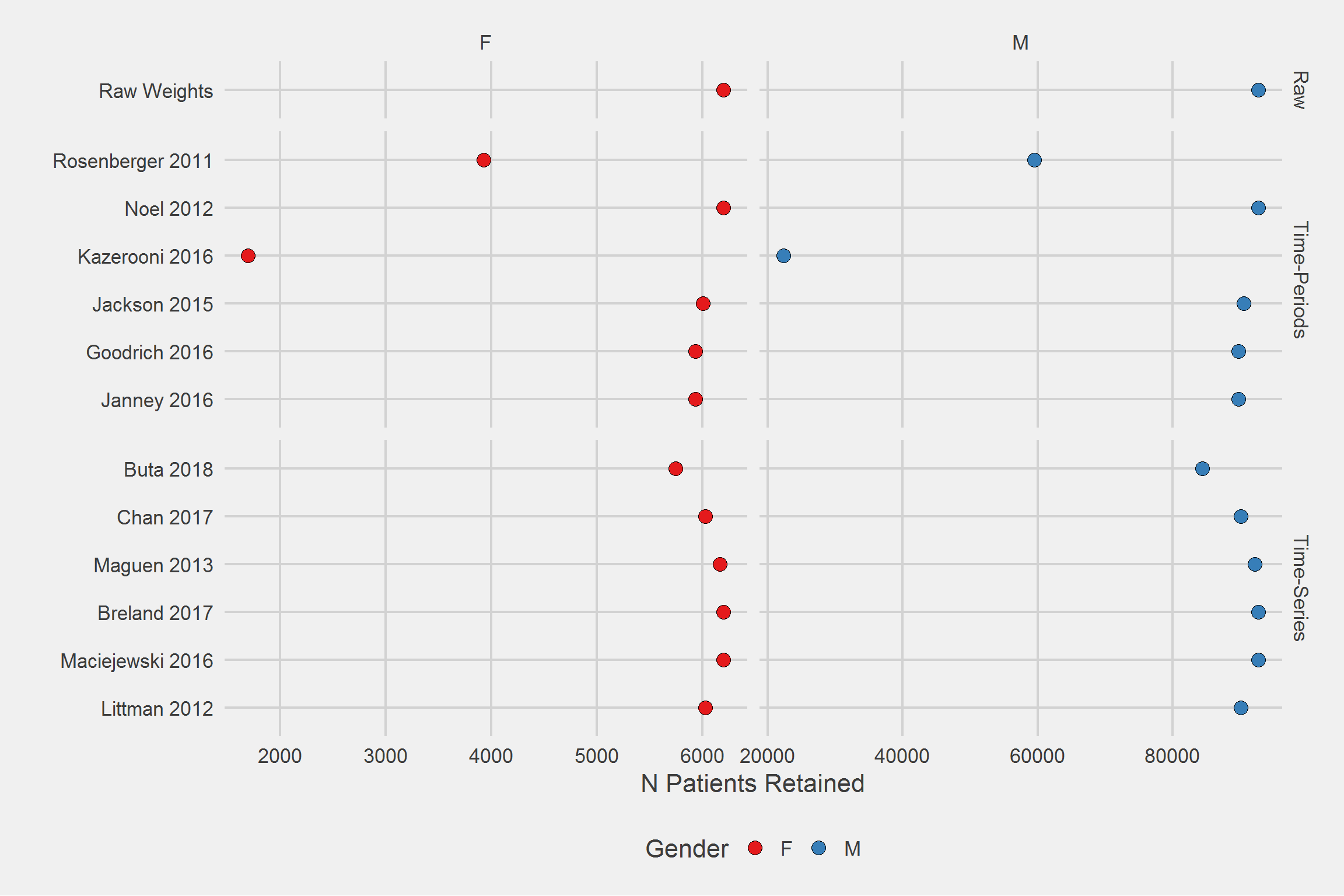
The relative differences in algorithmic effect between sexes are minute.
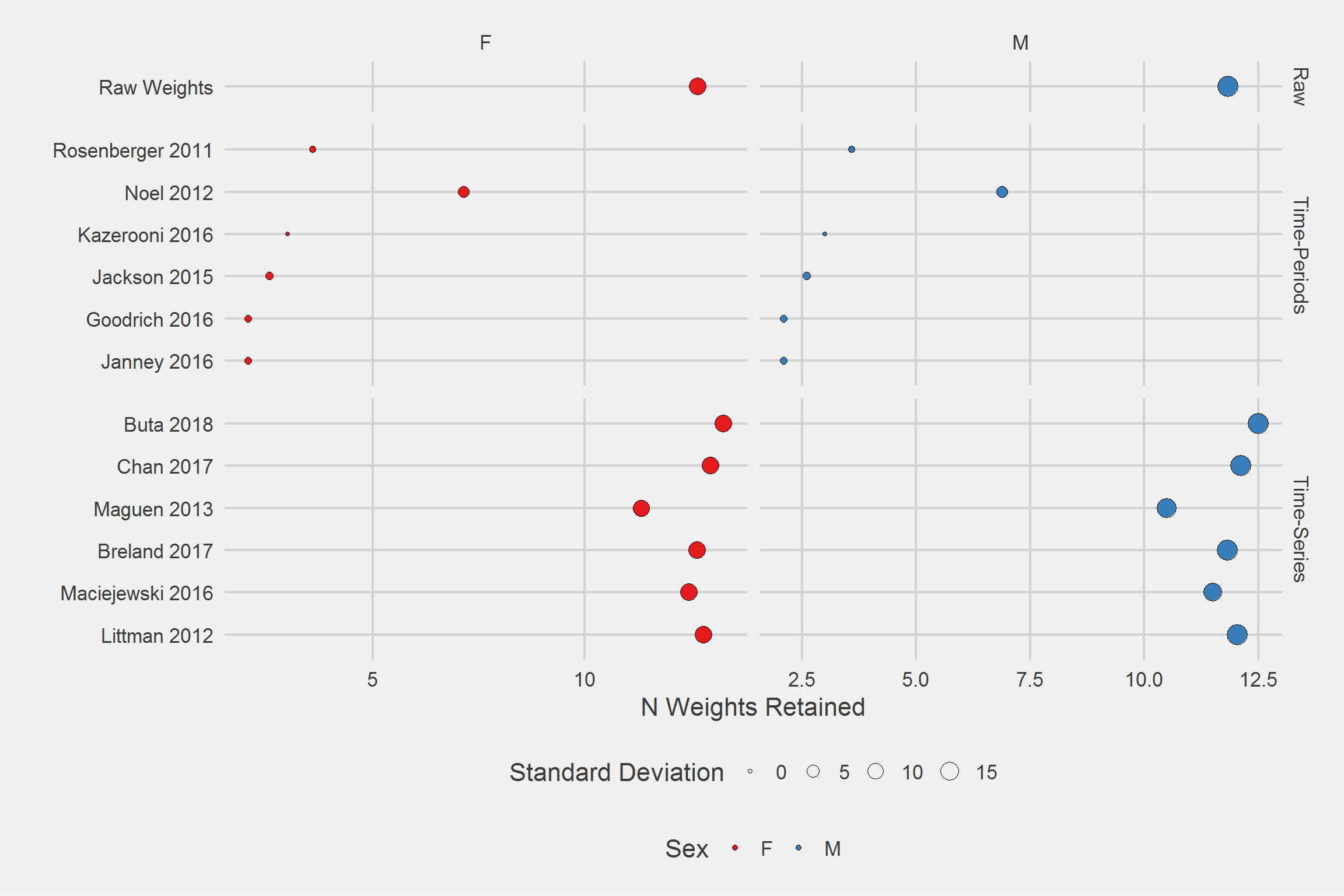
Number of weights retained, per person, by algorithm, by Sex,
tmp <- DF %>%
filter(!is.na(Weight)) %>%
group_by(Type, Algorithm, Gender, PatientICN) %>%
count() %>%
ungroup() %>%
group_by(Type, Algorithm, Gender) %>%
summarize(
mean = mean(n),
SD = sd(n),
min = min(n),
Q1 = fivenum(n)[2],
median = median(n),
Q3 = fivenum(n)[4],
max = max(n)
)
tableStyle(tmp)| Type | Algorithm | Gender | mean | SD | min | Q1 | median | Q3 | max |
|---|---|---|---|---|---|---|---|---|---|
| Raw | Raw Weights | F | 12.67 | 12.61 | 1 | 5 | 9 | 16 | 277 |
| Raw | Raw Weights | M | 11.83 | 19.05 | 1 | 4 | 8 | 14 | 1525 |
| Time-Periods | Janney 2016 | F | 2.07 | 0.78 | 1 | 1 | 2 | 3 | 3 |
| Time-Periods | Janney 2016 | M | 2.09 | 0.76 | 1 | 2 | 2 | 3 | 3 |
| Time-Periods | Goodrich 2016 | F | 2.07 | 0.78 | 1 | 1 | 2 | 3 | 3 |
| Time-Periods | Goodrich 2016 | M | 2.09 | 0.76 | 1 | 2 | 2 | 3 | 3 |
| Time-Periods | Jackson 2015 | F | 2.58 | 1.00 | 1 | 2 | 3 | 3 | 4 |
| Time-Periods | Jackson 2015 | M | 2.61 | 0.98 | 1 | 2 | 3 | 3 | 4 |
| Time-Periods | Kazerooni 2016 | F | 3.00 | 0.00 | 3 | 3 | 3 | 3 | 3 |
| Time-Periods | Kazerooni 2016 | M | 3.00 | 0.00 | 3 | 3 | 3 | 3 | 3 |
| Time-Periods | Noel 2012 | F | 7.15 | 3.75 | 1 | 4 | 7 | 10 | 17 |
| Time-Periods | Noel 2012 | M | 6.89 | 3.72 | 1 | 4 | 6 | 9 | 17 |
| Time-Periods | Rosenberger 2011 | F | 3.59 | 0.49 | 3 | 3 | 4 | 4 | 4 |
| Time-Periods | Rosenberger 2011 | M | 3.58 | 0.49 | 3 | 3 | 4 | 4 | 4 |
| Time-Series | Littman 2012 | F | 12.80 | 12.43 | 2 | 5 | 9 | 16 | 277 |
| Time-Series | Littman 2012 | M | 12.04 | 19.11 | 2 | 5 | 8 | 14 | 1524 |
| Time-Series | Maciejewski 2016 | F | 12.46 | 12.25 | 1 | 5 | 9 | 16 | 258 |
| Time-Series | Maciejewski 2016 | M | 11.51 | 14.43 | 1 | 4 | 8 | 14 | 637 |
| Time-Series | Breland 2017 | F | 12.66 | 12.61 | 1 | 5 | 9 | 16 | 277 |
| Time-Series | Breland 2017 | M | 11.82 | 19.02 | 1 | 4 | 8 | 14 | 1524 |
| Time-Series | Maguen 2013 | F | 11.33 | 10.89 | 1 | 5 | 8 | 14 | 192 |
| Time-Series | Maguen 2013 | M | 10.49 | 16.91 | 1 | 4 | 7 | 13 | 1516 |
| Time-Series | Chan 2017 | F | 12.97 | 12.56 | 2 | 5 | 9 | 16 | 269 |
| Time-Series | Chan 2017 | M | 12.12 | 19.11 | 2 | 5 | 8 | 14 | 1517 |
| Time-Series | Buta 2018 | F | 13.27 | 12.68 | 2 | 5 | 10 | 17 | 277 |
| Time-Series | Buta 2018 | M | 12.51 | 19.73 | 2 | 5 | 8 | 15 | 1524 |
Testing/modeling
test_mean <- mgcv::gam(
mean ~ Gender + Type + s(Algorithm, bs = 're'),
data = tmp,
family = gaussian,
method = 'REML'
)
summary(test_mean)
Family: gaussian
Link function: identity
Formula:
mean ~ Gender + Type + s(Algorithm, bs = "re")
Parametric coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 12.48003 1.40738 8.868 1.13e-06 ***
GenderM -0.46305 0.11716 -3.952 0.00185 **
TypeTime-Periods -8.85635 1.51882 -5.831 7.50e-05 ***
TypeTime-Series -0.08501 1.51882 -0.056 0.95627
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Approximate significance of smooth terms:
edf Ref.df F p-value
s(Algorithm) 9.774 10 43.33 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
R-sq.(adj) = 0.996 Deviance explained = 99.8%
-REML = 27.358 Scale est. = 0.089217 n = 26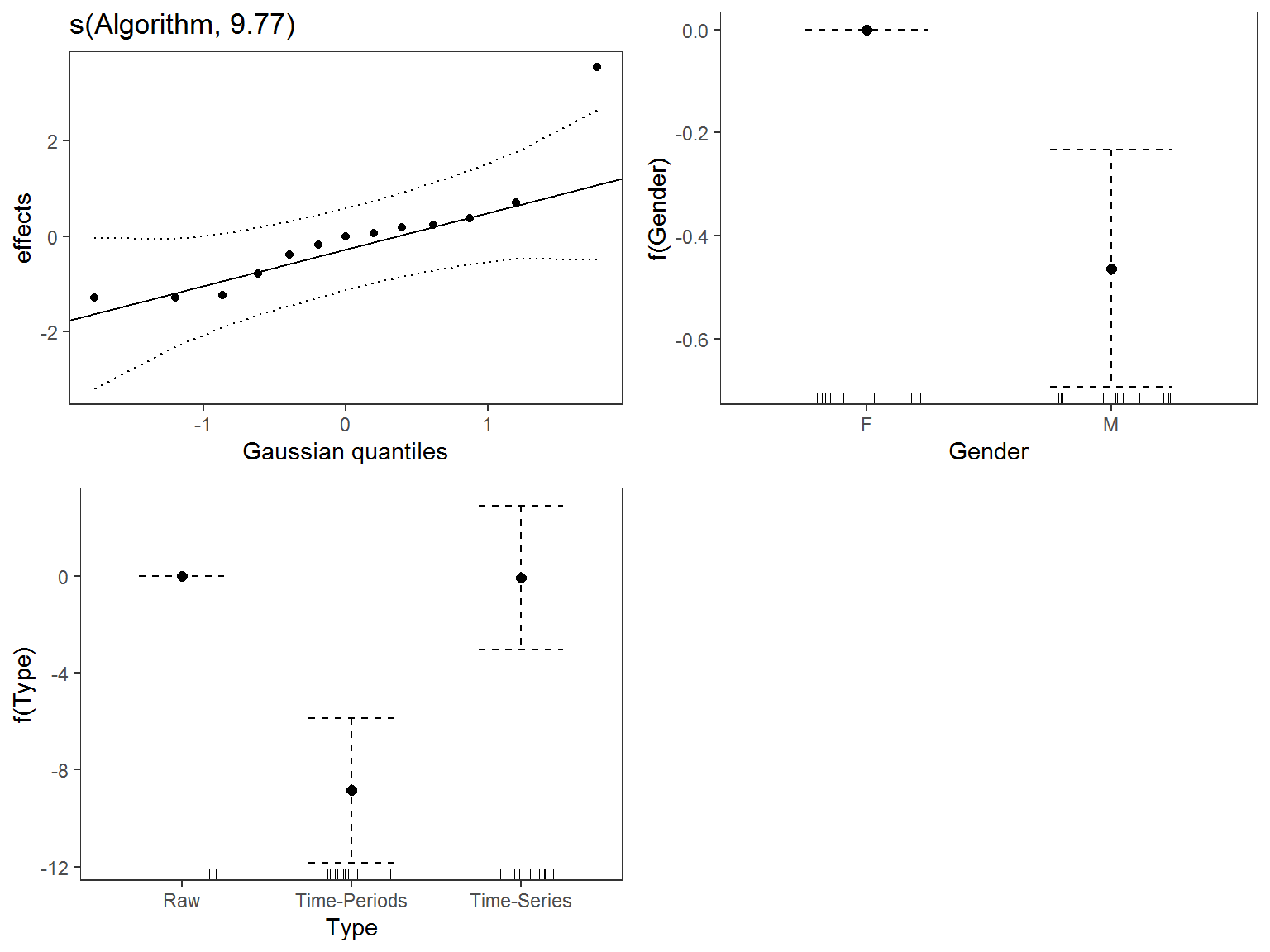
These results suggest there are differences between algorithms and sex, though not for algorithms that use all data, at least when compared to the “raw” data.
tmp %>%
ggplot(aes(x = Algorithm, y = mean)) %>%
add(geom_point(aes(fill = Gender, size = SD), pch = 21)) %>%
add(facet_grid(
cols = vars(Gender),
rows = vars(Type),
scales = "free",
space = "free_y"
)) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(scale_fill_brewer(palette = "Set1")) %>%
add(coord_flip()) %>%
add(labs(
x = "",
y = "N Weights Retained",
size = "Standard Deviation",
fill = "Sex"
))
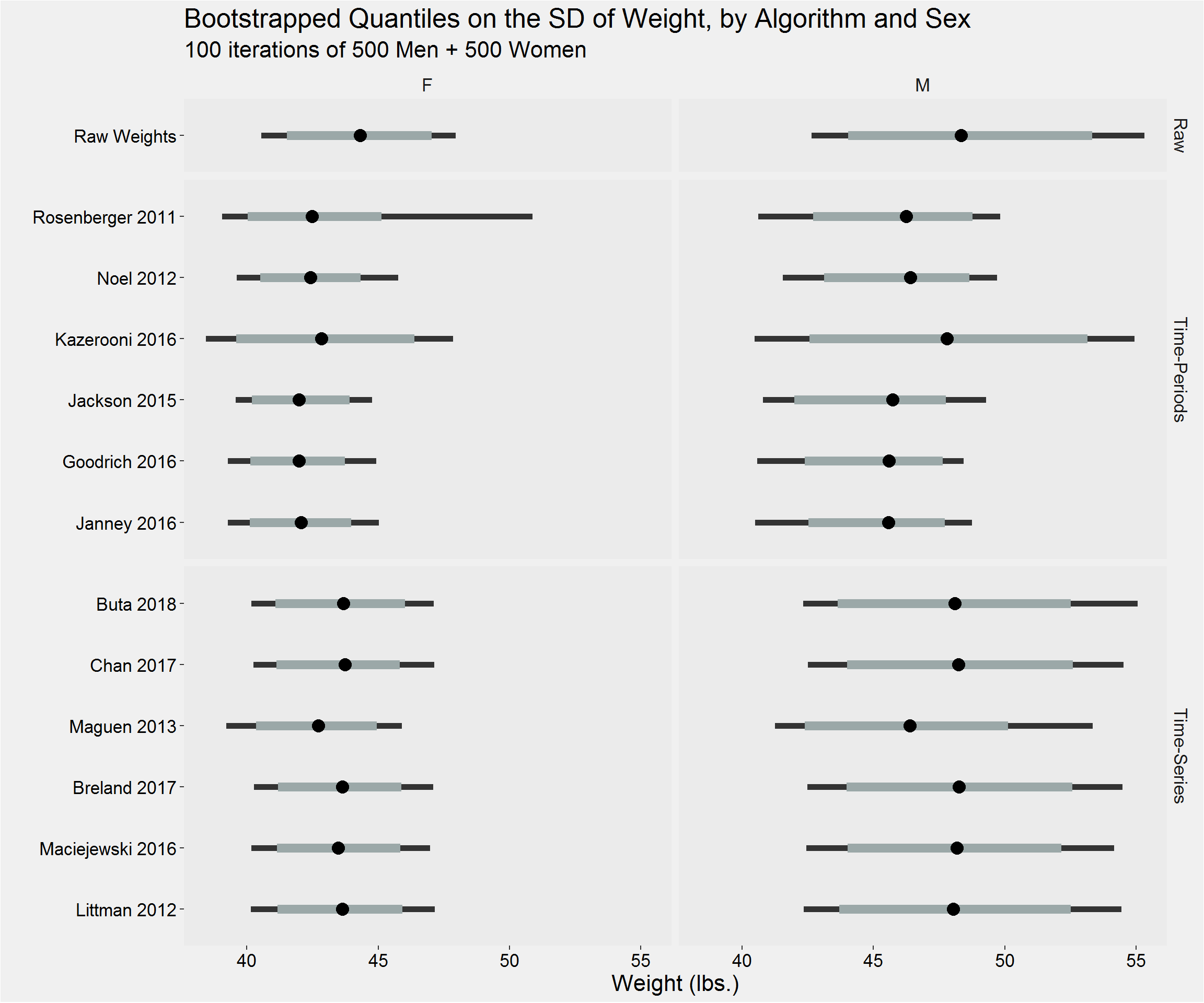
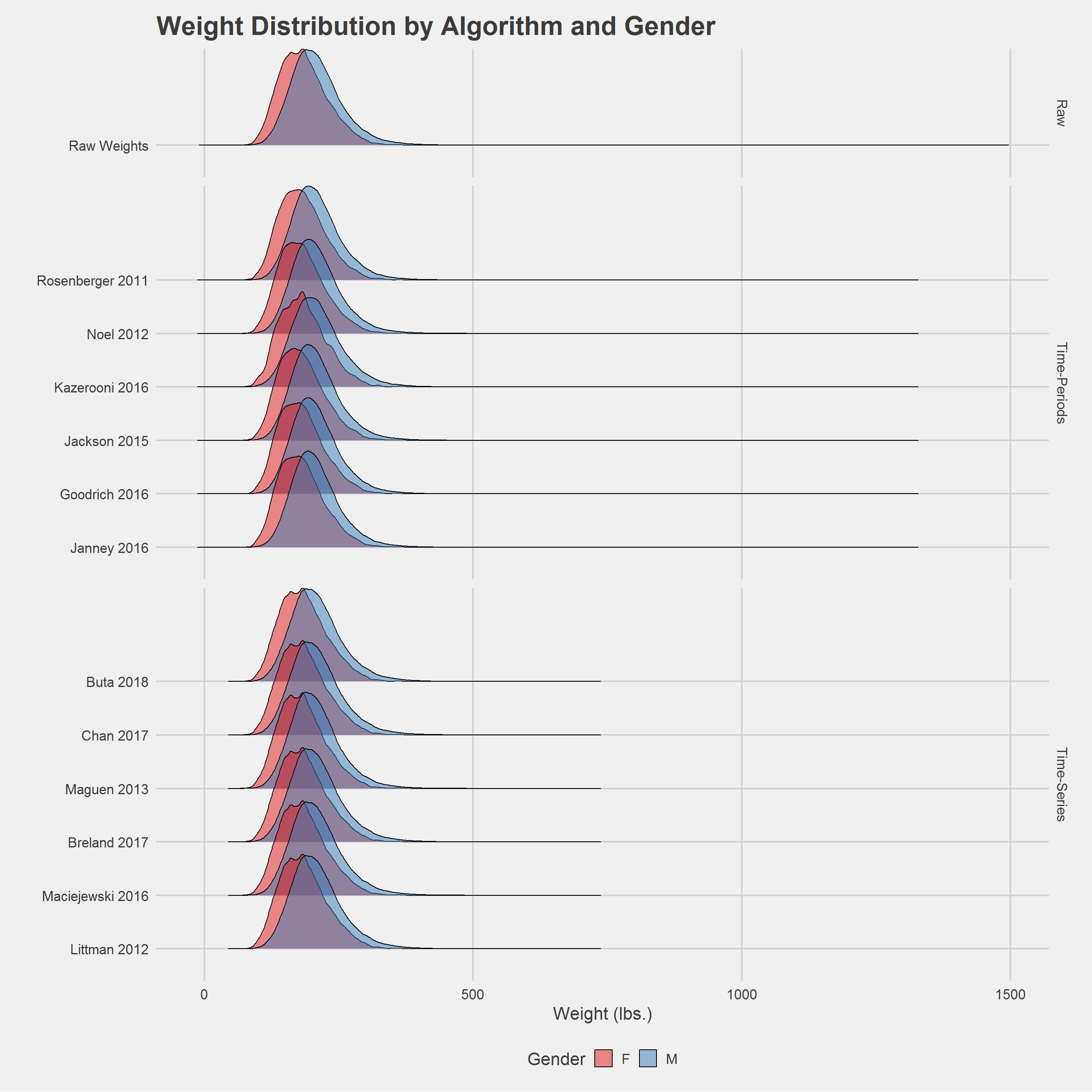
Bootstrapped Estimates
N <- 100
weight_samps <- vector("list", N)
key_cols <- c("Algorithm", "Gender")
pb <- txtProgressBar(min = 0, max = N, initial = 0, style = 3)
for (i in 1:N) {
setTxtProgressBar(pb, i)
sampM <- sample(
pts %>% filter(Gender == "M") %>% pull(PatientICN),
size = 500,
replace = TRUE
)
sampF <- sample(
pts %>% filter(Gender == "F") %>% pull(PatientICN),
size = 500,
replace = TRUE
)
samp <- c(sampM, sampF)
weight_samps[[i]] <- DF %>% filter(PatientICN %in% samp)
setDT(weight_samps[[i]])
setkeyv(weight_samps[[i]], key_cols)
weight_samps[[i]] <-
weight_samps[[i]][,
list(mean = mean(Weight), SD = sd(Weight)),
by = key_cols
]
}Visualization
Distribution of the mean
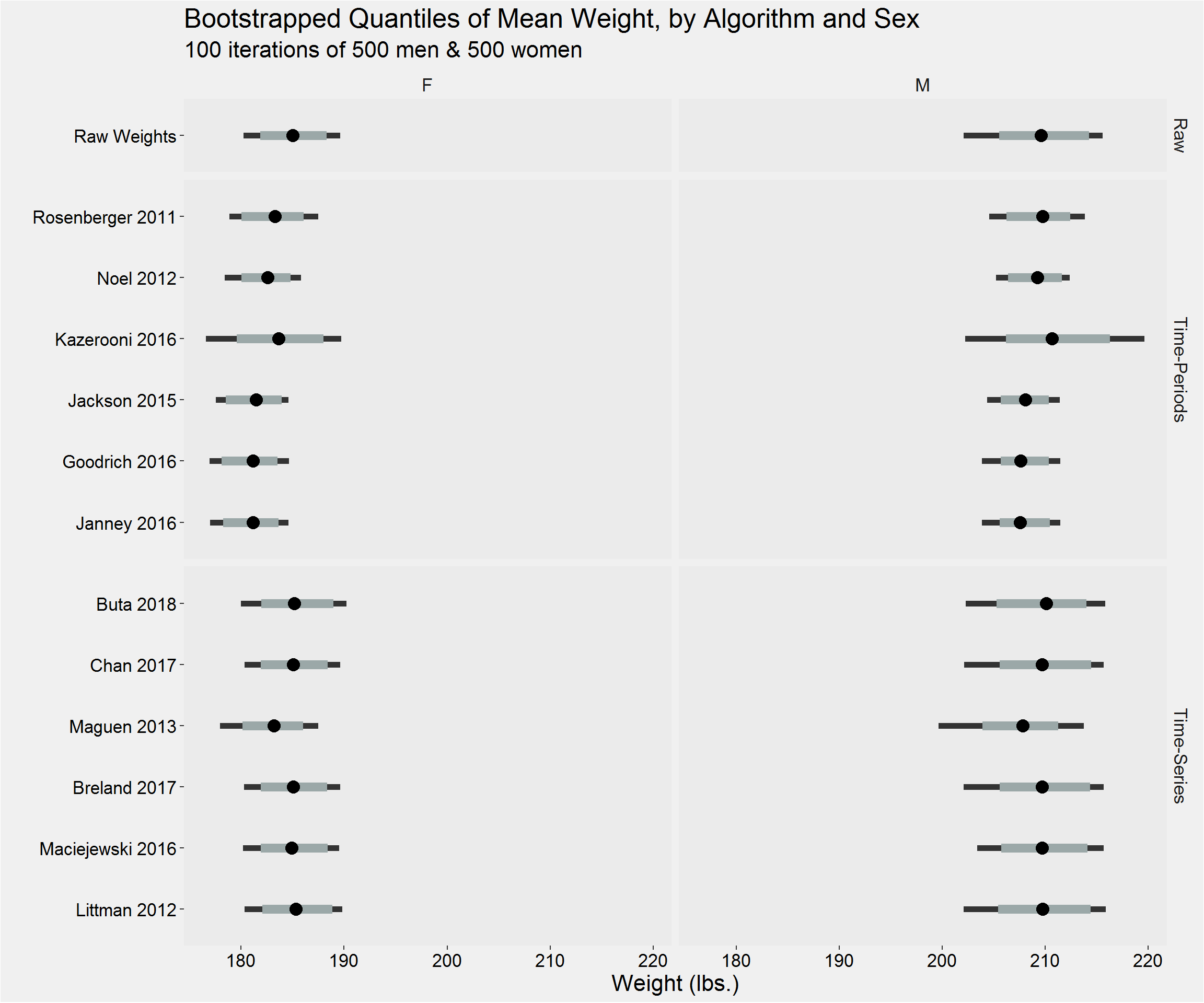
Distribution of the sample standard deviation
Alternative, ridgeline plot
Latent Trajectories
Sampling 500 men & 500 women from the PCP 2016 Cohort
library(lcmm)
# variable 'subject' must be numeric
for (i in 1:length(me)) {
if (class(me[[i]]$PatientICN) == "factor") {
me[[i]]$PatientICN <- as.integer(as.character(me[[i]]$PatientICN))
}
}
# initial values/model
B_M <- lcmm(
Weight ~ time,
random = ~time,
subject = "PatientICN",
ng = 1,
idiag = TRUE,
data = me[[1]] %>% filter(Gender == "M"),
verbose = TRUE
)
B_F <- lcmm(
Weight ~ time,
random = ~time,
subject = "PatientICN",
ng = 1,
idiag = TRUE,
data = me[[1]] %>% filter(Gender == "F"),
verbose = TRUE
)
lca.mods.M <- lapply(
me,
FUN = function(x) {
lcmm(
Weight ~ time,
random = ~time,
subject = "PatientICN",
mixture = ~time,
ng = 3,
idiag = TRUE,
data = x %>% filter(Gender == "M"),
B = B_M
)
}
)
lca.mods.F <- lapply(
me,
FUN = function(x) {
lcmm(
Weight ~ time,
random = ~time,
subject = "PatientICN",
mixture = ~time,
ng = 3,
idiag = TRUE,
data = x %>% filter(Gender == "F"),
B = B_F
)
}
)
names(lca.mods.M) <- names(me)
for (i in 1:length(lca.mods.M)) {
lca.mods.M[[i]]$data <- me[[i]] %>% filter(Gender == "M")
}
names(lca.mods.F) <- names(me)
for (i in 1:length(lca.mods.F)) {
lca.mods.F[[i]]$data <- me[[i]] %>% filter(Gender == "F")
}Predicted classes
So, there is no guarantee that each class within an algorithms LCMM, will be the same class across algorithms. We can get this information from the signs of the coefficients on “time” in each model.
concept_classes <- c("Loss", "Maintain", "Gain")
traj_edies_M <- sapply(
lca.mods.M,
function(x) {
time_coefs <- coef(x)
which_coef <- grepl("time", names(time_coefs))
time_coefs <- time_coefs[which_coef]
traj_class <- sign(round(time_coefs, 3))
traj_class
}
) %>%
as.data.frame() %>%
rownames_to_column("orig_class") %>%
pivot_longer(
!orig_class,
names_to = "Algorithm",
values_to = "value"
) %>%
mutate(
Algorithm = factor(Algorithm,
levels = names(me),
labels = names(me)),
concept_class = case_when(
value == -1 ~ concept_classes[1],
value == 0 ~ concept_classes[2],
value == 1 ~ concept_classes[3]
),
concept_class = factor(
concept_class,
levels = concept_classes,
labels = concept_classes
),
orig_class = case_when(
orig_class == "time class1" ~ "Class 1",
orig_class == "time class2" ~ "Class 2",
orig_class == "time class3" ~ "Class 3"
),
orig_class = factor(orig_class)
) %>%
select(-value) %>%
distinct(Algorithm, concept_class, .keep_all = TRUE)
traj_edies_F <- sapply(
lca.mods.F,
function(x) {
time_coefs <- coef(x)
which_coef <- grepl("time", names(time_coefs))
time_coefs <- time_coefs[which_coef]
traj_class <- sign(round(time_coefs, 3))
traj_class
}
) %>%
as.data.frame() %>%
rownames_to_column("orig_class") %>%
pivot_longer(
!orig_class,
names_to = "Algorithm",
values_to = "value"
) %>%
mutate(
Algorithm = factor(Algorithm,
levels = names(me),
labels = names(me)),
concept_class = case_when(
value == -1 ~ concept_classes[1],
value == 0 ~ concept_classes[2],
value == 1 ~ concept_classes[3]
),
concept_class = factor(
concept_class,
levels = concept_classes,
labels = concept_classes
),
orig_class = case_when(
orig_class == "time class1" ~ "Class 1",
orig_class == "time class2" ~ "Class 2",
orig_class == "time class3" ~ "Class 3"
),
orig_class = factor(orig_class)
) %>%
select(-value) %>%
distinct(Algorithm, concept_class, .keep_all = TRUE)newdata <- data.frame(time = seq(0, 365, 5))
Ypred_lcmm.M <- lapply(
lca.mods.M,
function(x) {
y <- predictY(x, newdata, var.time = "time")
cbind(y$pred, y$time)
}
) %>%
dplyr::bind_rows(.id = "Algorithm") %>%
pivot_longer(
!c(Algorithm, time),
names_to = "orig_class",
values_to = "Ypred"
) %>%
mutate(
Algorithm = factor(Algorithm,
levels = names(me),
labels = names(me)),
orig_class = factor(orig_class,
levels = c("Ypred_class1", "Ypred_class2", "Ypred_class3"),
labels = c("Class 1", "Class 2", "Class 3"))
) %>%
left_join(
traj_edies_M,
by = c("Algorithm", "orig_class")
) %>%
na.omit()
Ypred_lcmm.F <- lapply(
lca.mods.F,
function(x) {
y <- predictY(x, newdata, var.time = "time")
cbind(y$pred, y$time)
}
) %>%
dplyr::bind_rows(.id = "Algorithm") %>%
pivot_longer(
!c(Algorithm, time),
names_to = "orig_class",
values_to = "Ypred"
) %>%
mutate(
Algorithm = factor(Algorithm,
levels = names(me),
labels = names(me)),
orig_class = factor(orig_class,
levels = c("Ypred_class1", "Ypred_class2", "Ypred_class3"),
labels = c("Class 1", "Class 2", "Class 3"))
) %>%
left_join(
traj_edies_F,
by = c("Algorithm", "orig_class")
) %>%
na.omit()
Ypred_lcmm <- bind_rows(
`Males` = Ypred_lcmm.M,
`Females` = Ypred_lcmm.F,
.id = "Sex"
)
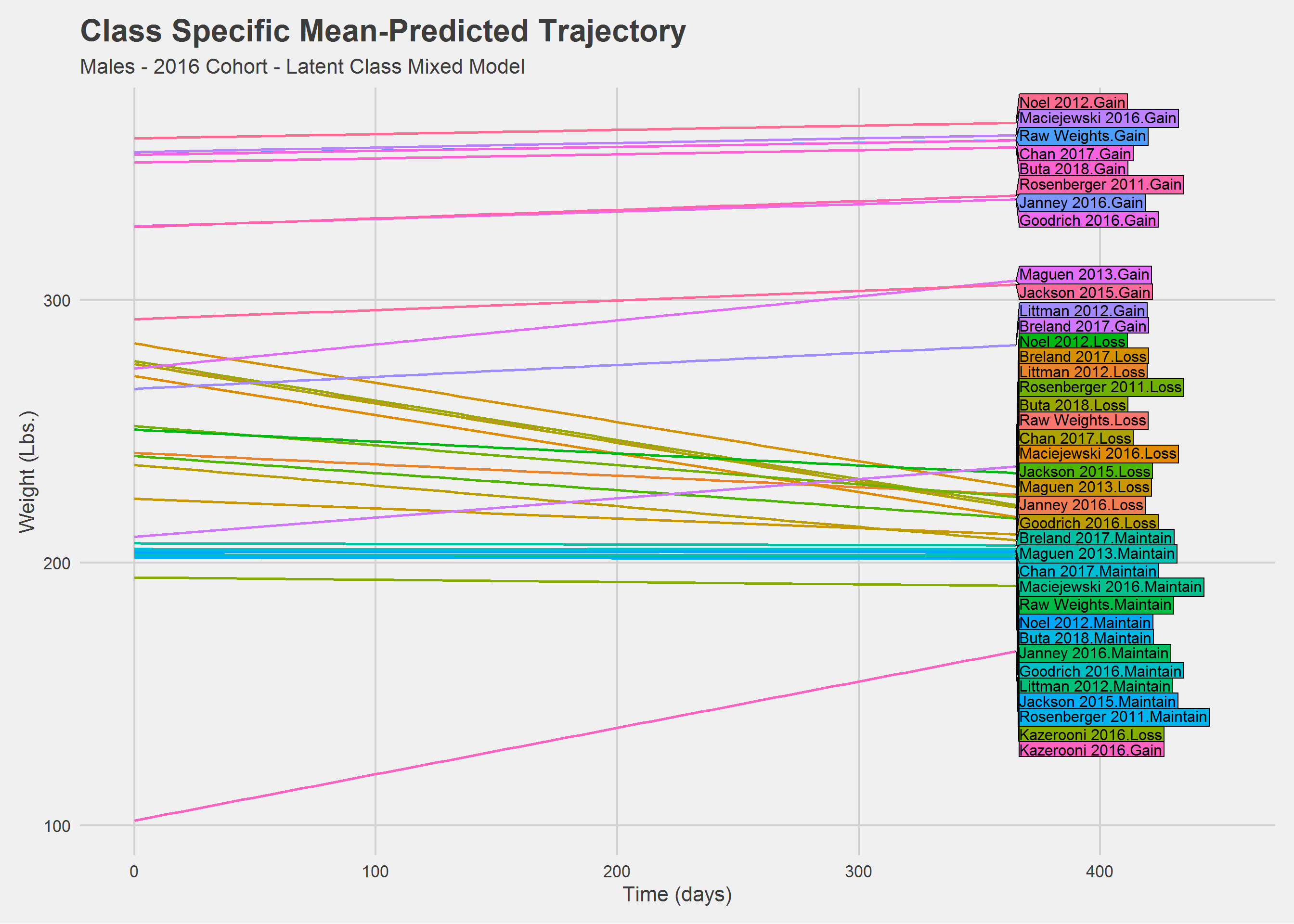
rm(Ypred_lcmm.M, Ypred_lcmm.F)Men
Ypred_lcmm %>%
filter(Sex == "Males") %>%
ggplot(
aes(
x = time, y = Ypred,
group = interaction(Algorithm, concept_class),
color = interaction(Algorithm, concept_class)
)
) %>%
add(geom_line(size = 1)) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text(), legend.position = "none")) %>%
add(xlim(0, 450)) %>%
add(directlabels::geom_dl(aes(label = interaction(Algorithm, concept_class)),
method = "last.polygons")) %>%
add(labs(
x = "Time (days)",
y = "Weight (Lbs.)",
title = "Class Specific Mean-Predicted Trajectory",
subtitle = "Males - 2016 Cohort - Latent Class Mixed Model",
color = ""
))
Women
Ypred_lcmm %>%
filter(Sex == "Females") %>%
ggplot(
aes(
x = time, y = Ypred,
group = interaction(Algorithm, concept_class),
color = interaction(Algorithm, concept_class)
)
) %>%
add(geom_line(size = 1)) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text(), legend.position = "none")) %>%
add(xlim(0, 450)) %>%
add(directlabels::geom_dl(aes(label = interaction(Algorithm, concept_class)),
method = "last.polygons")) %>%
add(labs(
x = "Time (days)",
y = "Weight (Lbs.)",
title = "Class Specific Mean-Predicted Trajectory",
subtitle = "Females - 2016 Cohort - Latent Class Mixed Model",
color = ""
))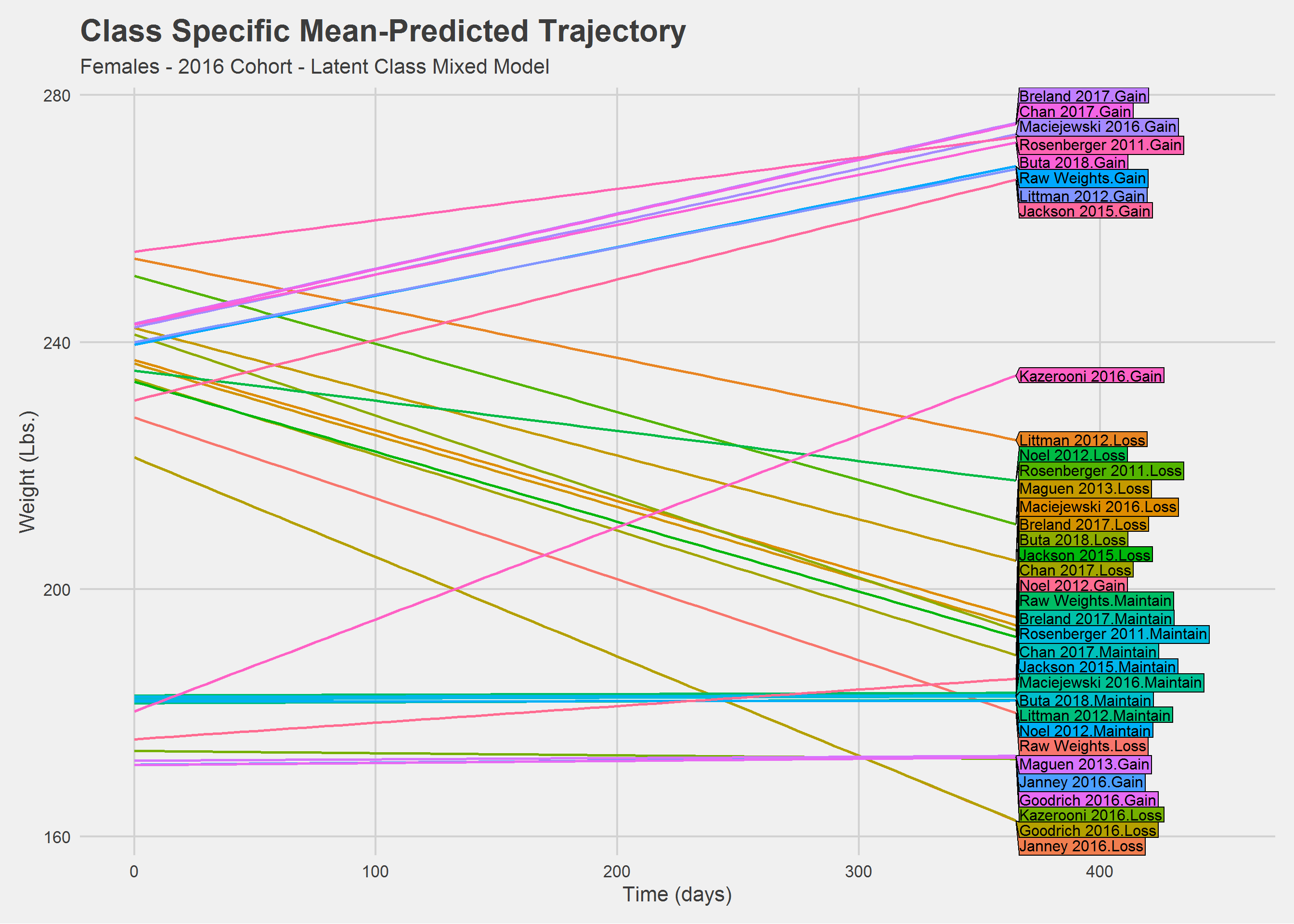
Still a mess. Let’s look at things a little differently.
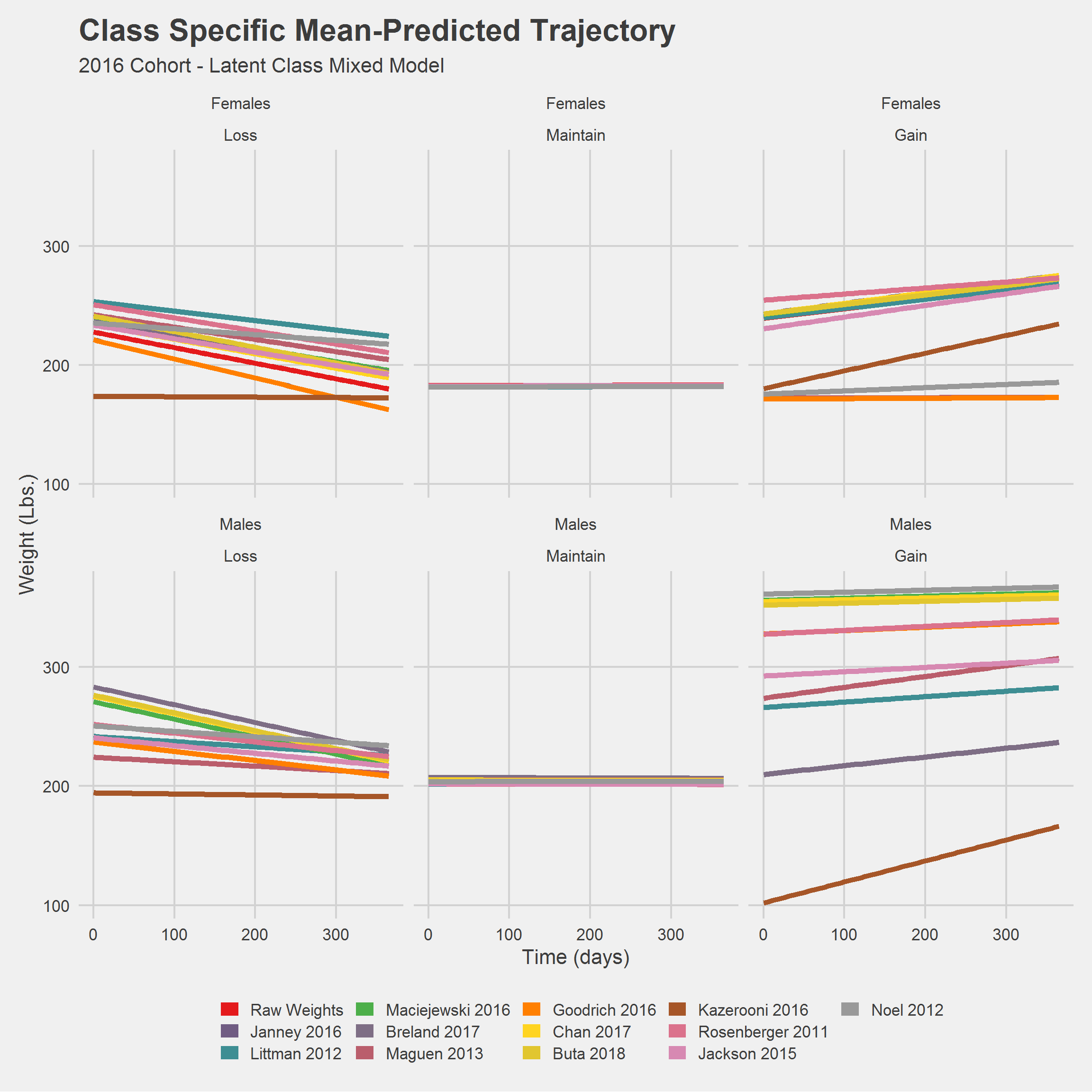
An Illustration:
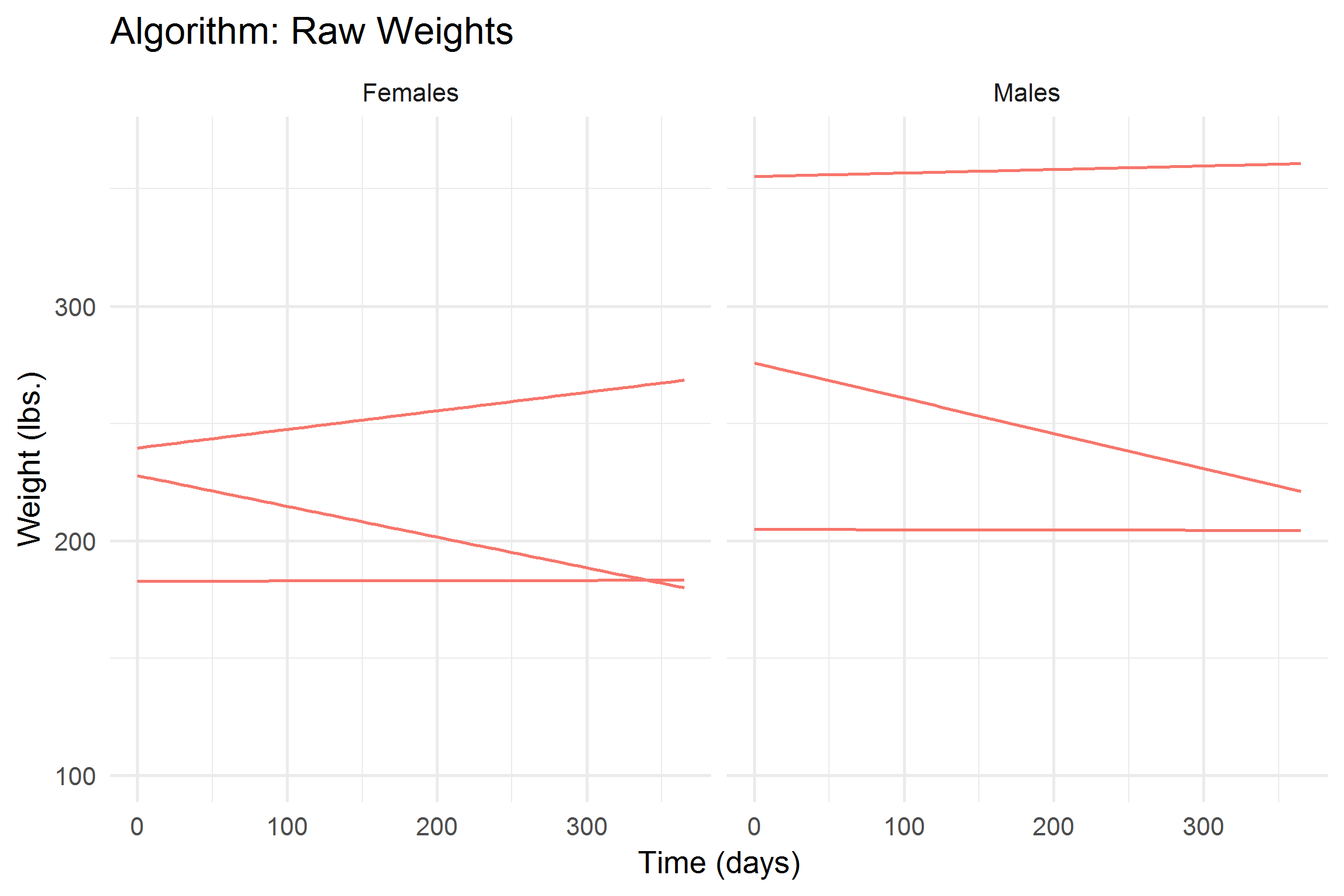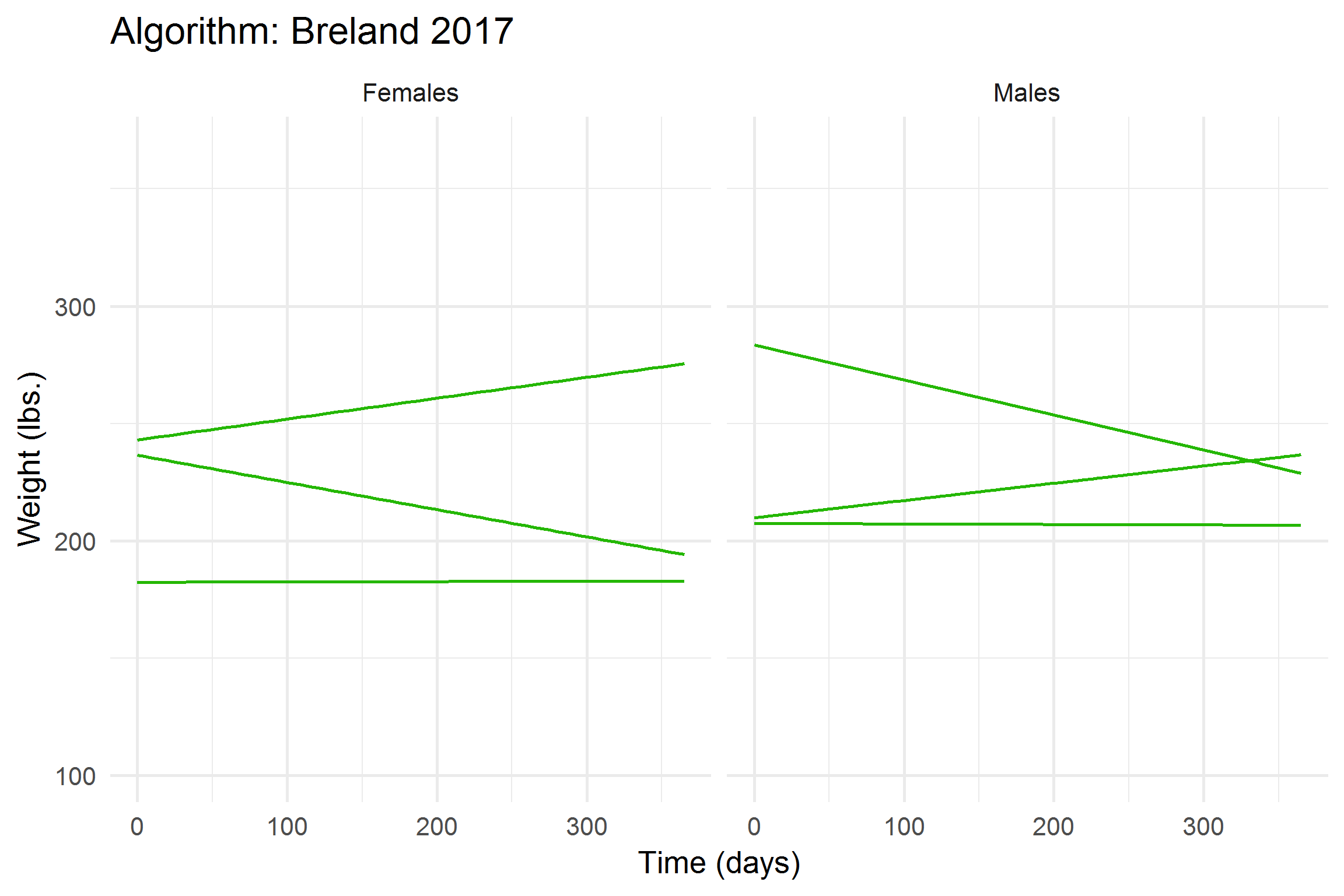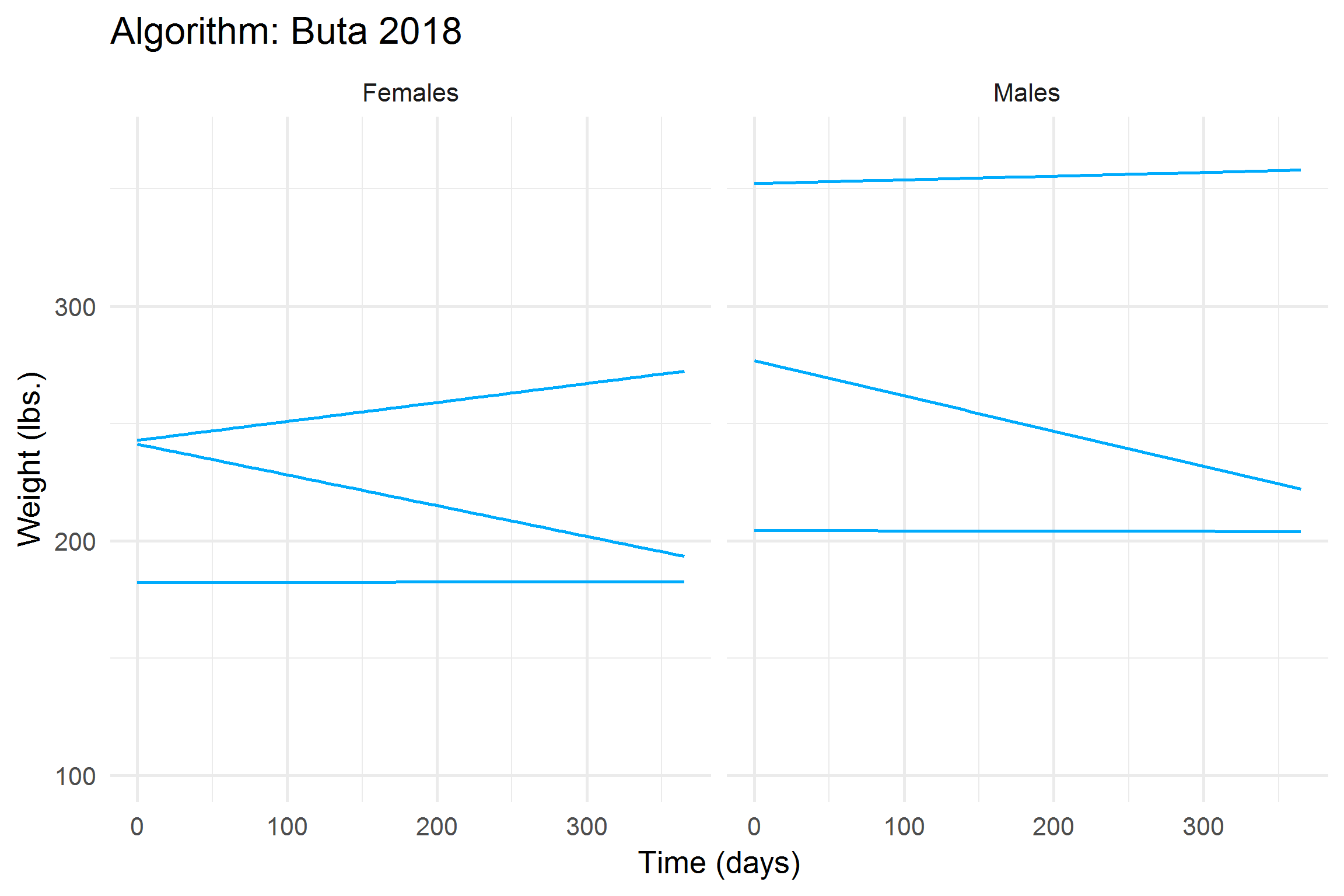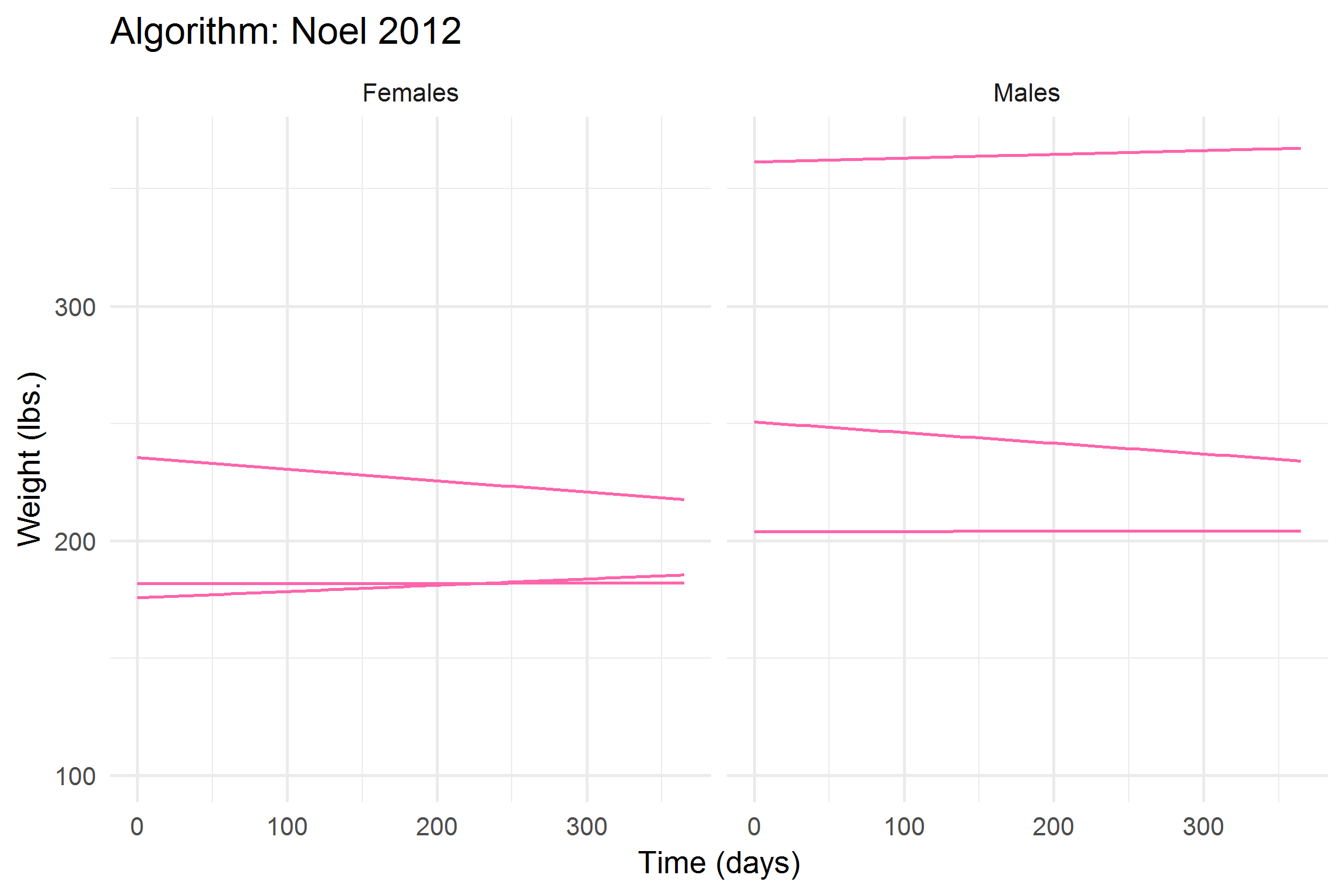
library(gganimate)
Ypred_lcmm %>%
ggplot(
aes(
x = time,
y = Ypred,
color = Algorithm,
group = interaction(Algorithm, concept_class)
)
) %>%
add(geom_line(size = 1)) %>%
add(facet_wrap(vars(Sex))) %>%
add(theme_minimal(20)) %>%
add(theme(legend.position = "none")) %>%
add(labs(
title = 'Algorithm: {closest_state}',
x = "Time (days)",
y = "Weight (lbs.)"
)) %>%
add(transition_states(
states = Algorithm,
transition_length = 1,
state_length = 2
))
Comparing slopes, numerically
slopes.M <- sapply(
lca.mods.M,
function(x) {
time_coefs <- coef(x)
which_coef <- grepl("time", names(time_coefs))
time_coefs <- time_coefs[which_coef]
time_coefs <- round(time_coefs, 5)
time_coefs
}
) %>%
reshape2::melt() %>%
mutate(
orig_class = case_when(
Var1 == "time class1" ~ "Class 1",
Var1 == "time class2" ~ "Class 2",
Var1 == "time class3" ~ "Class 3"
),
orig_class = factor(orig_class)
) %>%
select(-Var1) %>%
rename(Algorithm = Var2, Slope = value) %>%
full_join(traj_edies_M, by = c("Algorithm", "orig_class"))
slopes.F <- sapply(
lca.mods.F,
function(x) {
time_coefs <- coef(x)
which_coef <- grepl("time", names(time_coefs))
time_coefs <- time_coefs[which_coef]
time_coefs <- round(time_coefs, 5)
time_coefs
}
) %>%
reshape2::melt() %>%
mutate(
orig_class = case_when(
Var1 == "time class1" ~ "Class 1",
Var1 == "time class2" ~ "Class 2",
Var1 == "time class3" ~ "Class 3"
),
orig_class = factor(orig_class)
) %>%
select(-Var1) %>%
rename(Algorithm = Var2, Slope = value) %>%
full_join(traj_edies_F, by = c("Algorithm", "orig_class"))
slopes <- bind_rows(
`Males` = slopes.M,
`Females` = slopes.F,
.id = "Sex"
) %>%
na.omit()
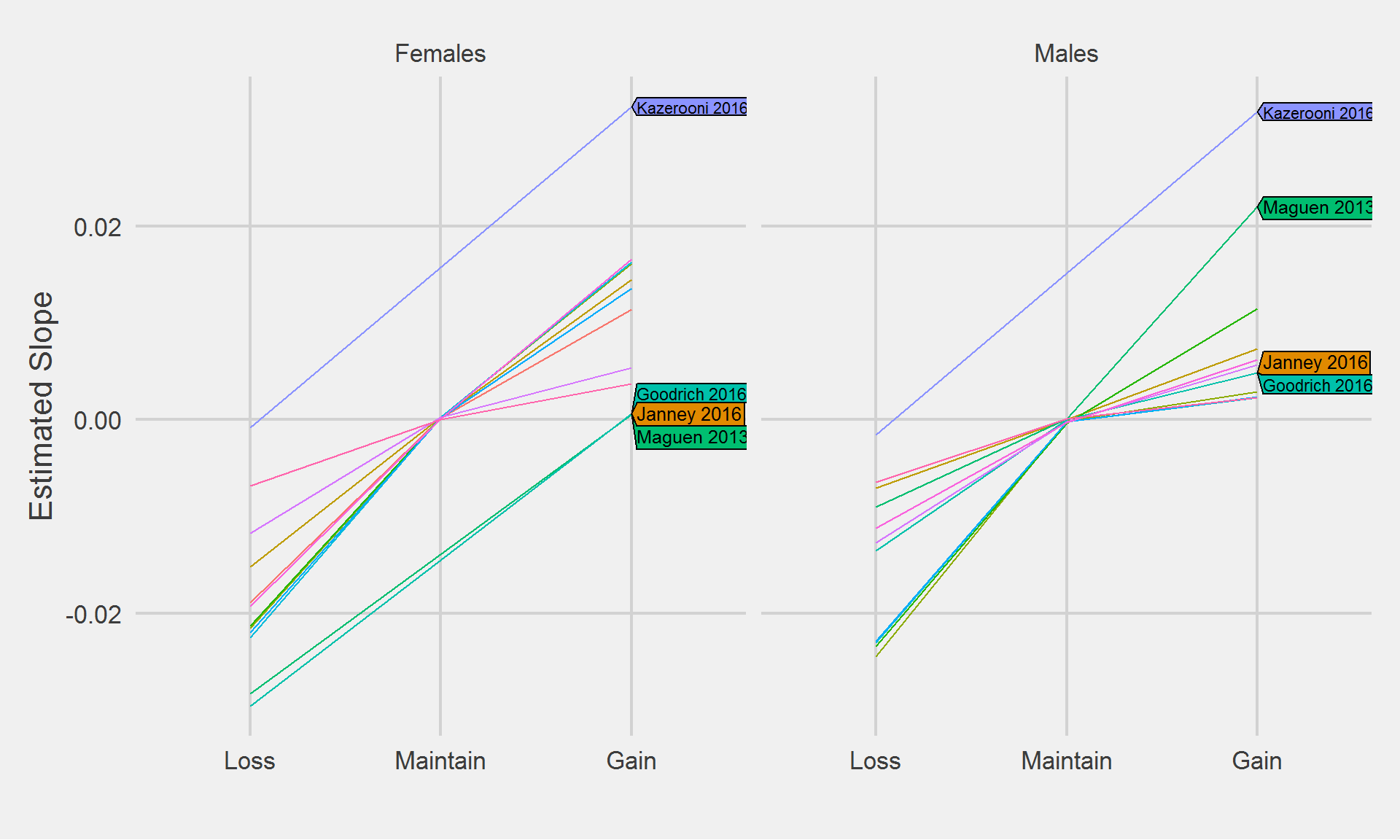
rm(slopes.M, slopes.F)algos <- c("Janney 2016", "Goodrich 2016", "Maguen 2013", "Kazerooni 2016")
slopes %>%
ggplot(aes(
x = concept_class,
y = Slope,
group = Algorithm,
color = Algorithm
)) %>%
add(geom_line()) %>%
add(facet_wrap(vars(Sex))) %>%
add(directlabels::geom_dl(
data = slopes %>%
filter(Algorithm %in% algos),
aes(label = Algorithm),
method = list("last.polygons")
)) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(legend.position = "none", axis.title = element_text())) %>%
add(labs(
x = "",
y = "Estimated Slope"
))
Weight Change Comparison
Add Gender/Sex
DF <- DF %>%
filter(Cohort == "PCP 2016") %>%
left_join(
weight.ls[["PCP2016"]] %>%
distinct(PatientICN, Gender),
by = "PatientICN"
)Individual Level
Let’s look at the proportions of weight loss \(\geq5\)% from baseline, by algorithm:
tmp <- DF %>%
count(Algorithm, Gender, gt5perc_weightloss) %>%
group_by(Algorithm, Gender) %>%
mutate(prop = prop.table(n)) %>%
filter(gt5perc_weightloss == 1) %>%
select(-gt5perc_weightloss)
tableStyle(tmp)| Algorithm | Gender | n | prop |
|---|---|---|---|
| Raw Weights | F | 535 | 0.15 |
| Raw Weights | M | 7627 | 0.13 |
| Janney 2016 | F | 509 | 0.15 |
| Janney 2016 | M | 7333 | 0.13 |
| Littman 2012 | F | 502 | 0.15 |
| Littman 2012 | M | 7349 | 0.13 |
| Maciejewski 2016 | F | 514 | 0.15 |
| Maciejewski 2016 | M | 7471 | 0.14 |
| Breland 2017 | F | 529 | 0.15 |
| Breland 2017 | M | 7595 | 0.13 |
| Maguen 2013 | F | 355 | 0.12 |
| Maguen 2013 | M | 4578 | 0.09 |
| Goodrich 2016 | F | 508 | 0.15 |
| Goodrich 2016 | M | 7320 | 0.13 |
| Chan 2017 | F | 523 | 0.15 |
| Chan 2017 | M | 7546 | 0.13 |
| Jackson 2015 | F | 503 | 0.15 |
| Jackson 2015 | M | 7470 | 0.13 |
| Buta 2018 | F | 515 | 0.15 |
| Buta 2018 | M | 7247 | 0.14 |
| Kazerooni 2016 | F | 228 | 0.13 |
| Kazerooni 2016 | M | 3127 | 0.14 |
| Noel 2012 | F | 515 | 0.15 |
| Noel 2012 | M | 7271 | 0.13 |
| Rosenberger 2011 | F | 356 | 0.16 |
| Rosenberger 2011 | M | 5069 | 0.14 |
Testing/modeling
test_prop <- mgcv::gam(
prop ~ Gender + s(Algorithm, bs = 're'),
data = tmp,
family = binomial,
method = 'REML'
)
summary(test_prop)
Family: binomial
Link function: logit
Formula:
prop ~ Gender + s(Algorithm, bs = "re")
Parametric coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -1.7528 0.7817 -2.242 0.0249 *
GenderM -0.1380 1.1340 -0.122 0.9032
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Approximate significance of smooth terms:
edf Ref.df Chi.sq p-value
s(Algorithm) 7.222e-06 12 0 1
R-sq.(adj) = 0.333 Deviance explained = 34.3%
-REML = 2.5266 Scale est. = 1 n = 26When factoring in Algorithm, the differences in weight loss proportion between sexes is insignificant
plot(sm(getViz(test_prop), 1)) %>%
add(l_fitLine(color = "red")) %>%
add(l_ciLine(mul = 5, color = "blue", linetype = 2)) %>%
add(l_points())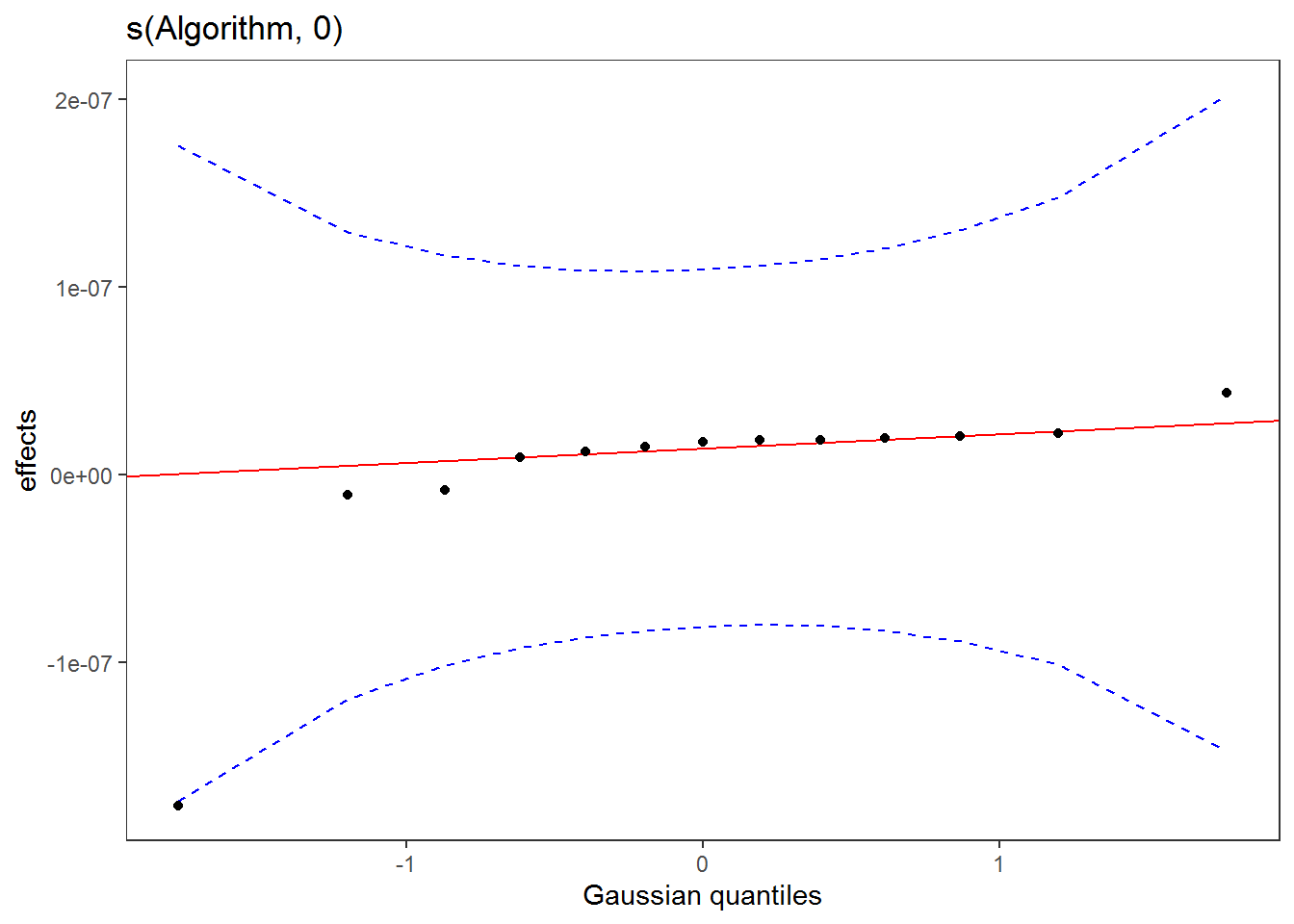
Let’s look at the proportions of weight gain \(\geq5\)% from baseline, by algorithm:
tmp <- DF %>%
count(Algorithm, Gender, gt5perc_weightgain) %>%
group_by(Algorithm, Gender) %>%
mutate(prop = prop.table(n)) %>%
filter(gt5perc_weightgain == 1) %>%
select(-gt5perc_weightgain)
tableStyle(tmp)| Algorithm | Gender | n | prop |
|---|---|---|---|
| Raw Weights | F | 626 | 0.18 |
| Raw Weights | M | 6351 | 0.11 |
| Janney 2016 | F | 590 | 0.18 |
| Janney 2016 | M | 6089 | 0.11 |
| Littman 2012 | F | 603 | 0.17 |
| Littman 2012 | M | 6184 | 0.11 |
| Maciejewski 2016 | F | 620 | 0.18 |
| Maciejewski 2016 | M | 6190 | 0.11 |
| Breland 2017 | F | 624 | 0.18 |
| Breland 2017 | M | 6312 | 0.11 |
| Maguen 2013 | F | 425 | 0.14 |
| Maguen 2013 | M | 3663 | 0.07 |
| Goodrich 2016 | F | 590 | 0.18 |
| Goodrich 2016 | M | 6078 | 0.11 |
| Chan 2017 | F | 624 | 0.18 |
| Chan 2017 | M | 6278 | 0.11 |
| Jackson 2015 | F | 574 | 0.18 |
| Jackson 2015 | M | 5920 | 0.10 |
| Buta 2018 | F | 606 | 0.18 |
| Buta 2018 | M | 6036 | 0.11 |
| Kazerooni 2016 | F | 247 | 0.15 |
| Kazerooni 2016 | M | 2256 | 0.10 |
| Noel 2012 | F | 615 | 0.18 |
| Noel 2012 | M | 6009 | 0.11 |
| Rosenberger 2011 | F | 426 | 0.19 |
| Rosenberger 2011 | M | 4299 | 0.12 |
Average weight change:
tmp <- DF %>%
filter(!is.na(diff)) %>%
select(Algorithm, Gender, diff) %>%
group_by(Algorithm, Gender) %>%
summarize(
mean = mean(diff),
SD = sd(diff),
min = min(diff),
Q1 = fivenum(diff)[2],
median = median(diff),
Q3 = fivenum(diff)[4],
max = max(diff)
)
tableStyle(tmp)| Algorithm | Gender | mean | SD | min | Q1 | median | Q3 | max |
|---|---|---|---|---|---|---|---|---|
| Raw Weights | F | -0.21 | 22.35 | -1000.00 | -5.20 | 0.56 | 6.40 | 185.00 |
| Raw Weights | M | -0.65 | 13.27 | -536.00 | -5.70 | -0.20 | 5.00 | 296.10 |
| Janney 2016 | F | 0.07 | 13.84 | -258.99 | -5.25 | 0.59 | 6.30 | 98.40 |
| Janney 2016 | M | -0.65 | 11.91 | -291.40 | -5.70 | -0.20 | 5.00 | 281.97 |
| Littman 2012 | F | 0.45 | 11.36 | -65.70 | -5.00 | 0.60 | 6.30 | 88.90 |
| Littman 2012 | M | -0.55 | 10.72 | -118.60 | -5.60 | -0.20 | 5.00 | 108.00 |
| Maciejewski 2016 | F | 0.37 | 12.03 | -95.30 | -5.40 | 0.70 | 6.70 | 66.40 |
| Maciejewski 2016 | M | -0.67 | 11.21 | -117.20 | -5.90 | -0.20 | 5.00 | 195.80 |
| Breland 2017 | F | 0.20 | 13.07 | -258.99 | -5.20 | 0.60 | 6.40 | 88.90 |
| Breland 2017 | M | -0.65 | 11.44 | -117.20 | -5.70 | -0.20 | 5.00 | 207.20 |
| Maguen 2013 | F | 0.45 | 8.38 | -42.00 | -4.30 | 0.51 | 5.40 | 41.20 |
| Maguen 2013 | M | -0.42 | 7.74 | -73.20 | -4.90 | -0.20 | 4.00 | 98.40 |
| Goodrich 2016 | F | 0.21 | 12.57 | -95.30 | -5.20 | 0.59 | 6.30 | 98.40 |
| Goodrich 2016 | M | -0.65 | 11.44 | -117.20 | -5.70 | -0.20 | 5.00 | 205.20 |
| Chan 2017 | F | 0.27 | 12.85 | -258.99 | -5.10 | 0.60 | 6.40 | 98.40 |
| Chan 2017 | M | -0.63 | 11.89 | -509.50 | -5.70 | -0.20 | 5.00 | 278.27 |
| Jackson 2015 | F | 0.21 | 12.26 | -133.50 | -5.30 | 0.78 | 6.30 | 98.40 |
| Jackson 2015 | M | -0.76 | 11.25 | -245.59 | -5.80 | -0.40 | 4.70 | 230.39 |
| Buta 2018 | F | 0.16 | 13.32 | -211.59 | -5.20 | 0.60 | 6.40 | 98.40 |
| Buta 2018 | M | -0.64 | 11.92 | -245.37 | -5.80 | -0.20 | 5.00 | 278.27 |
| Kazerooni 2016 | F | 0.48 | 10.63 | -84.00 | -4.30 | 0.80 | 6.00 | 92.90 |
| Kazerooni 2016 | M | -1.06 | 12.56 | -534.00 | -6.20 | -0.70 | 4.60 | 299.30 |
| Noel 2012 | F | 0.25 | 12.87 | -211.79 | -5.25 | 0.65 | 6.53 | 98.40 |
| Noel 2012 | M | -0.63 | 11.39 | -245.59 | -5.70 | -0.20 | 5.00 | 195.80 |
| Rosenberger 2011 | F | 0.03 | 25.07 | -1002.40 | -5.55 | 0.80 | 7.00 | 185.00 |
| Rosenberger 2011 | M | -0.72 | 13.17 | -538.00 | -6.00 | -0.30 | 5.00 | 298.70 |
Testing/modeling
test_mean <- mgcv::gam(
mean ~ Gender + s(Algorithm, bs = 're'),
data = tmp,
family = gaussian,
method = 'REML'
)
summary(test_mean)
Family: gaussian
Link function: identity
Formula:
mean ~ Gender + s(Algorithm, bs = "re")
Parametric coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.22586 0.04709 4.796 6.98e-05 ***
GenderM -0.89325 0.06660 -13.412 1.21e-12 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Approximate significance of smooth terms:
edf Ref.df F p-value
s(Algorithm) 4.182e-05 12 0 0.527
R-sq.(adj) = 0.877 Deviance explained = 88.2%
-REML = -5.9358 Scale est. = 0.028832 n = 26Obvious differences between sexes, but little to no difference between algorithm choice
Average Weight Change for those losing \(\ge 5\)% weight loss:
DF %>%
filter(!is.na(diff) & gt5perc_weightloss == 1) %>%
select(Algorithm, Gender, diff) %>%
group_by(Algorithm, Gender) %>%
summarize(
mean = mean(diff),
SD = sd(diff),
min = min(diff),
Q1 = fivenum(diff)[2],
median = median(diff),
Q3 = fivenum(diff)[4],
max = max(diff)
) %>%
tableStyle()| Algorithm | Gender | mean | SD | min | Q1 | median | Q3 | max |
|---|---|---|---|---|---|---|---|---|
| Raw Weights | F | -22.00 | 47.05 | -1000.00 | -20.80 | -15.00 | -11.10 | -5.90 |
| Raw Weights | M | -19.79 | 16.90 | -536.00 | -22.20 | -16.00 | -12.10 | -5.00 |
| Janney 2016 | F | -19.61 | 18.64 | -258.99 | -20.40 | -15.00 | -11.10 | -5.90 |
| Janney 2016 | M | -19.24 | 12.37 | -291.40 | -22.00 | -15.90 | -12.00 | -5.55 |
| Littman 2012 | F | -17.18 | 9.42 | -65.70 | -19.20 | -14.60 | -11.00 | -5.90 |
| Littman 2012 | M | -18.17 | 9.33 | -118.60 | -21.30 | -15.60 | -12.00 | -5.00 |
| Maciejewski 2016 | F | -17.96 | 11.48 | -95.30 | -20.00 | -14.75 | -11.00 | -5.90 |
| Maciejewski 2016 | M | -18.79 | 10.52 | -117.20 | -22.00 | -15.80 | -12.00 | -5.00 |
| Breland 2017 | F | -18.98 | 16.13 | -258.99 | -20.10 | -15.00 | -11.00 | -5.90 |
| Breland 2017 | M | -19.04 | 10.93 | -117.20 | -22.00 | -15.90 | -12.04 | -5.00 |
| Maguen 2013 | F | -13.79 | 5.45 | -42.00 | -16.02 | -12.80 | -10.08 | -5.90 |
| Maguen 2013 | M | -14.65 | 5.91 | -73.20 | -16.40 | -13.20 | -11.00 | -5.00 |
| Goodrich 2016 | F | -18.73 | 12.72 | -95.30 | -20.30 | -15.00 | -11.05 | -5.90 |
| Goodrich 2016 | M | -19.04 | 11.11 | -117.20 | -22.00 | -15.80 | -12.00 | -5.00 |
| Chan 2017 | F | -18.58 | 15.72 | -258.99 | -20.00 | -14.90 | -11.00 | -5.90 |
| Chan 2017 | M | -19.13 | 12.88 | -509.50 | -22.00 | -15.80 | -12.00 | -5.00 |
| Jackson 2015 | F | -18.13 | 12.38 | -133.50 | -19.95 | -14.61 | -11.20 | -5.90 |
| Jackson 2015 | M | -18.69 | 11.22 | -245.59 | -21.65 | -15.40 | -12.00 | -5.00 |
| Buta 2018 | F | -19.46 | 15.96 | -211.59 | -20.50 | -15.00 | -11.05 | -5.90 |
| Buta 2018 | M | -19.31 | 12.02 | -245.37 | -22.25 | -16.00 | -12.10 | -5.00 |
| Kazerooni 2016 | F | -16.46 | 9.34 | -84.00 | -18.30 | -14.20 | -11.00 | -4.80 |
| Kazerooni 2016 | M | -19.01 | 14.85 | -534.00 | -22.00 | -15.50 | -12.00 | -6.00 |
| Noel 2012 | F | -18.69 | 15.55 | -211.79 | -20.00 | -14.65 | -11.00 | -5.90 |
| Noel 2012 | M | -18.89 | 11.29 | -245.59 | -22.00 | -15.60 | -12.10 | -4.40 |
| Rosenberger 2011 | F | -21.57 | 53.54 | -1002.40 | -21.70 | -15.30 | -11.25 | -5.90 |
| Rosenberger 2011 | M | -20.01 | 17.14 | -538.00 | -22.80 | -16.00 | -12.10 | -5.00 |
Average Weight Change for those gaining \(\ge 5\)% weight loss:
DF %>%
filter(!is.na(diff) & gt5perc_weightgain == 1) %>%
select(Algorithm, Gender, diff) %>%
group_by(Algorithm, Gender) %>%
summarize(
mean = mean(diff),
SD = sd(diff),
min = min(diff),
Q1 = fivenum(diff)[2],
median = median(diff),
Q3 = fivenum(diff)[4],
max = max(diff)
) %>%
tableStyle()| Algorithm | Gender | mean | SD | min | Q1 | median | Q3 | max |
|---|---|---|---|---|---|---|---|---|
| Raw Weights | F | 16.76 | 11.59 | 5.00 | 10.90 | 13.80 | 19.00 | 185.00 |
| Raw Weights | M | 18.51 | 15.44 | 5.00 | 11.80 | 15.00 | 20.45 | 296.10 |
| Janney 2016 | F | 16.48 | 9.55 | 5.00 | 10.80 | 13.80 | 19.00 | 98.40 |
| Janney 2016 | M | 17.90 | 12.03 | 5.00 | 11.80 | 15.00 | 20.30 | 281.97 |
| Littman 2012 | F | 16.00 | 8.53 | 5.00 | 10.84 | 13.60 | 18.58 | 88.90 |
| Littman 2012 | M | 17.11 | 8.46 | 5.20 | 11.70 | 14.90 | 20.00 | 108.00 |
| Maciejewski 2016 | F | 16.09 | 8.08 | 5.00 | 10.90 | 13.80 | 18.90 | 66.40 |
| Maciejewski 2016 | M | 17.21 | 8.88 | 5.20 | 11.80 | 14.95 | 20.00 | 195.80 |
| Breland 2017 | F | 16.36 | 8.85 | 5.00 | 10.90 | 13.80 | 19.00 | 88.90 |
| Breland 2017 | M | 17.64 | 10.20 | 5.00 | 11.80 | 15.00 | 20.20 | 207.20 |
| Maguen 2013 | F | 13.24 | 5.12 | 5.00 | 10.00 | 12.00 | 15.00 | 41.20 |
| Maguen 2013 | M | 13.74 | 4.95 | 5.20 | 10.70 | 12.80 | 15.70 | 98.40 |
| Goodrich 2016 | F | 16.48 | 9.55 | 5.00 | 10.80 | 13.80 | 19.00 | 98.40 |
| Goodrich 2016 | M | 17.65 | 10.12 | 5.00 | 11.80 | 15.00 | 20.20 | 205.20 |
| Chan 2017 | F | 16.26 | 8.80 | 5.00 | 10.90 | 13.80 | 19.00 | 98.40 |
| Chan 2017 | M | 17.79 | 11.76 | 5.00 | 11.80 | 15.00 | 20.20 | 278.27 |
| Jackson 2015 | F | 16.00 | 9.26 | 4.90 | 10.33 | 13.50 | 18.50 | 98.40 |
| Jackson 2015 | M | 17.46 | 10.48 | 5.74 | 11.70 | 14.80 | 20.00 | 230.39 |
| Buta 2018 | F | 16.57 | 9.54 | 5.00 | 10.90 | 13.80 | 19.00 | 98.40 |
| Buta 2018 | M | 17.97 | 11.84 | 5.00 | 11.80 | 15.00 | 20.40 | 278.27 |
| Kazerooni 2016 | F | 15.95 | 8.89 | 6.20 | 10.75 | 13.50 | 18.90 | 92.90 |
| Kazerooni 2016 | M | 18.06 | 14.85 | 5.80 | 11.60 | 14.80 | 19.60 | 299.30 |
| Noel 2012 | F | 16.00 | 8.62 | 5.00 | 10.62 | 13.60 | 18.23 | 98.40 |
| Noel 2012 | M | 17.55 | 10.33 | 5.45 | 11.80 | 15.00 | 20.00 | 195.80 |
| Rosenberger 2011 | F | 17.26 | 12.72 | 5.00 | 11.00 | 14.00 | 19.00 | 185.00 |
| Rosenberger 2011 | M | 18.11 | 12.47 | 5.20 | 12.00 | 15.00 | 20.50 | 298.70 |
Visual summaries
p <- DF %>%
filter(!is.na(diff)) %>%
ggplot(aes(x = diff, y = Algorithm, fill = Algorithm)) %>%
add(ggridges::geom_density_ridges(
jittered_points = TRUE,
point_shape = "|",
position = ggridges::position_points_jitter(height = 0),
scale = 3,
size = 0.25,
rel_min_height = 10^-5
)) %>%
add(facet_grid(Type ~ Gender, scales = "free_y", space = "free_y")) %>%
add(scale_fill_viridis_d(option = "A")) %>%
add(theme(
legend.position = "none",
text = element_text(size = 20),
axis.text = element_text(color = "black"),
panel.grid = element_blank(),
strip.background = element_blank()
)) %>%
add(labs(
x = "Weight Change (Lbs.)",
y = "",
title = "Weight Change 1-year Post Baseline"
))
p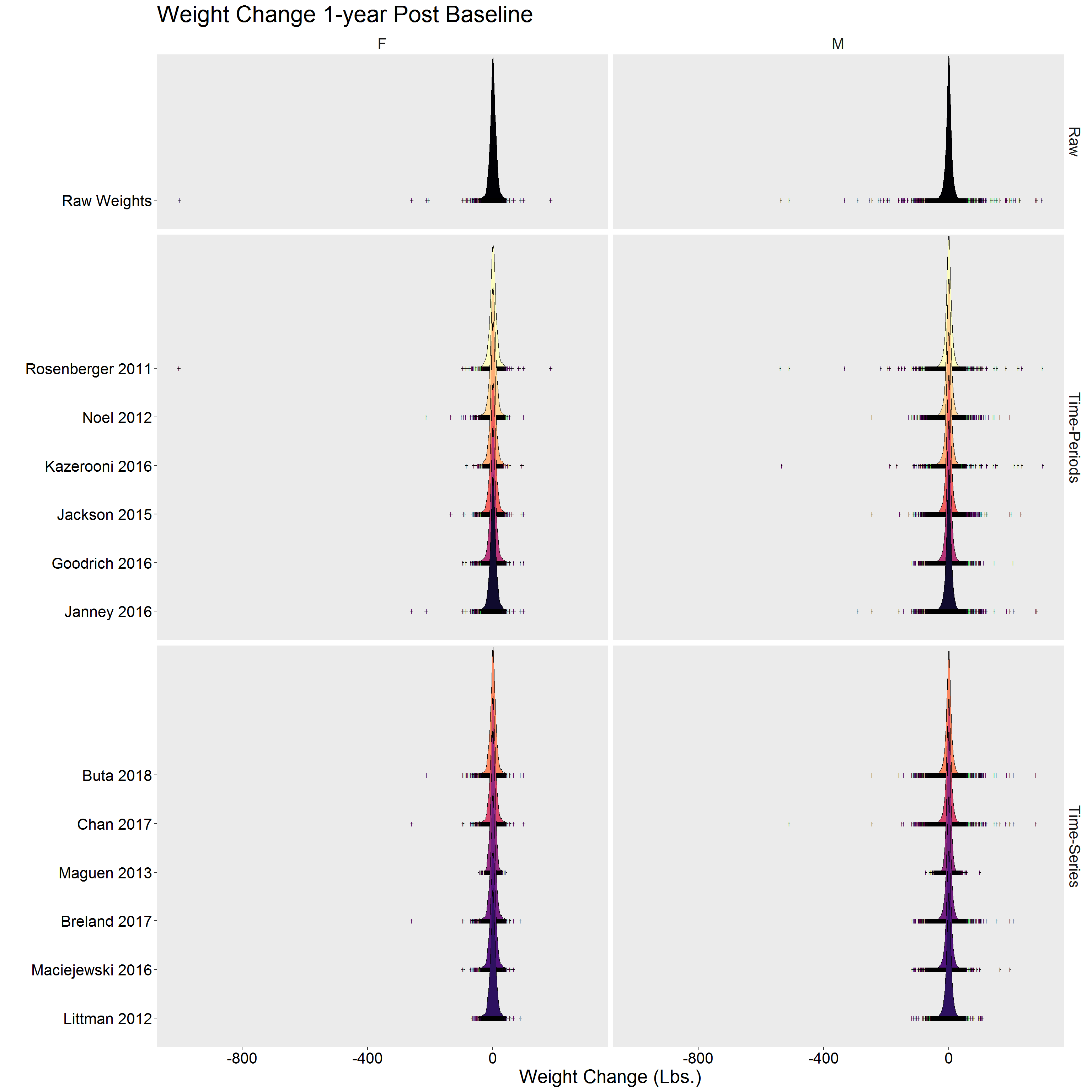
Alternative view
p <- DF %>%
filter(!is.na(diff)) %>%
group_by(Algorithm) %>%
mutate(dens = approxfun(density(diff))(diff)) %>%
ggplot(aes(y = Algorithm, x = diff, color = dens)) %>%
add(geom_jitter(size = 2)) %>%
add(scale_color_viridis_c()) %>%
add(facet_grid(Type ~ Gender, scales = "free_y", space = "free_y")) %>%
add(theme(
legend.position = "none",
text = element_text(size = 20),
axis.text = element_text(color = "black"),
panel.grid = element_blank(),
strip.background = element_blank()
)) %>%
add(labs(
x = "Weight Change (Lbs.)",
y = "",
title = ""
))
p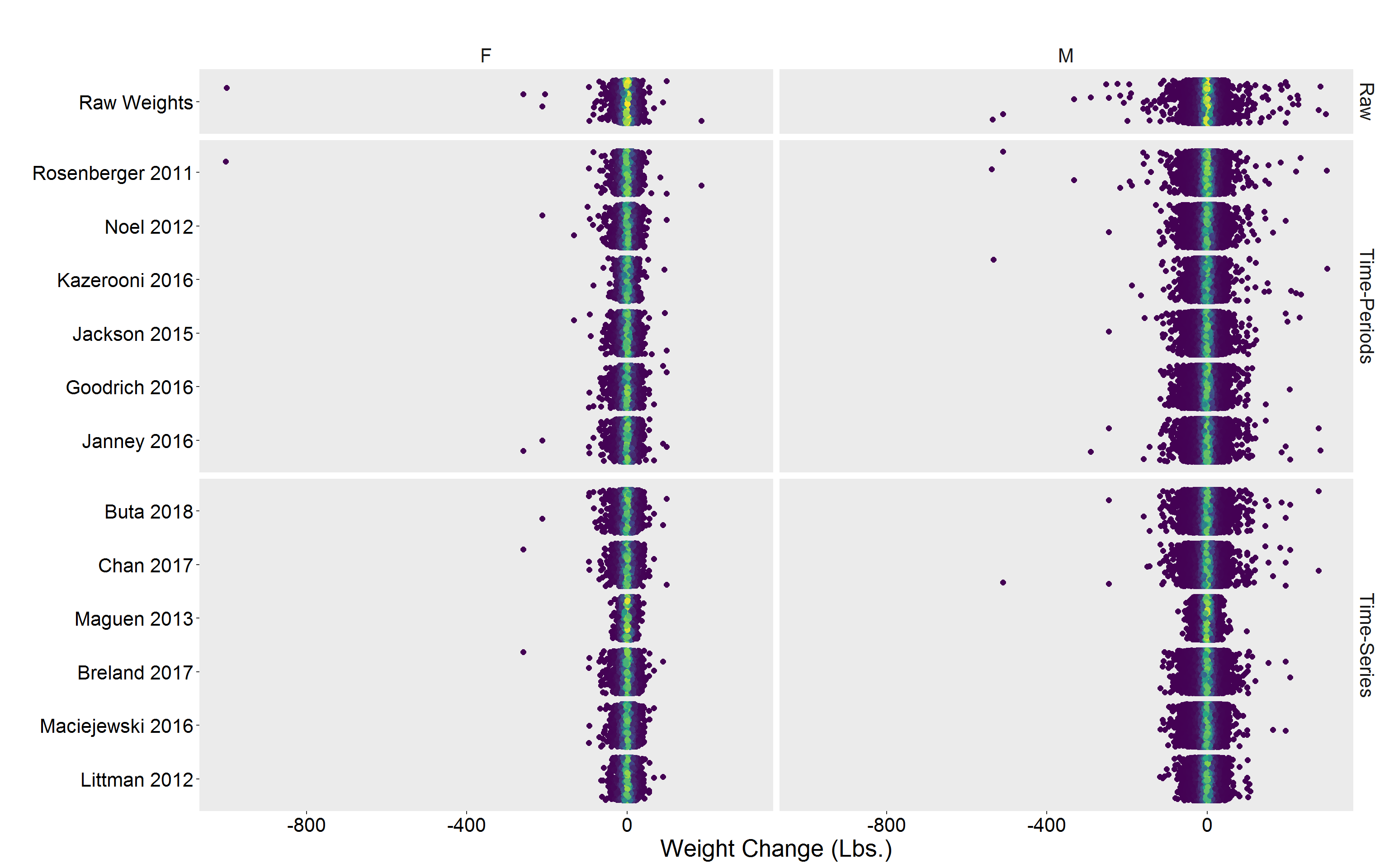
Site/Facility Level Analyses
Sta3n_wtdx_diff <- DF %>%
select(Algorithm, Gender, Sta3n, diff, diff_perc) %>%
reshape2::melt(id.vars = c("Gender", "Algorithm", "Sta3n")) %>%
group_by(Gender, Algorithm, Sta3n, variable) %>%
na.omit() %>%
summarize(
n = n(),
mean = mean(value),
median = median(value)
) %>%
pivot_wider(
names_from = variable,
values_from = c("n", "mean", "median")
)
Sta3n_wtpct <- DF %>%
select(Gender, Algorithm, Sta3n, contains("weight")) %>%
reshape2::melt(id.vars = c("Gender", "Algorithm", "Sta3n")) %>%
group_by(Gender, Algorithm, Sta3n, variable) %>%
summarize(prop = sum(value == 1) / n()) %>%
pivot_wider(names_from = variable, values_from = "prop")
Sta3n_wtdx <- Sta3n_wtdx_diff %>%
left_join(Sta3n_wtpct, by = c("Gender", "Algorithm", "Sta3n")) %>%
left_join(
DF %>%
distinct(Algorithm, Type),
by = "Algorithm"
) %>%
select(
Type,
Gender,
Sta3n,
n_diff,
mean_diff,
median_diff,
mean_diff_perc,
median_diff_perc,
gt5perc_weightloss,
gt5perc_weightgain
) %>%
ungroup()
rm(Sta3n_wtdx_diff, Sta3n_wtpct)
COLS <- c("#2E364F", "#2D5D7C", "#F3F0E2", "#EF5939")
p <- Sta3n_wtdx %>%
ggplot(aes(mean_diff, group = Gender, fill = Gender)) %>%
add(geom_histogram(bins = 100, alpha = 0.5)) %>%
add(theme_minimal(16)) %>%
add(theme(legend.position = c(0.2, 0.8))) %>%
add(scale_fill_manual(values = COLS[c(2, 4)])) %>%
add(labs(
x = "Weight Change (lbs.)",
y = "Frequency",
fill = ""
))
q <- Sta3n_wtdx %>%
filter(!is.infinite(mean_diff_perc)) %>%
ggplot(aes(mean_diff_perc, group = Gender, fill = Gender)) %>%
add(geom_histogram(bins = 100, alpha = 0.5)) %>%
add(theme_minimal(16)) %>%
add(theme(legend.position = "none")) %>%
add(scale_fill_manual(values = COLS[c(2, 4)])) %>%
add(labs(
x = "Weight Change %",
y = "Frequency",
fill = ""
))
r <- Sta3n_wtdx %>%
ggplot(aes(gt5perc_weightloss, group = Gender, fill = Gender)) %>%
add(geom_histogram(bins = 100, alpha = 0.5)) %>%
add(theme_minimal(16)) %>%
add(theme(legend.position = "none")) %>%
add(scale_fill_manual(values = COLS[c(2, 4)])) %>%
add(labs(
x = ">= 5% Weight Loss",
y = "Frequency"
))
s <- Sta3n_wtdx %>%
filter(!is.na(gt5perc_weightgain)) %>%
ggplot(aes(gt5perc_weightgain, group = Gender, fill = Gender)) %>%
add(geom_histogram(bins = 100, alpha = 0.5)) %>%
add(theme_minimal(16)) %>%
add(theme(legend.position = "none")) %>%
add(scale_fill_manual(values = COLS[c(2, 4)])) %>%
add(labs(
x = ">= 5% Weight Gain",
y = "Frequency"
))
library(patchwork)
p + q + r + s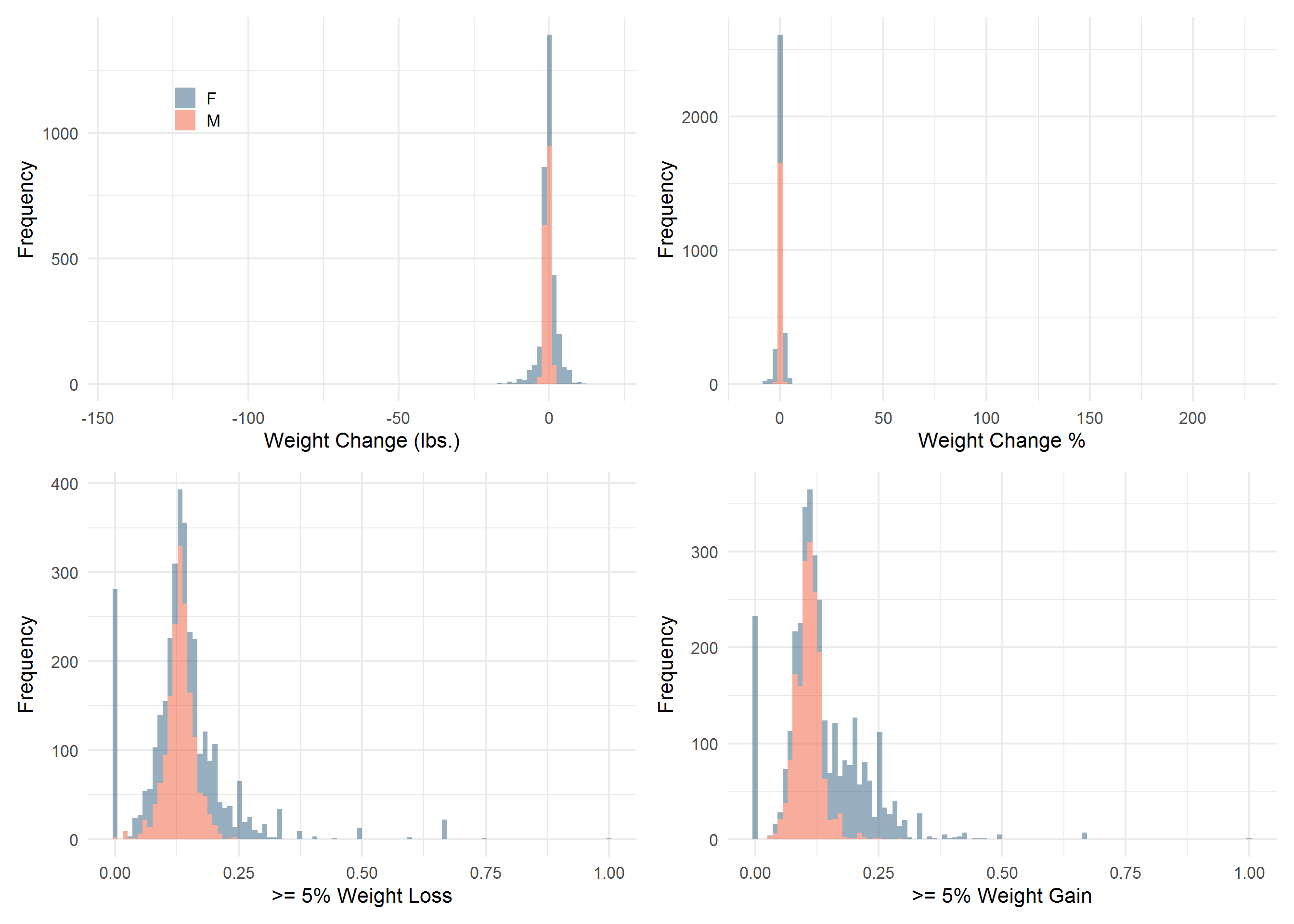
library(gridExtra)
tmp <- Sta3n_wtdx %>%
select(Gender, Type, Algorithm, Sta3n, gt5perc_weightloss)
raw <- tmp %>%
filter(Algorithm == "Raw Weights") %>%
group_by(Gender) %>%
arrange(gt5perc_weightloss) %>%
mutate(Sta3n_rank = row_number()) %>%
select(-Type, -Algorithm) %>%
rename(raw_wtloss = gt5perc_weightloss)
tp <- tmp %>%
filter(Type == "Time-Periods") %>%
left_join(raw, by = c("Sta3n", "Gender")) %>%
mutate(loss_diff = gt5perc_weightloss - raw_wtloss)
ts <- tmp %>%
filter(Type == "Time-Series") %>%
left_join(raw, by = c("Sta3n", "Gender")) %>%
mutate(loss_diff = gt5perc_weightloss - raw_wtloss)
p1 <- tp %>%
ggplot(aes(x = Sta3n_rank, y = raw_wtloss)) %>%
add(facet_wrap(vars(Type, Gender))) %>%
add(geom_line(size = 1)) %>%
add(geom_segment(
aes(
x = Sta3n_rank,
y = raw_wtloss,
xend = Sta3n_rank,
yend = gt5perc_weightloss
),
alpha = 0.3
)) %>%
add(geom_point(
aes(
x = Sta3n_rank,
y = gt5perc_weightloss,
color = Algorithm
),
size = 2,
alpha = 0.7
)) %>%
add(expand_limits(y = c(0.0, 0.3))) %>%
add(theme_minimal(16)) %>%
add(theme(
legend.position = "bottom",
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank()
)) %>%
add(guides(color = guide_legend(override.aes = list(size = 5)))) %>%
add(scale_color_brewer(palette = "Dark2")) %>%
add(labs(
x = "Sta3n",
y = "Proportion Achieving 5% Weight Loss",
color = ""
))
p2 <- p1 %+% ts
p2 <- p2 %>% add(labs(y = ""))
grid.arrange(p1, p2, ncol = 2)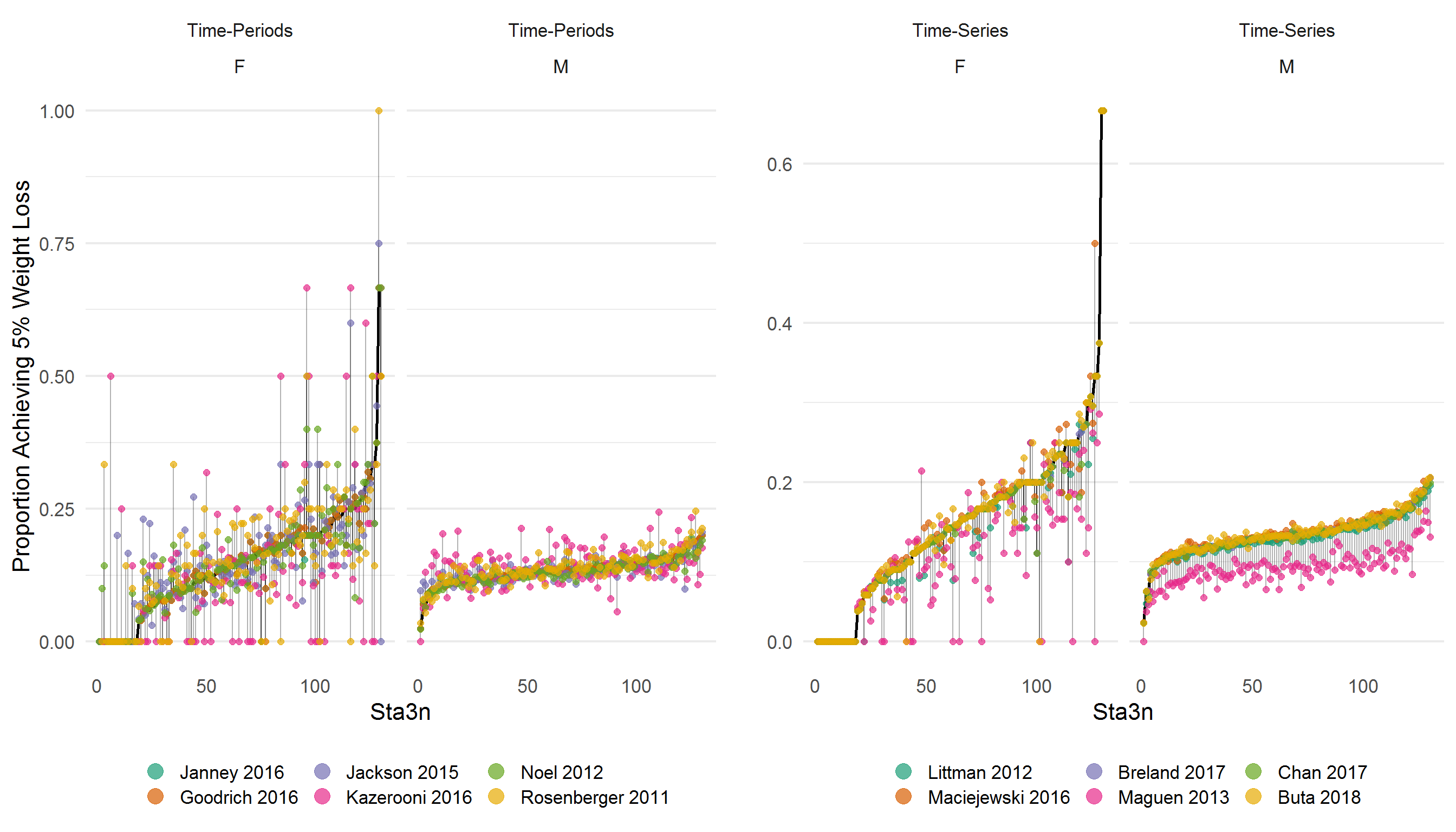
Sites are ordered/ranked along the x-axis using the percent of patients with at least 5% weight loss using the raw data. The left hand side shows the “time-period” algorithms, while the right displays the “time-series” algorithms. Results suggest that generally, time-series algorithms may result in more similar rates by site, save for Maguen 2013. Results are more mixed using the “time-period” algorithms.
By BMI Category
BMI works differently for Men and Women, so I’ll use only the Males for this analysis.
Total number of weight samples, by BMI category
bmi_cohort <- weight.ls[[2]] %>%
filter(Gender == "M", !is.na(BMI)) %>%
group_by(PatientICN) %>%
arrange(PatientICN, WeightDateTime) %>%
filter(row_number() == 1) %>%
mutate(
BMI_cat = case_when(
BMI < 18.5 ~ 0,
BMI >= 18.5 & BMI < 24.9 ~ 1,
BMI >= 24.9 & BMI < 30 ~ 2,
BMI >= 30 ~ 3
),
BMI_cat = factor(
BMI_cat,
0:3,
c("Underweight", "Normal", "Overweight", "Obese")
)
) %>%
select(PatientICN, BMI_cat) %>%
left_join(weight.ls[[2]], by = "PatientICN") %>%
relocate(BMI_cat, .after = last_col()) %>%
ungroup()
bmi_cohort %>%
janitor::tabyl(BMI_cat) %>%
arrange(desc(n)) %>%
adorn_pct_formatting() %>%
tableStyle()| BMI_cat | n | percent |
|---|---|---|
| Obese | 505717 | 47.8% |
| Overweight | 371623 | 35.1% |
| Normal | 171864 | 16.2% |
| Underweight | 9174 | 0.9% |
Total number of people
bmi_cohort %>%
distinct(PatientICN, BMI_cat) %>%
tabyl(BMI_cat) %>%
adorn_pct_formatting() %>%
tableStyle()| BMI_cat | n | percent |
|---|---|---|
| Underweight | 629 | 0.7% |
| Normal | 14662 | 16.9% |
| Overweight | 33243 | 38.4% |
| Obese | 38103 | 44.0% |
Association between number of weight samples collected, and BMI category,
bmi_cohort %>%
group_by(BMI_cat) %>%
count(PatientICN) %>%
summarise(
mean = mean(n),
SD = sd(n),
median = median(n),
min = min(n),
max = max(n)
) %>%
tableStyle()| BMI_cat | mean | SD | median | min | max |
|---|---|---|---|---|---|
| Underweight | 14.59 | 16.35 | 10 | 1 | 148 |
| Normal | 11.72 | 26.84 | 7 | 1 | 1525 |
| Overweight | 11.18 | 16.93 | 7 | 1 | 1254 |
| Obese | 13.27 | 18.36 | 9 | 1 | 1196 |
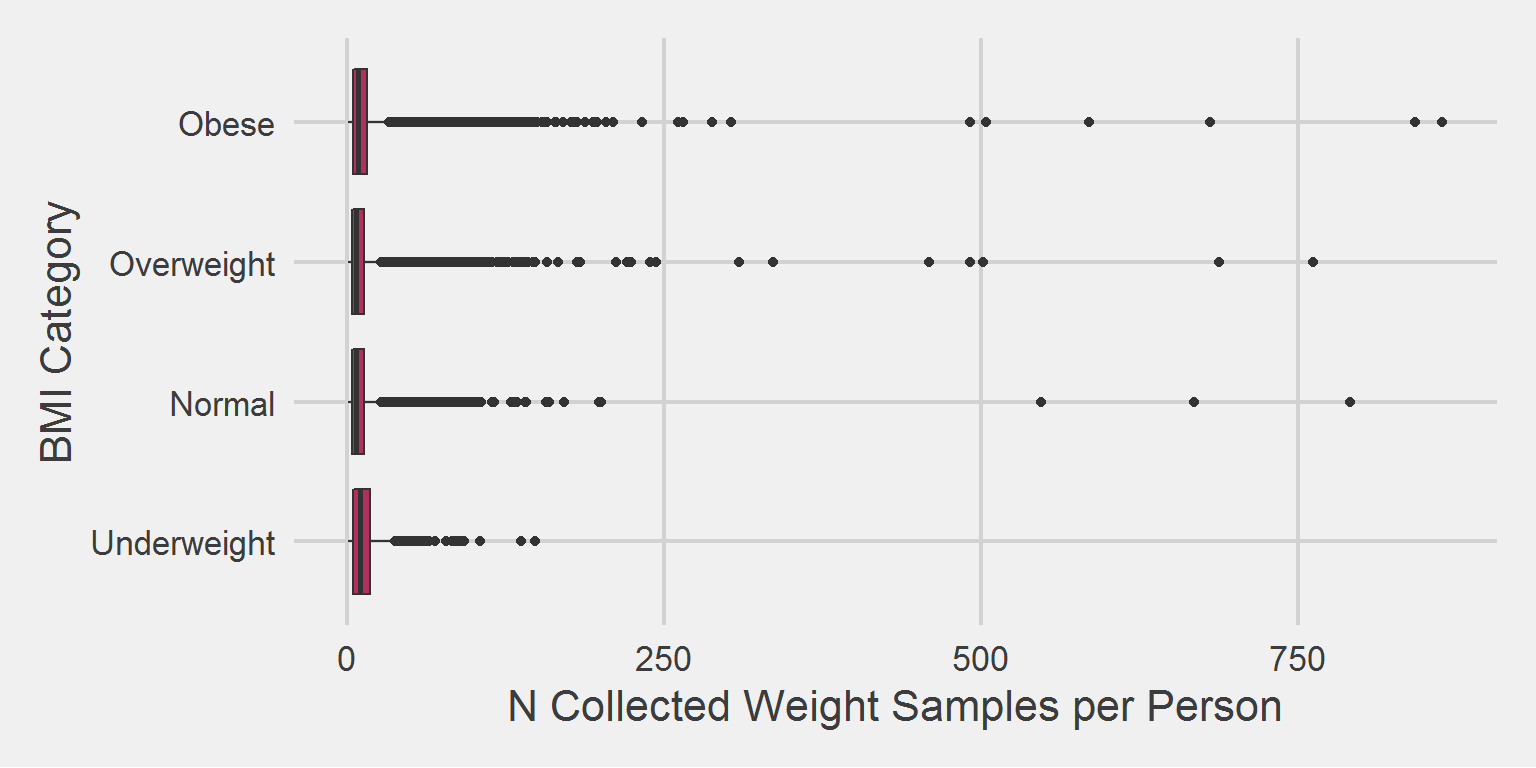
bmi_cohort %>%
group_by(BMI_cat) %>%
count(PatientICN) %>%
filter(n < 1000) %>%
ggplot(aes(x = BMI_cat, y = n)) %>%
add(geom_boxplot(fill = "maroon")) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(coord_flip()) %>%
add(labs(
x = "BMI Category",
y = "N Collected Weight Samples per Person"
))
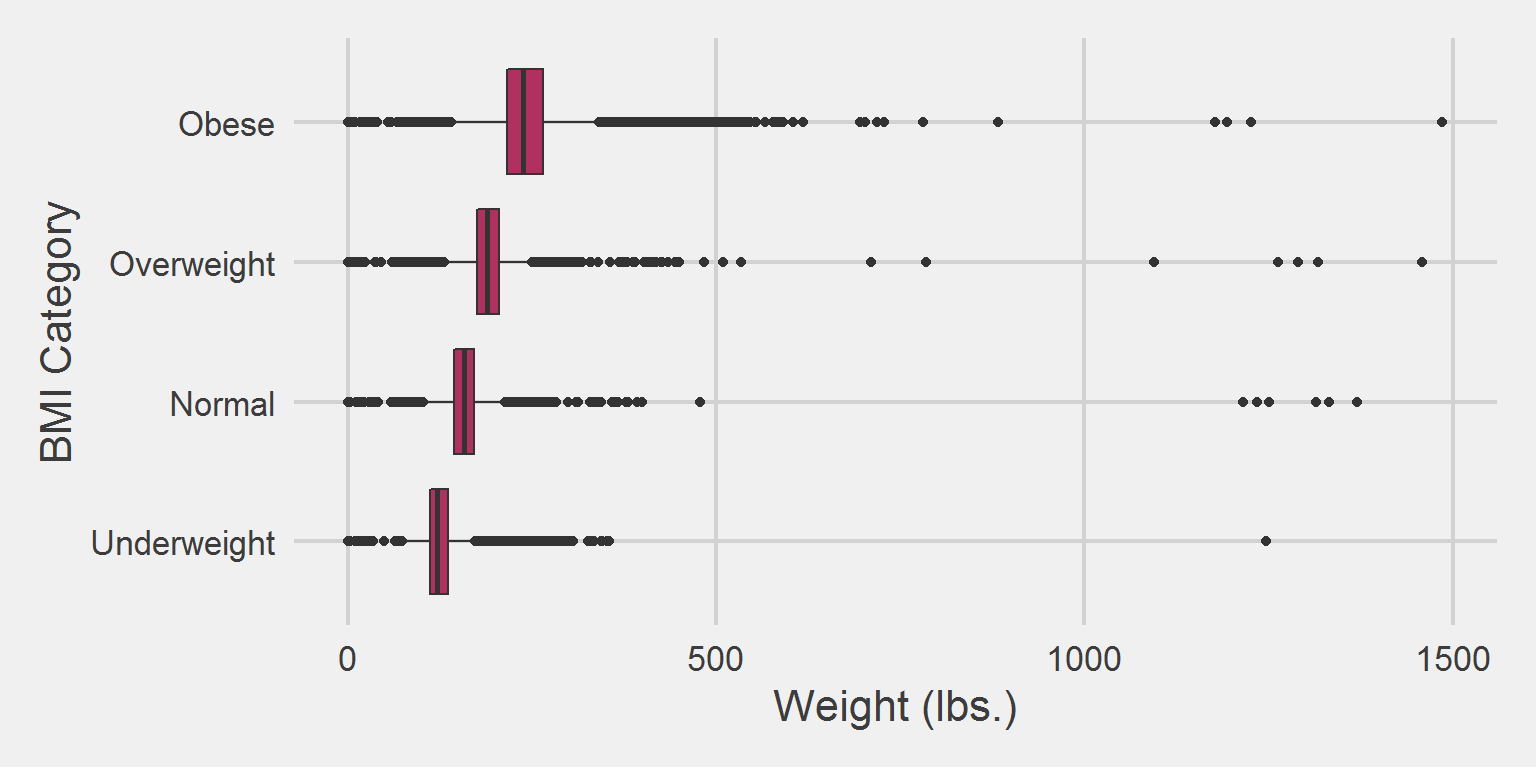
All weight in lbs. by BMI category, for the 2016 cohort only
bmi_cohort %>%
group_by(BMI_cat) %>%
summarise(
mean = mean(Weight),
SD = sd(Weight),
median = median(Weight),
min = min(Weight),
max = max(Weight)
) %>%
tableStyle()| BMI_cat | mean | SD | median | min | max |
|---|---|---|---|---|---|
| Underweight | 126.11 | 29.42 | 121.0 | 0 | 1246.00 |
| Normal | 157.37 | 22.39 | 156.7 | 0 | 1370.04 |
| Overweight | 189.66 | 23.60 | 189.0 | 0 | 1458.15 |
| Obese | 243.42 | 42.90 | 236.6 | 0 | 1486.20 |
It would also make sense to look at this from the perspective of BMI ranges,
bmi_cohort %>%
group_by(BMI_cat) %>%
summarise(
mean = mean(BMI),
SD = sd(BMI),
median = median(BMI),
min = min(BMI),
max = max(BMI)
) %>%
tableStyle()| BMI_cat | mean | SD | median | min | max |
|---|---|---|---|---|---|
| Underweight | 18.15 | 3.96 | 17.62 | 0 | 178.76 |
| Normal | 22.80 | 2.62 | 22.96 | 0 | 227.96 |
| Overweight | 27.40 | 2.49 | 27.44 | 0 | 215.31 |
| Obese | 35.15 | 8.32 | 33.98 | 0 | 1525.51 |
It’s illuminating in the context of outliers, but we already know this from analyzing the weight samples themselves.
bmi_cohort %>%
ggplot(aes(x = BMI_cat, y = Weight)) %>%
add(geom_boxplot(fill = "maroon")) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(coord_flip()) %>%
add(labs(
x = "BMI Category",
y = "Weight (lbs.)"
))
By Algorithm
raw <- bmi_cohort %>%
select(PatientICN, Weight, BMI_cat)
#---- apply Janney et al. 2016 ----#
Janney2016.df <- Janney2016.f(
DF = bmi_cohort,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime",
startPoint = "VisitDateTime"
) %>%
select(PatientICN, Weight_OR, BMI_cat) %>%
rename(Weight = Weight_OR) %>%
na.omit()
#---- apply Littman et al. 2012 ----#
Littman2012.df <- Littman2012.f(
DF = bmi_cohort,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime"
) %>%
select(PatientICN, OutputMeasurement, BMI_cat) %>%
rename(Weight = OutputMeasurement) %>%
na.omit()
#---- Coerce Maciejewski et al. 2016 to a workable format ----#
Maciejewski2016.df <- maciejewski %>%
filter(IO == "Output", SampleYear == "2016") %>%
mutate(PatientICN = as.character(PatientICN)) %>%
left_join(
raw %>%
distinct(PatientICN, BMI_cat),
by = "PatientICN"
) %>%
select(PatientICN, Weight, BMI_cat) %>%
na.omit()
#---- Apply Breland et al. 2017 ----#
Breland2017.df <- Breland2017.f(
DF = bmi_cohort,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime"
) %>%
select(PatientICN, measures_aug_, BMI_cat) %>%
rename(Weight = measures_aug_) %>%
na.omit()
#---- Apply Maguen et al. 2013 ----#
Maguen2013.df <- Maguen2013.f(
DF = bmi_cohort,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime",
variables = "AgeAtVisit"
) %>%
select(PatientICN, Output, BMI_cat) %>%
rename(Weight = Output) %>%
na.omit()
#---- Apply Goodrich 2016 ----#
Goodrich2016.df <- Goodrich2016.f(
DF = bmi_cohort,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime",
startPoint = "VisitDateTime"
) %>%
select(PatientICN, output, BMI_cat) %>%
rename(Weight = output) %>%
na.omit()
#---- Apply Chan & Raffa 2017 ----#
Chan2017.df <- Chan2017.f(
DF = bmi_cohort,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime"
) %>%
select(PatientICN, measures_aug_, BMI_cat) %>%
rename(Weight = measures_aug_) %>%
na.omit()
#---- Apply Jackson et al. 2015 ----#
Jackson2015.df <- Jackson2015.f(
DF = bmi_cohort,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime",
startPoint = "VisitDateTime"
) %>%
left_join(
bmi_cohort %>%
distinct(PatientICN, BMI_cat),
by = "PatientICN"
) %>%
select(PatientICN, BMI_cat, output) %>%
rename(Weight = output) %>%
na.omit()
#---- Apply Buta et al., 2018 ----#
Buta2018.df <- Buta2018.f(
DF = bmi_cohort,
id = "PatientICN",
measures = "BMI",
tmeasures = "WeightDateTime"
) %>%
select(PatientICN, BMI_cat, Weight) %>%
na.omit()
#---- Apply Kazerooni & Lim, 2016 ----#
Kazerooni2016.df <- Kazerooni2016.f(
DF = bmi_cohort,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime",
startPoint = "VisitDateTime"
) %>%
select(PatientICN, BMI_cat, Weight) %>%
na.omit()
#---- Apply Noel et al., 2012 ----#
Noel2012.df <- Noel2012.f(
DF = bmi_cohort,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime"
) %>%
distinct(PatientICN, BMI_cat, FYQ, Qmedian) %>%
select(-FYQ) %>%
rename(Weight = Qmedian) %>%
na.omit()
#---- Apply Rosenberger et al., 2011 ----#
Rosenberger2011.df <- Rosenberger2011.f(
DF = bmi_cohort,
id = "PatientICN",
tmeasures = "WeightDateTime",
startPoint = "VisitDateTime",
pad = 1
) %>%
select(PatientICN, BMI_cat, Weight) %>%
na.omit()
# stack original (raw), janney 2016, littman 2012, maciejewski 2016,
# Breland 2017, Maguen 2013, Goodrich 2016, Chan & Raffa 2017,
# Jackson 2015, Buta 2018, Kazerooni & Lim 2016, Noel 2012,
# and Rosenberger 2011 processed weights
algos.fac <- c("Raw Weights",
"Janney 2016",
"Littman 2012",
"Maciejewski 2016",
"Breland 2017",
"Maguen 2013",
"Goodrich 2016",
"Chan 2017",
"Jackson 2015",
"Buta 2018",
"Kazerooni 2016",
"Noel 2012",
"Rosenberger 2011")
DF <- bind_rows(
list(`Raw Weights` = raw,
`Janney 2016` = Janney2016.df,
`Littman 2012` = Littman2012.df,
`Maciejewski 2016` = Maciejewski2016.df,
`Breland 2017` = Breland2017.df,
`Maguen 2013` = Maguen2013.df,
`Goodrich 2016` = Goodrich2016.df,
`Chan 2017` = Chan2017.df,
`Jackson 2015` = Jackson2015.df,
`Buta 2018` = Buta2018.df,
`Kazerooni 2016` = Kazerooni2016.df,
`Noel 2012` = Noel2012.df,
`Rosenberger 2011` = Rosenberger2011.df),
.id = "Algorithm"
)
rm(
Buta2018.df,
Kazerooni2016.df,
Noel2012.df,
Rosenberger2011.df,
Breland2017.df,
Chan2017.df,
Goodrich2016.df,
Jackson2015.df,
Janney2016.df,
Littman2012.df,
Maciejewski2016.df,
Maguen2013.df,
raw
)
DF$Algorithm <- factor(DF$Algorithm, levels = algos.fac, labels = algos.fac)
DF$Type <- case_when(
DF$Algorithm == "Raw Weights" ~ 1,
DF$Algorithm %in% algos.fac[c(2, 7, 9, 11:13)] ~ 2,
DF$Algorithm %in% algos.fac[c(3:6, 8, 10)] ~ 3
)
DF$Type <- factor(DF$Type, 1:3, c("Raw", "Time-Periods", "Time-Series"))Total weight measurements, by algorithm and Sex.
DF %>%
tabyl(Algorithm, BMI_cat) %>%
adorn_percentages() %>%
adorn_pct_formatting() %>%
adorn_ns() %>%
tableStyle()| Algorithm | Underweight | Normal | Overweight | Obese |
|---|---|---|---|---|
| Raw Weights | 0.9% (9174) | 16.2% (171864) | 35.1% (371623) | 47.8% (505717) |
| Janney 2016 | 0.7% (1256) | 16.2% (28727) | 37.9% (67155) | 45.1% (79837) |
| Littman 2012 | 0.9% (8915) | 16.2% (169459) | 35.2% (367844) | 47.8% (499858) |
| Maciejewski 2016 | 0.9% (8838) | 16.0% (164857) | 35.2% (362454) | 47.9% (492648) |
| Breland 2017 | 0.9% (9135) | 16.2% (171726) | 35.1% (371437) | 47.8% (505338) |
| Maguen 2013 | 0.9% (8339) | 17.0% (158002) | 36.3% (338323) | 45.8% (426941) |
| Goodrich 2016 | 0.7% (1265) | 16.2% (28713) | 37.9% (67136) | 45.1% (79818) |
| Chan 2017 | 0.9% (9103) | 16.2% (170995) | 35.1% (369876) | 47.8% (503380) |
| Jackson 2015 | 0.7% (1538) | 16.1% (35908) | 37.8% (84435) | 45.4% (101254) |
| Buta 2018 | 0.9% (9113) | 16.2% (171333) | 35.1% (370629) | 47.8% (504637) |
| Kazerooni 2016 | 0.9% (576) | 15.3% (9909) | 35.4% (22992) | 48.4% (31461) |
| Noel 2012 | 0.7% (4411) | 15.9% (96637) | 36.9% (224645) | 46.5% (282526) |
| Rosenberger 2011 | 0.7% (1336) | 15.4% (31400) | 37.2% (75674) | 46.8% (95247) |
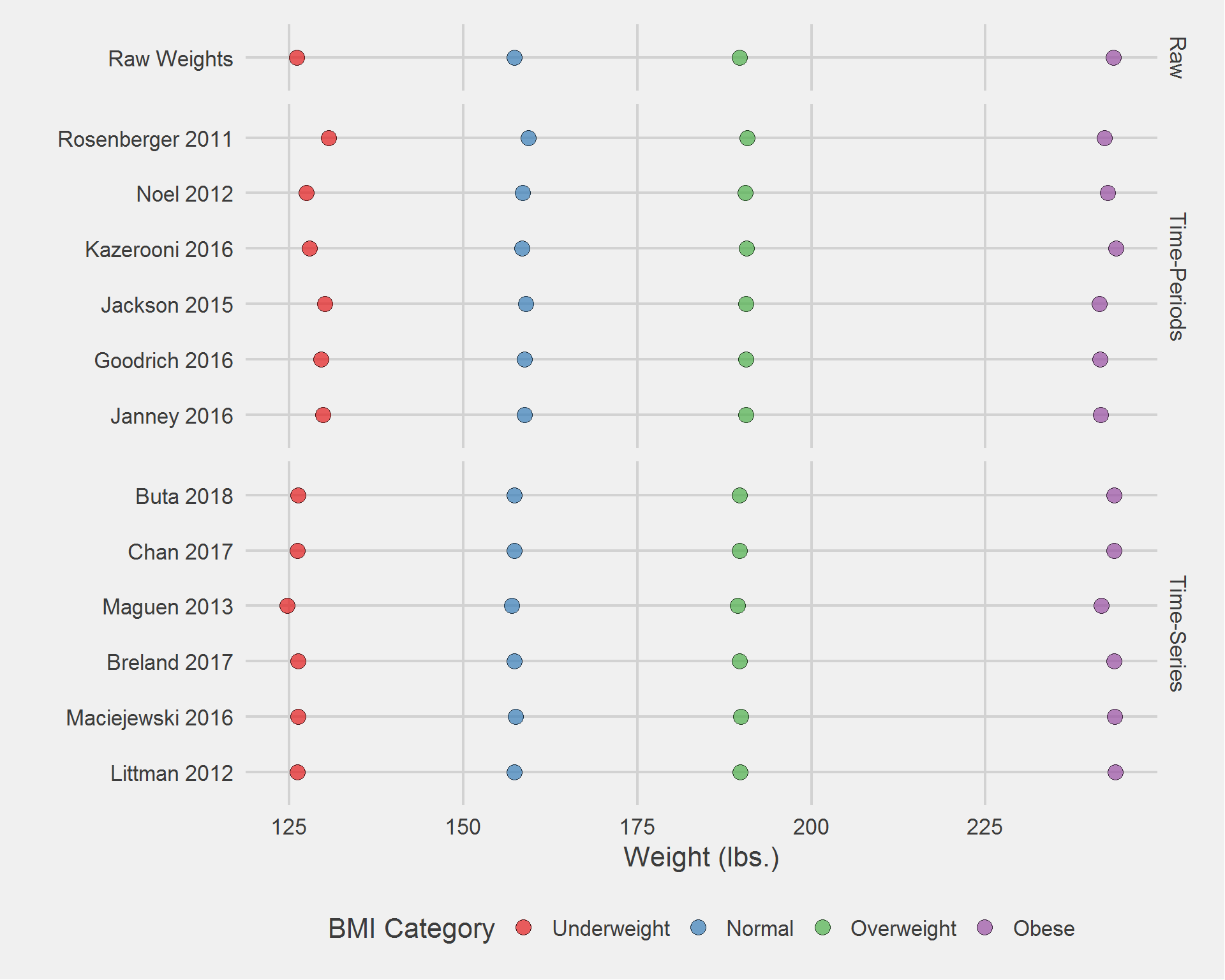
Distribution of weight measurements, by algorithm and BMI category
tmp <- DF %>%
filter(!is.na(Weight)) %>%
group_by(Type, Algorithm, BMI_cat) %>%
dplyr::summarise(
weight_n = n(),
mean = mean(Weight),
SD = sd(Weight),
median = median(Weight),
IQR = IQR(Weight),
min = min(Weight),
max = max(Weight)
)
tableStyle(tmp)| Type | Algorithm | BMI_cat | weight_n | mean | SD | median | IQR | min | max |
|---|---|---|---|---|---|---|---|---|---|
| Raw | Raw Weights | Underweight | 9174 | 126.11 | 29.42 | 121.00 | 24.50 | 0.00 | 1246.00 |
| Raw | Raw Weights | Normal | 171864 | 157.37 | 22.39 | 156.70 | 28.00 | 0.00 | 1370.04 |
| Raw | Raw Weights | Overweight | 371623 | 189.66 | 23.60 | 189.00 | 30.00 | 0.00 | 1458.15 |
| Raw | Raw Weights | Obese | 505717 | 243.42 | 42.90 | 236.60 | 50.00 | 0.00 | 1486.20 |
| Time-Periods | Janney 2016 | Underweight | 1256 | 129.92 | 32.02 | 123.00 | 25.03 | 82.60 | 351.60 |
| Time-Periods | Janney 2016 | Normal | 28727 | 158.83 | 20.73 | 158.10 | 27.00 | 80.00 | 478.00 |
| Time-Periods | Janney 2016 | Overweight | 67155 | 190.59 | 22.19 | 190.00 | 29.40 | 92.20 | 533.30 |
| Time-Periods | Janney 2016 | Obese | 79837 | 241.51 | 40.08 | 235.10 | 47.80 | 98.40 | 546.00 |
| Time-Periods | Goodrich 2016 | Underweight | 1265 | 129.59 | 32.13 | 123.00 | 25.40 | 80.20 | 351.60 |
| Time-Periods | Goodrich 2016 | Normal | 28713 | 158.82 | 20.68 | 158.00 | 27.00 | 80.00 | 478.00 |
| Time-Periods | Goodrich 2016 | Overweight | 67136 | 190.58 | 22.12 | 190.00 | 29.40 | 96.00 | 368.17 |
| Time-Periods | Goodrich 2016 | Obese | 79818 | 241.49 | 40.01 | 235.10 | 47.80 | 87.00 | 500.00 |
| Time-Periods | Jackson 2015 | Underweight | 1538 | 130.19 | 33.51 | 123.00 | 25.61 | 78.00 | 354.50 |
| Time-Periods | Jackson 2015 | Normal | 35908 | 158.96 | 20.90 | 158.30 | 27.05 | 83.38 | 365.97 |
| Time-Periods | Jackson 2015 | Overweight | 84435 | 190.57 | 22.46 | 189.80 | 29.73 | 95.30 | 449.74 |
| Time-Periods | Jackson 2015 | Obese | 101254 | 241.36 | 40.25 | 235.00 | 48.00 | 87.00 | 553.00 |
| Time-Periods | Kazerooni 2016 | Underweight | 576 | 127.98 | 32.19 | 121.00 | 26.35 | 82.50 | 342.80 |
| Time-Periods | Kazerooni 2016 | Normal | 9909 | 158.46 | 24.09 | 157.90 | 28.00 | 16.50 | 1233.70 |
| Time-Periods | Kazerooni 2016 | Overweight | 22992 | 190.71 | 23.25 | 189.90 | 30.25 | 16.50 | 710.00 |
| Time-Periods | Kazerooni 2016 | Obese | 31461 | 243.75 | 41.66 | 237.20 | 49.90 | 0.00 | 517.44 |
| Time-Periods | Noel 2012 | Underweight | 4411 | 127.49 | 28.53 | 122.15 | 23.30 | 70.00 | 354.50 |
| Time-Periods | Noel 2012 | Normal | 96637 | 158.55 | 20.30 | 158.00 | 26.50 | 74.00 | 365.97 |
| Time-Periods | Noel 2012 | Overweight | 224645 | 190.53 | 21.90 | 189.80 | 28.90 | 70.00 | 449.74 |
| Time-Periods | Noel 2012 | Obese | 282526 | 242.52 | 40.40 | 236.00 | 48.00 | 71.25 | 588.90 |
| Time-Periods | Rosenberger 2011 | Underweight | 1336 | 130.68 | 34.40 | 123.50 | 26.00 | 22.90 | 354.50 |
| Time-Periods | Rosenberger 2011 | Normal | 31400 | 159.34 | 22.17 | 159.00 | 27.80 | 11.02 | 1314.50 |
| Time-Periods | Rosenberger 2011 | Overweight | 75674 | 190.76 | 22.99 | 190.00 | 29.90 | 1.00 | 710.00 |
| Time-Periods | Rosenberger 2011 | Obese | 95247 | 242.09 | 40.83 | 235.80 | 48.49 | 0.00 | 727.50 |
| Time-Series | Littman 2012 | Underweight | 8915 | 126.17 | 26.27 | 121.00 | 24.10 | 75.20 | 354.50 |
| Time-Series | Littman 2012 | Normal | 169459 | 157.33 | 20.84 | 156.70 | 28.00 | 75.30 | 365.97 |
| Time-Series | Littman 2012 | Overweight | 367844 | 189.74 | 22.82 | 189.00 | 29.80 | 78.00 | 356.00 |
| Time-Series | Littman 2012 | Obese | 499858 | 243.61 | 42.36 | 236.80 | 50.00 | 88.00 | 546.00 |
| Time-Series | Maciejewski 2016 | Underweight | 8838 | 126.31 | 26.54 | 121.10 | 24.20 | 66.00 | 354.50 |
| Time-Series | Maciejewski 2016 | Normal | 164857 | 157.53 | 21.03 | 156.97 | 28.00 | 64.00 | 365.97 |
| Time-Series | Maciejewski 2016 | Overweight | 362454 | 189.84 | 22.78 | 189.00 | 29.89 | 87.00 | 318.00 |
| Time-Series | Maciejewski 2016 | Obese | 492648 | 243.58 | 42.46 | 236.90 | 50.00 | 93.00 | 546.00 |
| Time-Series | Breland 2017 | Underweight | 9135 | 126.26 | 26.45 | 121.03 | 24.25 | 75.20 | 354.50 |
| Time-Series | Breland 2017 | Normal | 171726 | 157.35 | 21.12 | 156.70 | 28.00 | 77.00 | 365.97 |
| Time-Series | Breland 2017 | Overweight | 371437 | 189.67 | 23.00 | 189.00 | 30.00 | 82.50 | 379.20 |
| Time-Series | Breland 2017 | Obese | 505338 | 243.48 | 42.56 | 236.60 | 50.00 | 93.00 | 694.46 |
| Time-Series | Maguen 2013 | Underweight | 8339 | 124.71 | 24.05 | 120.50 | 23.48 | 74.60 | 345.00 |
| Time-Series | Maguen 2013 | Normal | 158002 | 157.00 | 20.24 | 156.31 | 27.38 | 74.00 | 343.10 |
| Time-Series | Maguen 2013 | Overweight | 338323 | 189.44 | 22.16 | 188.80 | 29.20 | 95.70 | 309.00 |
| Time-Series | Maguen 2013 | Obese | 426941 | 241.65 | 40.07 | 235.07 | 47.20 | 103.80 | 541.10 |
| Time-Series | Chan 2017 | Underweight | 9103 | 126.20 | 26.54 | 121.00 | 24.40 | 64.00 | 354.50 |
| Time-Series | Chan 2017 | Normal | 170995 | 157.34 | 21.13 | 156.70 | 28.00 | 58.60 | 365.97 |
| Time-Series | Chan 2017 | Overweight | 369876 | 189.67 | 22.99 | 189.00 | 30.00 | 59.00 | 509.27 |
| Time-Series | Chan 2017 | Obese | 503380 | 243.49 | 42.57 | 236.60 | 50.00 | 54.00 | 727.50 |
| Time-Series | Buta 2018 | Underweight | 9113 | 126.29 | 26.52 | 121.04 | 24.20 | 66.80 | 354.50 |
| Time-Series | Buta 2018 | Normal | 171333 | 157.38 | 21.26 | 156.70 | 28.00 | 67.46 | 478.00 |
| Time-Series | Buta 2018 | Overweight | 370629 | 189.67 | 23.09 | 189.00 | 30.00 | 74.50 | 509.27 |
| Time-Series | Buta 2018 | Obese | 504637 | 243.42 | 42.55 | 236.60 | 50.00 | 66.50 | 540.00 |
tmp %>%
ggplot(aes(mean, Algorithm)) %>%
add(geom_point(aes(fill = BMI_cat), size = 4, pch = 21, alpha = 0.7)) %>%
add(facet_grid(rows = vars(Type), scales = "free", space = "free")) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(scale_fill_brewer(palette = "Set1")) %>%
add(labs(
x = "Weight (lbs.)",
y = "",
fill = "BMI Category"
))
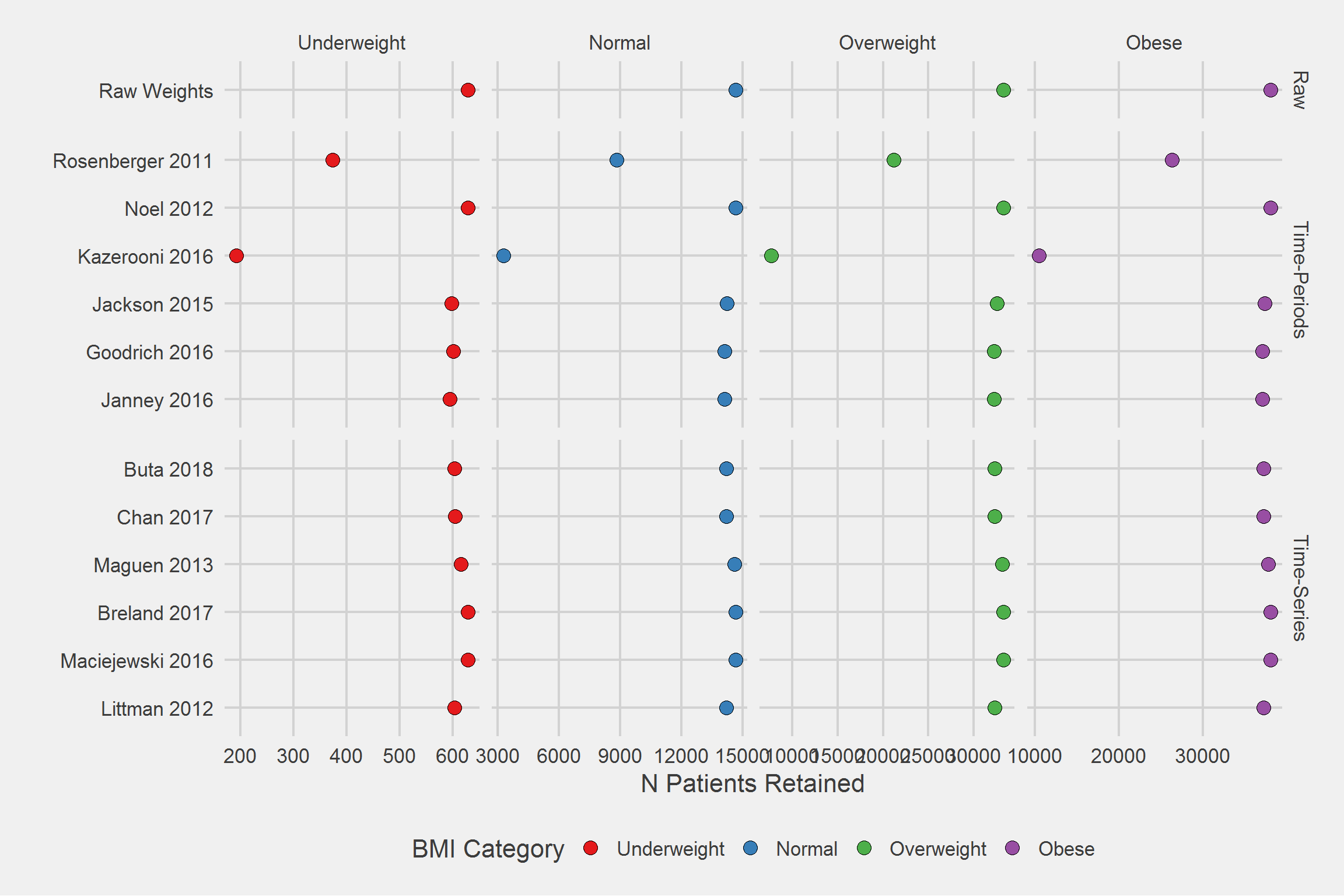
Number of Patients/Veterans Retained, by algorithm, by BMI category
tmp <- DF %>%
filter(!is.na(Weight)) %>%
group_by(Type, Algorithm, BMI_cat) %>%
distinct(PatientICN) %>%
count()
tableStyle(tmp)| Type | Algorithm | BMI_cat | n |
|---|---|---|---|
| Raw | Raw Weights | Underweight | 629 |
| Raw | Raw Weights | Normal | 14662 |
| Raw | Raw Weights | Overweight | 33243 |
| Raw | Raw Weights | Obese | 38103 |
| Time-Periods | Janney 2016 | Underweight | 595 |
| Time-Periods | Janney 2016 | Normal | 14116 |
| Time-Periods | Janney 2016 | Overweight | 32250 |
| Time-Periods | Janney 2016 | Obese | 37102 |
| Time-Periods | Goodrich 2016 | Underweight | 601 |
| Time-Periods | Goodrich 2016 | Normal | 14117 |
| Time-Periods | Goodrich 2016 | Overweight | 32247 |
| Time-Periods | Goodrich 2016 | Obese | 37100 |
| Time-Periods | Jackson 2015 | Underweight | 598 |
| Time-Periods | Jackson 2015 | Normal | 14235 |
| Time-Periods | Jackson 2015 | Overweight | 32533 |
| Time-Periods | Jackson 2015 | Obese | 37395 |
| Time-Periods | Kazerooni 2016 | Underweight | 192 |
| Time-Periods | Kazerooni 2016 | Normal | 3303 |
| Time-Periods | Kazerooni 2016 | Overweight | 7664 |
| Time-Periods | Kazerooni 2016 | Obese | 10487 |
| Time-Periods | Noel 2012 | Underweight | 629 |
| Time-Periods | Noel 2012 | Normal | 14662 |
| Time-Periods | Noel 2012 | Overweight | 33243 |
| Time-Periods | Noel 2012 | Obese | 38103 |
| Time-Periods | Rosenberger 2011 | Underweight | 374 |
| Time-Periods | Rosenberger 2011 | Normal | 8839 |
| Time-Periods | Rosenberger 2011 | Overweight | 21208 |
| Time-Periods | Rosenberger 2011 | Obese | 26324 |
| Time-Series | Littman 2012 | Underweight | 604 |
| Time-Series | Littman 2012 | Normal | 14201 |
| Time-Series | Littman 2012 | Overweight | 32339 |
| Time-Series | Littman 2012 | Obese | 37280 |
| Time-Series | Maciejewski 2016 | Underweight | 629 |
| Time-Series | Maciejewski 2016 | Normal | 14662 |
| Time-Series | Maciejewski 2016 | Overweight | 33243 |
| Time-Series | Maciejewski 2016 | Obese | 38103 |
| Time-Series | Breland 2017 | Underweight | 629 |
| Time-Series | Breland 2017 | Normal | 14662 |
| Time-Series | Breland 2017 | Overweight | 33243 |
| Time-Series | Breland 2017 | Obese | 38103 |
| Time-Series | Maguen 2013 | Underweight | 616 |
| Time-Series | Maguen 2013 | Normal | 14604 |
| Time-Series | Maguen 2013 | Overweight | 33128 |
| Time-Series | Maguen 2013 | Obese | 37781 |
| Time-Series | Chan 2017 | Underweight | 605 |
| Time-Series | Chan 2017 | Normal | 14201 |
| Time-Series | Chan 2017 | Overweight | 32339 |
| Time-Series | Chan 2017 | Obese | 37280 |
| Time-Series | Buta 2018 | Underweight | 603 |
| Time-Series | Buta 2018 | Normal | 14201 |
| Time-Series | Buta 2018 | Overweight | 32339 |
| Time-Series | Buta 2018 | Obese | 37267 |
tmp %>%
ggplot(aes(x = Algorithm, y = n)) %>%
add(geom_point(aes(fill = BMI_cat), size = 4, pch = 21)) %>%
add(facet_grid(
cols = vars(BMI_cat),
rows = vars(Type),
scales = "free",
space = "free_y"
)) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(scale_fill_brewer(palette = "Set1")) %>%
add(coord_flip()) %>%
add(labs(
x = "",
y = "N Patients Retained",
fill = "BMI Category"
))
Number of weights retained, per person, by algorithm, by BMI Category,
tmp <- DF %>%
filter(!is.na(Weight)) %>%
group_by(Type, Algorithm, BMI_cat, PatientICN) %>%
count() %>%
ungroup() %>%
group_by(Type, Algorithm, BMI_cat) %>%
summarize(
mean = mean(n),
SD = sd(n),
min = min(n),
Q1 = fivenum(n)[2],
median = median(n),
Q3 = fivenum(n)[4],
max = max(n)
)
tableStyle(tmp)| Type | Algorithm | BMI_cat | mean | SD | min | Q1 | median | Q3 | max |
|---|---|---|---|---|---|---|---|---|---|
| Raw | Raw Weights | Underweight | 14.59 | 16.35 | 1 | 5 | 10 | 18.0 | 148 |
| Raw | Raw Weights | Normal | 11.72 | 26.84 | 1 | 4 | 7 | 13.0 | 1525 |
| Raw | Raw Weights | Overweight | 11.18 | 16.93 | 1 | 4 | 7 | 13.0 | 1254 |
| Raw | Raw Weights | Obese | 13.27 | 18.36 | 1 | 5 | 9 | 16.0 | 1196 |
| Time-Periods | Janney 2016 | Underweight | 2.11 | 0.79 | 1 | 1 | 2 | 3.0 | 3 |
| Time-Periods | Janney 2016 | Normal | 2.04 | 0.76 | 1 | 1 | 2 | 3.0 | 3 |
| Time-Periods | Janney 2016 | Overweight | 2.08 | 0.75 | 1 | 2 | 2 | 3.0 | 3 |
| Time-Periods | Janney 2016 | Obese | 2.15 | 0.76 | 1 | 2 | 2 | 3.0 | 3 |
| Time-Periods | Goodrich 2016 | Underweight | 2.10 | 0.79 | 1 | 1 | 2 | 3.0 | 3 |
| Time-Periods | Goodrich 2016 | Normal | 2.03 | 0.76 | 1 | 1 | 2 | 3.0 | 3 |
| Time-Periods | Goodrich 2016 | Overweight | 2.08 | 0.75 | 1 | 2 | 2 | 3.0 | 3 |
| Time-Periods | Goodrich 2016 | Obese | 2.15 | 0.76 | 1 | 2 | 2 | 3.0 | 3 |
| Time-Periods | Jackson 2015 | Underweight | 2.57 | 1.01 | 1 | 2 | 3 | 3.0 | 4 |
| Time-Periods | Jackson 2015 | Normal | 2.52 | 0.99 | 1 | 2 | 3 | 3.0 | 4 |
| Time-Periods | Jackson 2015 | Overweight | 2.60 | 0.97 | 1 | 2 | 3 | 3.0 | 4 |
| Time-Periods | Jackson 2015 | Obese | 2.71 | 0.98 | 1 | 2 | 3 | 3.0 | 4 |
| Time-Periods | Kazerooni 2016 | Underweight | 3.00 | 0.00 | 3 | 3 | 3 | 3.0 | 3 |
| Time-Periods | Kazerooni 2016 | Normal | 3.00 | 0.00 | 3 | 3 | 3 | 3.0 | 3 |
| Time-Periods | Kazerooni 2016 | Overweight | 3.00 | 0.00 | 3 | 3 | 3 | 3.0 | 3 |
| Time-Periods | Kazerooni 2016 | Obese | 3.00 | 0.00 | 3 | 3 | 3 | 3.0 | 3 |
| Time-Periods | Noel 2012 | Underweight | 7.01 | 3.81 | 1 | 4 | 7 | 9.0 | 17 |
| Time-Periods | Noel 2012 | Normal | 6.59 | 3.62 | 1 | 4 | 6 | 9.0 | 17 |
| Time-Periods | Noel 2012 | Overweight | 6.76 | 3.63 | 1 | 4 | 6 | 9.0 | 17 |
| Time-Periods | Noel 2012 | Obese | 7.41 | 3.81 | 1 | 4 | 7 | 10.0 | 17 |
| Time-Periods | Rosenberger 2011 | Underweight | 3.57 | 0.50 | 3 | 3 | 4 | 4.0 | 4 |
| Time-Periods | Rosenberger 2011 | Normal | 3.55 | 0.50 | 3 | 3 | 4 | 4.0 | 4 |
| Time-Periods | Rosenberger 2011 | Overweight | 3.57 | 0.50 | 3 | 3 | 4 | 4.0 | 4 |
| Time-Periods | Rosenberger 2011 | Obese | 3.62 | 0.49 | 3 | 3 | 4 | 4.0 | 4 |
| Time-Series | Littman 2012 | Underweight | 14.76 | 16.16 | 2 | 5 | 10 | 18.0 | 148 |
| Time-Series | Littman 2012 | Normal | 11.93 | 27.10 | 2 | 4 | 8 | 14.0 | 1524 |
| Time-Series | Littman 2012 | Overweight | 11.37 | 16.97 | 2 | 4 | 8 | 13.0 | 1253 |
| Time-Series | Littman 2012 | Obese | 13.41 | 18.31 | 2 | 5 | 9 | 16.0 | 1193 |
| Time-Series | Maciejewski 2016 | Underweight | 14.05 | 15.39 | 1 | 5 | 10 | 17.0 | 139 |
| Time-Series | Maciejewski 2016 | Normal | 11.24 | 17.29 | 1 | 4 | 7 | 13.0 | 629 |
| Time-Series | Maciejewski 2016 | Overweight | 10.90 | 13.16 | 1 | 4 | 7 | 13.0 | 637 |
| Time-Series | Maciejewski 2016 | Obese | 12.93 | 14.93 | 1 | 5 | 9 | 16.0 | 615 |
| Time-Series | Breland 2017 | Underweight | 14.52 | 16.33 | 1 | 5 | 10 | 18.0 | 148 |
| Time-Series | Breland 2017 | Normal | 11.71 | 26.79 | 1 | 4 | 7 | 13.0 | 1524 |
| Time-Series | Breland 2017 | Overweight | 11.17 | 16.91 | 1 | 4 | 7 | 13.0 | 1251 |
| Time-Series | Breland 2017 | Obese | 13.26 | 18.34 | 1 | 5 | 9 | 16.0 | 1192 |
| Time-Series | Maguen 2013 | Underweight | 13.54 | 14.66 | 1 | 5 | 9 | 16.5 | 146 |
| Time-Series | Maguen 2013 | Normal | 10.82 | 25.45 | 1 | 4 | 7 | 13.0 | 1516 |
| Time-Series | Maguen 2013 | Overweight | 10.21 | 15.32 | 1 | 4 | 7 | 12.0 | 1160 |
| Time-Series | Maguen 2013 | Obese | 11.30 | 15.11 | 1 | 4 | 8 | 14.0 | 1083 |
| Time-Series | Chan 2017 | Underweight | 15.05 | 16.36 | 2 | 5 | 10 | 18.0 | 147 |
| Time-Series | Chan 2017 | Normal | 12.04 | 27.04 | 2 | 4 | 8 | 14.0 | 1517 |
| Time-Series | Chan 2017 | Overweight | 11.44 | 16.98 | 2 | 4 | 8 | 14.0 | 1246 |
| Time-Series | Chan 2017 | Obese | 13.50 | 18.32 | 2 | 5 | 9 | 16.0 | 1188 |
| Time-Series | Buta 2018 | Underweight | 15.11 | 16.45 | 2 | 5 | 10 | 18.0 | 148 |
| Time-Series | Buta 2018 | Normal | 12.06 | 27.17 | 2 | 4 | 8 | 14.0 | 1524 |
| Time-Series | Buta 2018 | Overweight | 11.46 | 17.07 | 2 | 4 | 8 | 14.0 | 1253 |
| Time-Series | Buta 2018 | Obese | 13.54 | 18.46 | 2 | 5 | 9 | 16.0 | 1193 |
tmp %>%
ggplot(aes(x = Algorithm, y = mean)) %>%
add(geom_point(aes(fill = BMI_cat, size = SD), pch = 21)) %>%
add(facet_grid(
cols = vars(BMI_cat),
rows = vars(Type),
scales = "free",
space = "free_y"
)) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(scale_fill_brewer(palette = "Set1")) %>%
add(coord_flip()) %>%
add(labs(
x = "",
y = "N Weights Retained",
size = "Standard Deviation",
fill = "BMI Category"
))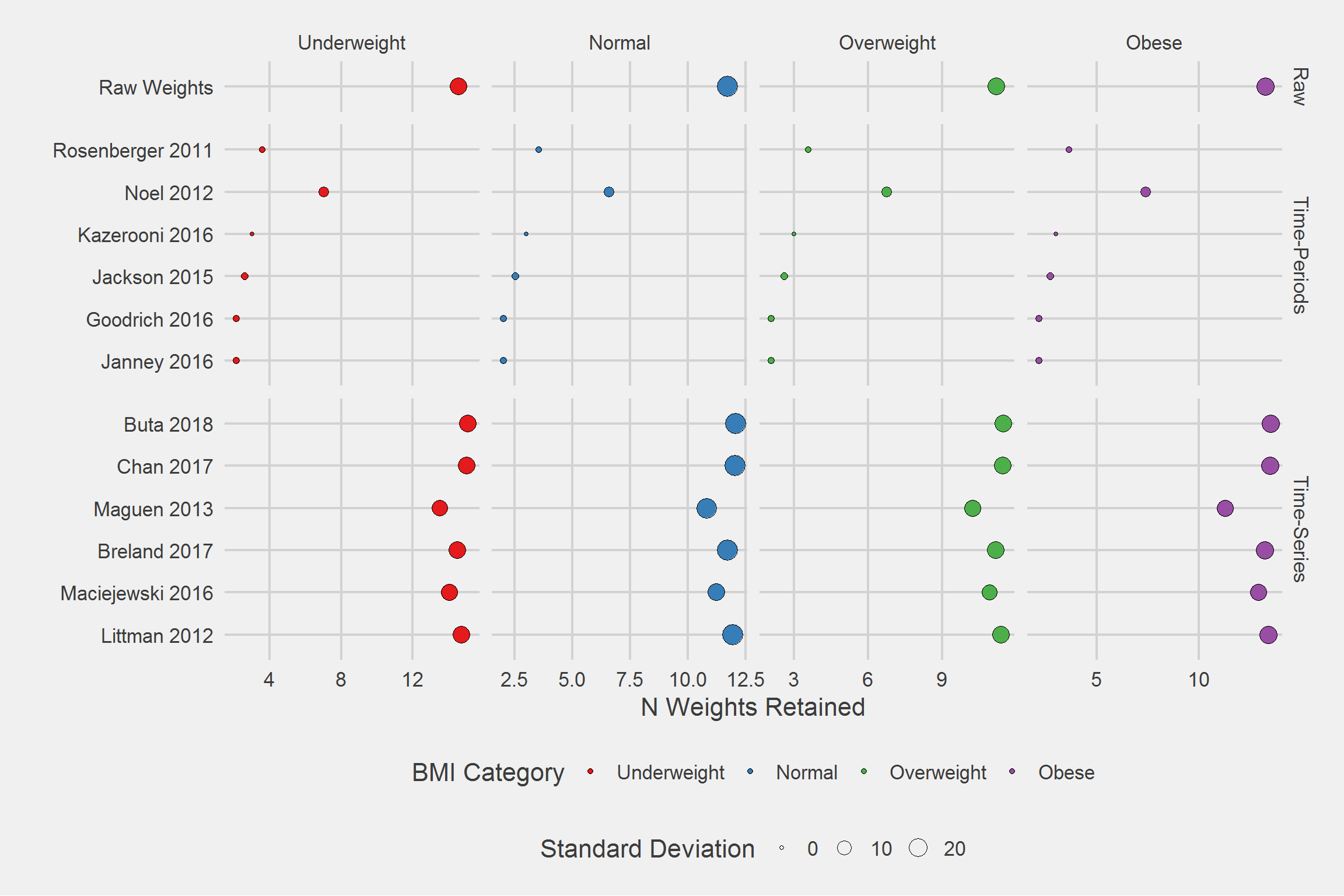
N <- 100
weight_samps <- vector("list", N)
key_cols <- c("Algorithm", "BMI_cat")
pb <- txtProgressBar(min = 0, max = N, initial = 0, style = 3)
for (i in 1:N) {
setTxtProgressBar(pb, i)
sampU <- sample(
pts %>% filter(BMI_cat == "Underweight") %>% pull(PatientICN),
size = 500,
replace = TRUE
)
sampN <- sample(
pts %>% filter(BMI_cat == "Normal") %>% pull(PatientICN),
size = 500,
replace = TRUE
)
sampO <- sample(
pts %>% filter(BMI_cat == "Overweight") %>% pull(PatientICN),
size = 500,
replace = TRUE
)
sampOb <- sample(
pts %>% filter(BMI_cat == "Obese") %>% pull(PatientICN),
size = 500,
replace = TRUE
)
samp <- c(sampU, sampN, sampO, sampOb)
weight_samps[[i]] <- DF %>% filter(PatientICN %in% samp)
setDT(weight_samps[[i]])
setkeyv(weight_samps[[i]], key_cols)
weight_samps[[i]] <-
weight_samps[[i]][,
list(mean = mean(Weight), SD = sd(Weight)),
by = key_cols
]
}Visualization
Distribution of the mean
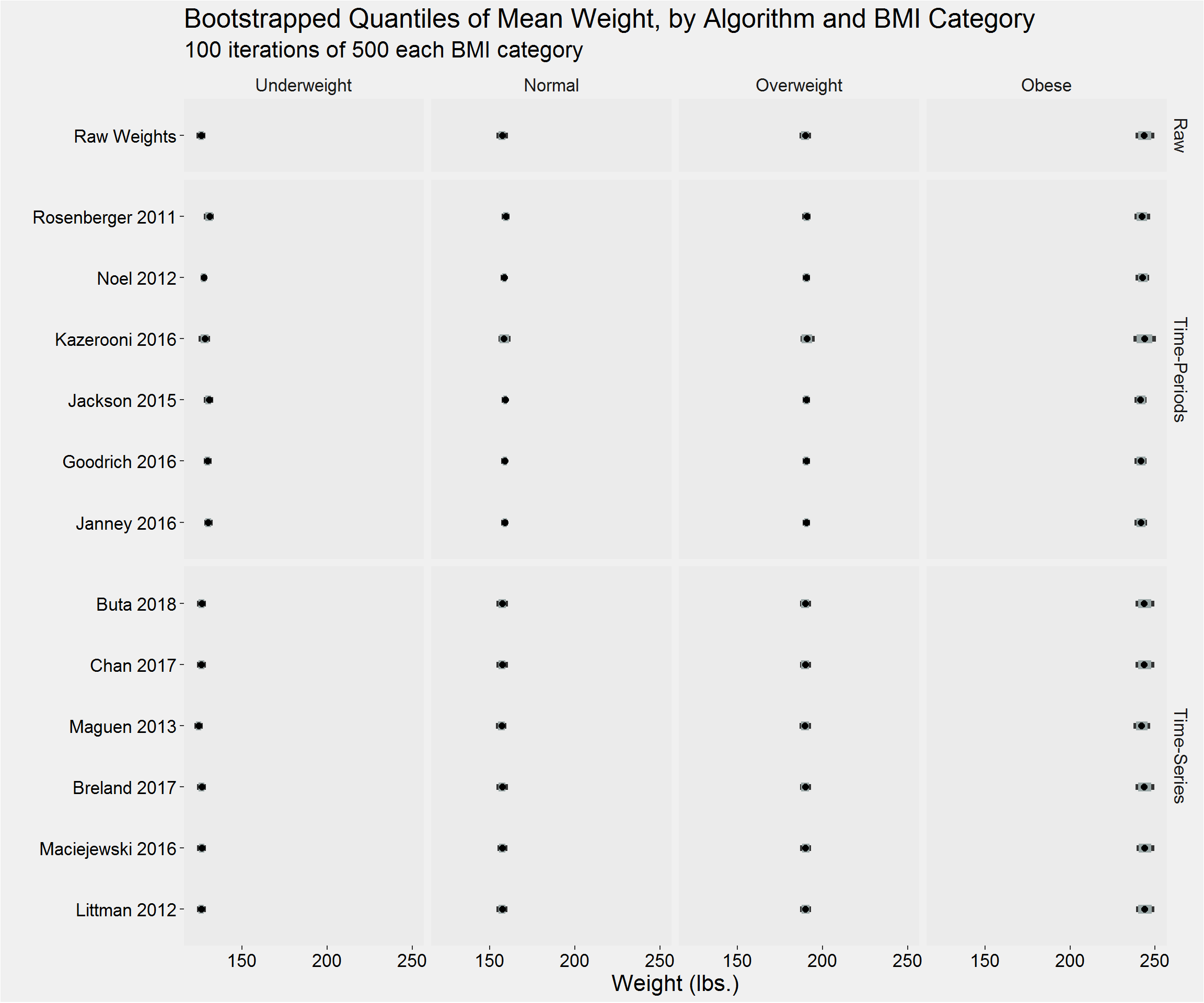
Mean weight does not change much
Distribution of the sample standard deviation,
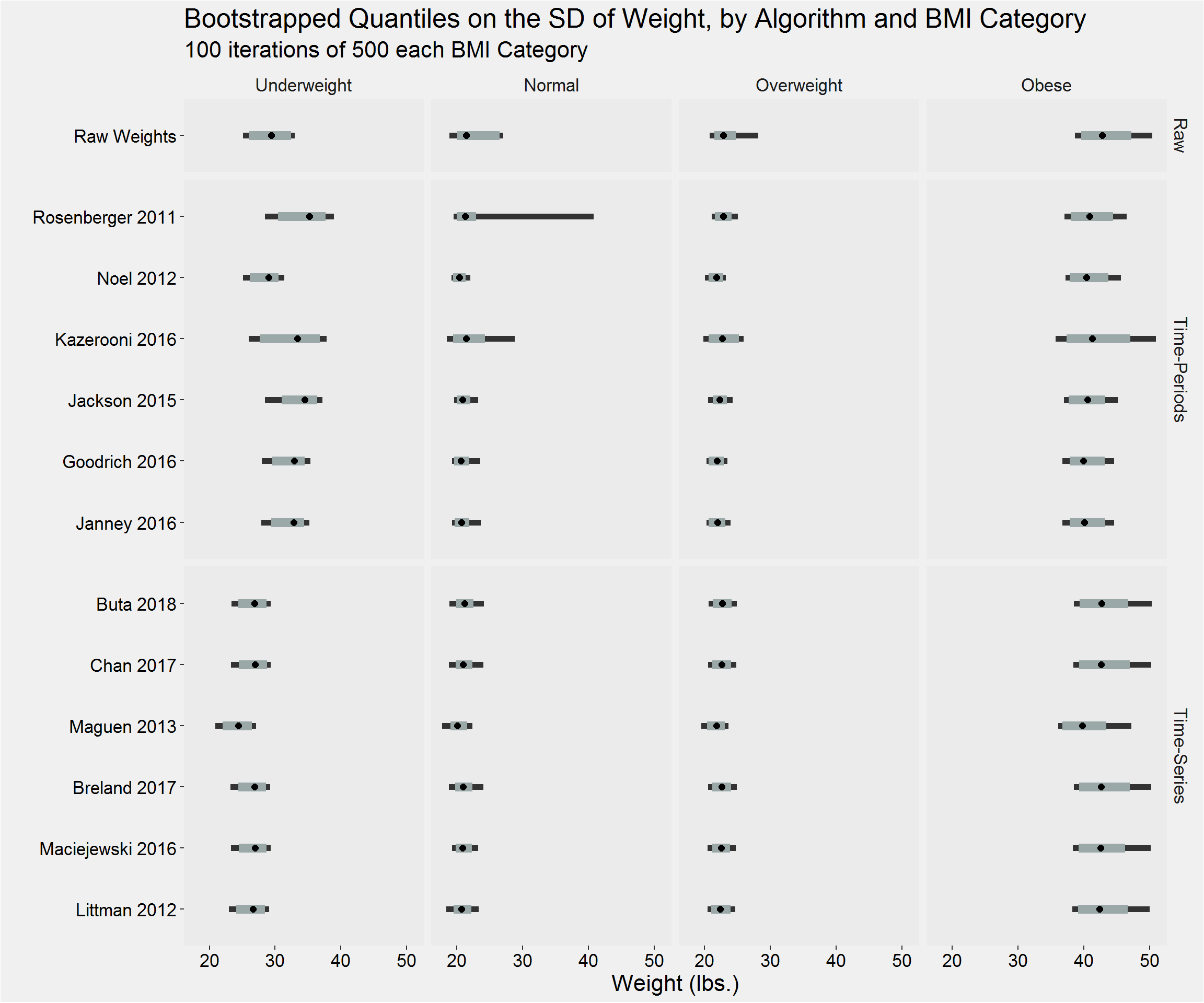
Weight Change Comparison
Add BMI category
DF <- DF %>%
filter(Cohort == "PCP 2016") %>%
left_join(
bmi_cohort %>%
distinct(PatientICN, BMI_cat),
by = "PatientICN"
) %>%
na.omit()Individual Level
Let’s look at the proportions of weight loss \(\geq5\)% from baseline, by algorithm:
DF %>%
count(Algorithm, BMI_cat, gt5perc_weightloss) %>%
group_by(Algorithm, BMI_cat) %>%
mutate(prop = prop.table(n)) %>%
filter(gt5perc_weightloss == 1) %>%
select(-gt5perc_weightloss) %>%
tableStyle()| Algorithm | BMI_cat | n | prop |
|---|---|---|---|
| Raw Weights | Underweight | 72 | 0.20 |
| Raw Weights | Normal | 1223 | 0.15 |
| Raw Weights | Overweight | 2595 | 0.13 |
| Raw Weights | Obese | 3378 | 0.14 |
| Janney 2016 | Underweight | 69 | 0.20 |
| Janney 2016 | Normal | 1176 | 0.14 |
| Janney 2016 | Overweight | 2502 | 0.13 |
| Janney 2016 | Obese | 3237 | 0.14 |
| Littman 2012 | Underweight | 70 | 0.20 |
| Littman 2012 | Normal | 1194 | 0.14 |
| Littman 2012 | Overweight | 2518 | 0.12 |
| Littman 2012 | Obese | 3223 | 0.13 |
| Maciejewski 2016 | Underweight | 71 | 0.21 |
| Maciejewski 2016 | Normal | 1197 | 0.15 |
| Maciejewski 2016 | Overweight | 2556 | 0.13 |
| Maciejewski 2016 | Obese | 3297 | 0.14 |
| Breland 2017 | Underweight | 72 | 0.20 |
| Breland 2017 | Normal | 1221 | 0.14 |
| Breland 2017 | Overweight | 2584 | 0.13 |
| Breland 2017 | Obese | 3360 | 0.14 |
| Maguen 2013 | Underweight | 55 | 0.18 |
| Maguen 2013 | Normal | 908 | 0.12 |
| Maguen 2013 | Overweight | 1736 | 0.09 |
| Maguen 2013 | Obese | 1677 | 0.08 |
| Goodrich 2016 | Underweight | 70 | 0.20 |
| Goodrich 2016 | Normal | 1175 | 0.14 |
| Goodrich 2016 | Overweight | 2497 | 0.13 |
| Goodrich 2016 | Obese | 3229 | 0.14 |
| Chan 2017 | Underweight | 71 | 0.20 |
| Chan 2017 | Normal | 1212 | 0.14 |
| Chan 2017 | Overweight | 2572 | 0.13 |
| Chan 2017 | Obese | 3336 | 0.14 |
| Jackson 2015 | Underweight | 67 | 0.20 |
| Jackson 2015 | Normal | 1208 | 0.14 |
| Jackson 2015 | Overweight | 2525 | 0.12 |
| Jackson 2015 | Obese | 3292 | 0.14 |
| Buta 2018 | Underweight | 72 | 0.20 |
| Buta 2018 | Normal | 1222 | 0.15 |
| Buta 2018 | Overweight | 2588 | 0.13 |
| Buta 2018 | Obese | 3365 | 0.14 |
| Kazerooni 2016 | Underweight | 58 | 0.30 |
| Kazerooni 2016 | Normal | 526 | 0.16 |
| Kazerooni 2016 | Overweight | 979 | 0.13 |
| Kazerooni 2016 | Obese | 1464 | 0.14 |
| Noel 2012 | Underweight | 71 | 0.21 |
| Noel 2012 | Normal | 1183 | 0.15 |
| Noel 2012 | Overweight | 2460 | 0.12 |
| Noel 2012 | Obese | 3218 | 0.14 |
| Rosenberger 2011 | Underweight | 53 | 0.22 |
| Rosenberger 2011 | Normal | 775 | 0.15 |
| Rosenberger 2011 | Overweight | 1732 | 0.13 |
| Rosenberger 2011 | Obese | 2335 | 0.14 |
Let’s look at the proportions of weight gain \(\geq5\)% from baseline, by algorithm:
DF %>%
count(Algorithm, BMI_cat, gt5perc_weightgain) %>%
group_by(Algorithm, BMI_cat) %>%
mutate(prop = prop.table(n)) %>%
filter(gt5perc_weightgain == 1) %>%
select(-gt5perc_weightgain) %>%
tableStyle()| Algorithm | BMI_cat | n | prop |
|---|---|---|---|
| Raw Weights | Underweight | 65 | 0.18 |
| Raw Weights | Normal | 1158 | 0.14 |
| Raw Weights | Overweight | 2173 | 0.11 |
| Raw Weights | Obese | 2662 | 0.11 |
| Janney 2016 | Underweight | 59 | 0.17 |
| Janney 2016 | Normal | 1120 | 0.14 |
| Janney 2016 | Overweight | 2090 | 0.11 |
| Janney 2016 | Obese | 2534 | 0.11 |
| Littman 2012 | Underweight | 55 | 0.16 |
| Littman 2012 | Normal | 1113 | 0.13 |
| Littman 2012 | Overweight | 2134 | 0.10 |
| Littman 2012 | Obese | 2595 | 0.11 |
| Maciejewski 2016 | Underweight | 60 | 0.18 |
| Maciejewski 2016 | Normal | 1132 | 0.14 |
| Maciejewski 2016 | Overweight | 2131 | 0.11 |
| Maciejewski 2016 | Obese | 2583 | 0.11 |
| Breland 2017 | Underweight | 60 | 0.17 |
| Breland 2017 | Normal | 1153 | 0.14 |
| Breland 2017 | Overweight | 2168 | 0.11 |
| Breland 2017 | Obese | 2640 | 0.11 |
| Maguen 2013 | Underweight | 44 | 0.14 |
| Maguen 2013 | Normal | 858 | 0.11 |
| Maguen 2013 | Overweight | 1408 | 0.08 |
| Maguen 2013 | Obese | 1182 | 0.06 |
| Goodrich 2016 | Underweight | 58 | 0.17 |
| Goodrich 2016 | Normal | 1119 | 0.14 |
| Goodrich 2016 | Overweight | 2086 | 0.10 |
| Goodrich 2016 | Obese | 2530 | 0.11 |
| Chan 2017 | Underweight | 63 | 0.18 |
| Chan 2017 | Normal | 1147 | 0.14 |
| Chan 2017 | Overweight | 2154 | 0.10 |
| Chan 2017 | Obese | 2624 | 0.11 |
| Jackson 2015 | Underweight | 51 | 0.15 |
| Jackson 2015 | Normal | 1117 | 0.13 |
| Jackson 2015 | Overweight | 2013 | 0.10 |
| Jackson 2015 | Obese | 2430 | 0.10 |
| Buta 2018 | Underweight | 60 | 0.17 |
| Buta 2018 | Normal | 1155 | 0.14 |
| Buta 2018 | Overweight | 2172 | 0.11 |
| Buta 2018 | Obese | 2649 | 0.11 |
| Kazerooni 2016 | Underweight | 24 | 0.12 |
| Kazerooni 2016 | Normal | 448 | 0.14 |
| Kazerooni 2016 | Overweight | 742 | 0.10 |
| Kazerooni 2016 | Obese | 989 | 0.09 |
| Noel 2012 | Underweight | 58 | 0.17 |
| Noel 2012 | Normal | 1086 | 0.13 |
| Noel 2012 | Overweight | 2054 | 0.10 |
| Noel 2012 | Obese | 2528 | 0.11 |
| Rosenberger 2011 | Underweight | 34 | 0.14 |
| Rosenberger 2011 | Normal | 756 | 0.14 |
| Rosenberger 2011 | Overweight | 1463 | 0.11 |
| Rosenberger 2011 | Obese | 1884 | 0.12 |
Average weight change:
DF %>%
filter(!is.na(diff)) %>%
select(Algorithm, BMI_cat, diff) %>%
group_by(Algorithm, BMI_cat) %>%
summarize(
mean = mean(diff),
SD = sd(diff),
min = min(diff),
Q1 = fivenum(diff)[2],
median = median(diff),
Q3 = fivenum(diff)[4],
max = max(diff)
) %>%
tableStyle()| Algorithm | BMI_cat | mean | SD | min | Q1 | median | Q3 | max |
|---|---|---|---|---|---|---|---|---|
| Raw Weights | Underweight | 2.24 | 23.78 | -57.00 | -4.60 | -0.10 | 3.85 | 195.80 |
| Raw Weights | Normal | -0.21 | 10.07 | -148.00 | -4.80 | -0.37 | 4.00 | 185.20 |
| Raw Weights | Overweight | -0.54 | 11.64 | -536.00 | -5.10 | -0.20 | 4.40 | 281.97 |
| Raw Weights | Obese | -0.95 | 15.44 | -509.50 | -6.80 | -0.20 | 5.80 | 296.10 |
| Janney 2016 | Underweight | 0.21 | 15.44 | -57.00 | -4.60 | -0.25 | 3.60 | 195.80 |
| Janney 2016 | Normal | -0.20 | 9.60 | -118.60 | -4.80 | -0.37 | 4.00 | 185.20 |
| Janney 2016 | Overweight | -0.48 | 10.68 | -291.40 | -5.00 | -0.20 | 4.31 | 281.97 |
| Janney 2016 | Obese | -0.97 | 13.65 | -159.20 | -6.70 | -0.20 | 5.80 | 145.50 |
| Littman 2012 | Underweight | -0.65 | 8.78 | -34.40 | -4.60 | -0.30 | 3.40 | 31.80 |
| Littman 2012 | Normal | -0.31 | 8.68 | -118.60 | -4.80 | -0.40 | 4.00 | 54.70 |
| Littman 2012 | Overweight | -0.43 | 9.25 | -67.20 | -5.00 | -0.20 | 4.40 | 70.50 |
| Littman 2012 | Obese | -0.75 | 12.55 | -111.80 | -6.60 | -0.10 | 5.80 | 108.00 |
| Maciejewski 2016 | Underweight | 0.78 | 17.64 | -46.00 | -5.00 | -0.50 | 3.70 | 195.80 |
| Maciejewski 2016 | Normal | -0.23 | 9.09 | -55.20 | -4.90 | -0.40 | 4.10 | 70.30 |
| Maciejewski 2016 | Overweight | -0.52 | 9.59 | -80.74 | -5.20 | -0.20 | 4.50 | 90.00 |
| Maciejewski 2016 | Obese | -0.99 | 13.07 | -117.20 | -7.00 | -0.20 | 6.00 | 99.80 |
| Breland 2017 | Underweight | 0.16 | 15.27 | -57.00 | -4.65 | -0.30 | 3.45 | 195.80 |
| Breland 2017 | Normal | -0.22 | 9.18 | -58.80 | -4.80 | -0.38 | 4.00 | 78.80 |
| Breland 2017 | Overweight | -0.49 | 9.80 | -80.86 | -5.07 | -0.20 | 4.40 | 207.20 |
| Breland 2017 | Obese | -0.95 | 13.36 | -117.20 | -6.70 | -0.20 | 5.80 | 152.60 |
| Maguen 2013 | Underweight | -0.30 | 7.07 | -22.45 | -4.35 | -0.35 | 3.10 | 30.70 |
| Maguen 2013 | Normal | -0.24 | 7.07 | -32.00 | -4.20 | -0.30 | 3.70 | 57.00 |
| Maguen 2013 | Overweight | -0.38 | 7.16 | -44.50 | -4.60 | -0.12 | 4.00 | 45.00 |
| Maguen 2013 | Obese | -0.54 | 8.59 | -73.20 | -5.40 | -0.10 | 4.70 | 98.40 |
| Goodrich 2016 | Underweight | -0.37 | 11.25 | -57.00 | -4.65 | -0.30 | 3.45 | 79.00 |
| Goodrich 2016 | Normal | -0.21 | 9.28 | -95.10 | -4.80 | -0.37 | 4.00 | 78.80 |
| Goodrich 2016 | Overweight | -0.48 | 9.82 | -82.30 | -5.00 | -0.20 | 4.30 | 205.20 |
| Goodrich 2016 | Obese | -0.95 | 13.43 | -117.20 | -6.70 | -0.20 | 5.80 | 145.50 |
| Chan 2017 | Underweight | 1.62 | 20.83 | -46.00 | -4.60 | -0.15 | 3.70 | 195.80 |
| Chan 2017 | Normal | -0.19 | 9.26 | -58.80 | -4.80 | -0.37 | 4.00 | 98.12 |
| Chan 2017 | Overweight | -0.49 | 10.16 | -245.37 | -5.00 | -0.20 | 4.31 | 278.27 |
| Chan 2017 | Obese | -0.96 | 13.88 | -509.50 | -6.70 | -0.20 | 5.80 | 182.15 |
| Jackson 2015 | Underweight | -0.13 | 15.40 | -55.75 | -5.00 | -1.00 | 3.25 | 195.80 |
| Jackson 2015 | Normal | -0.22 | 9.42 | -156.50 | -4.90 | -0.40 | 4.00 | 199.97 |
| Jackson 2015 | Overweight | -0.56 | 9.68 | -245.59 | -5.17 | -0.38 | 4.20 | 230.39 |
| Jackson 2015 | Obese | -1.15 | 13.07 | -126.70 | -7.00 | -0.40 | 5.50 | 122.60 |
| Buta 2018 | Underweight | 0.17 | 15.29 | -57.00 | -4.70 | -0.30 | 3.50 | 195.80 |
| Buta 2018 | Normal | -0.21 | 9.61 | -118.60 | -4.80 | -0.40 | 4.00 | 185.20 |
| Buta 2018 | Overweight | -0.48 | 10.27 | -245.37 | -5.07 | -0.20 | 4.40 | 278.27 |
| Buta 2018 | Obese | -0.94 | 13.75 | -159.20 | -6.70 | -0.20 | 5.80 | 152.60 |
| Kazerooni 2016 | Underweight | -1.29 | 14.04 | -28.70 | -7.45 | -1.20 | 3.10 | 142.37 |
| Kazerooni 2016 | Normal | -0.40 | 9.47 | -103.30 | -5.20 | -0.70 | 4.20 | 86.20 |
| Kazerooni 2016 | Overweight | -0.79 | 11.58 | -534.00 | -5.50 | -0.60 | 4.10 | 154.30 |
| Kazerooni 2016 | Obese | -1.40 | 14.08 | -165.97 | -7.20 | -0.80 | 5.00 | 299.30 |
| Noel 2012 | Underweight | 1.03 | 18.70 | -57.00 | -5.00 | -0.55 | 3.50 | 195.80 |
| Noel 2012 | Normal | -0.27 | 9.13 | -128.05 | -4.90 | -0.40 | 4.00 | 84.85 |
| Noel 2012 | Overweight | -0.48 | 9.75 | -245.59 | -5.00 | -0.20 | 4.40 | 142.13 |
| Noel 2012 | Obese | -0.92 | 13.26 | -119.60 | -6.75 | -0.20 | 5.90 | 145.00 |
| Rosenberger 2011 | Underweight | -0.36 | 12.58 | -57.00 | -5.20 | -0.80 | 3.20 | 102.80 |
| Rosenberger 2011 | Normal | -0.22 | 9.62 | -141.00 | -5.00 | -0.50 | 4.20 | 78.20 |
| Rosenberger 2011 | Overweight | -0.61 | 11.61 | -538.00 | -5.30 | -0.31 | 4.50 | 154.30 |
| Rosenberger 2011 | Obese | -1.02 | 15.42 | -509.50 | -7.00 | -0.20 | 6.00 | 298.70 |
Average Weight Change for those losing \(\ge 5\)% weight loss:
DF %>%
filter(!is.na(diff) & gt5perc_weightloss == 1) %>%
select(Algorithm, BMI_cat, diff) %>%
group_by(Algorithm, BMI_cat) %>%
summarize(
mean = mean(diff),
SD = sd(diff),
min = min(diff),
Q1 = fivenum(diff)[2],
median = median(diff),
Q3 = fivenum(diff)[4],
max = max(diff)
) %>%
tableStyle()| Algorithm | BMI_cat | mean | SD | min | Q1 | median | Q3 | max |
|---|---|---|---|---|---|---|---|---|
| Raw Weights | Underweight | -13.64 | 8.86 | -57.00 | -16.00 | -11.05 | -7.75 | -5.00 |
| Raw Weights | Normal | -14.47 | 9.26 | -148.00 | -16.45 | -12.10 | -9.60 | -6.00 |
| Raw Weights | Overweight | -17.43 | 16.44 | -536.00 | -19.20 | -14.10 | -11.30 | -7.20 |
| Raw Weights | Obese | -23.85 | 18.99 | -509.50 | -27.00 | -19.00 | -14.80 | -9.00 |
| Janney 2016 | Underweight | -13.88 | 8.96 | -57.00 | -16.00 | -11.10 | -8.00 | -5.55 |
| Janney 2016 | Normal | -14.20 | 7.61 | -118.60 | -16.30 | -12.05 | -9.60 | -6.00 |
| Janney 2016 | Overweight | -16.94 | 11.31 | -291.40 | -19.00 | -14.00 | -11.29 | -7.20 |
| Janney 2016 | Obese | -23.14 | 13.69 | -159.20 | -26.80 | -19.00 | -14.70 | -9.00 |
| Littman 2012 | Underweight | -12.52 | 6.06 | -34.40 | -15.80 | -10.91 | -7.50 | -5.00 |
| Littman 2012 | Normal | -13.77 | 6.45 | -118.60 | -16.00 | -12.00 | -9.60 | -6.00 |
| Littman 2012 | Overweight | -16.08 | 7.06 | -67.20 | -18.60 | -14.00 | -11.20 | -7.20 |
| Littman 2012 | Obese | -21.71 | 10.65 | -111.80 | -25.30 | -18.60 | -14.60 | -9.00 |
| Maciejewski 2016 | Underweight | -13.02 | 7.23 | -46.00 | -16.00 | -11.00 | -7.75 | -5.00 |
| Maciejewski 2016 | Normal | -13.93 | 6.22 | -55.20 | -16.30 | -12.00 | -9.60 | -6.00 |
| Maciejewski 2016 | Overweight | -16.53 | 7.98 | -80.74 | -19.00 | -14.00 | -11.25 | -7.20 |
| Maciejewski 2016 | Obese | -22.60 | 12.28 | -117.20 | -26.20 | -18.80 | -14.60 | -9.00 |
| Breland 2017 | Underweight | -13.64 | 8.86 | -57.00 | -16.00 | -11.05 | -7.75 | -5.00 |
| Breland 2017 | Normal | -14.08 | 6.54 | -58.80 | -16.40 | -12.10 | -9.60 | -6.00 |
| Breland 2017 | Overweight | -16.67 | 8.27 | -80.86 | -19.05 | -14.10 | -11.30 | -7.20 |
| Breland 2017 | Obese | -22.93 | 12.74 | -117.20 | -26.85 | -19.00 | -14.70 | -9.00 |
| Maguen 2013 | Underweight | -10.10 | 3.70 | -22.45 | -11.65 | -9.50 | -7.30 | -5.00 |
| Maguen 2013 | Normal | -11.94 | 4.00 | -32.00 | -13.80 | -11.00 | -9.00 | -6.00 |
| Maguen 2013 | Overweight | -13.71 | 4.81 | -44.50 | -15.20 | -12.50 | -10.70 | -7.20 |
| Maguen 2013 | Obese | -17.40 | 6.89 | -73.20 | -19.30 | -15.40 | -13.00 | -9.00 |
| Goodrich 2016 | Underweight | -13.75 | 8.96 | -57.00 | -16.00 | -11.05 | -7.50 | -5.00 |
| Goodrich 2016 | Normal | -14.11 | 6.97 | -95.10 | -16.30 | -12.00 | -9.60 | -6.00 |
| Goodrich 2016 | Overweight | -16.66 | 8.39 | -82.30 | -19.00 | -14.00 | -11.20 | -7.20 |
| Goodrich 2016 | Obese | -22.95 | 12.98 | -117.20 | -26.60 | -19.00 | -14.70 | -9.00 |
| Chan 2017 | Underweight | -13.03 | 7.24 | -46.00 | -16.00 | -11.00 | -7.75 | -5.00 |
| Chan 2017 | Normal | -14.01 | 6.53 | -58.80 | -16.25 | -12.00 | -9.60 | -6.00 |
| Chan 2017 | Overweight | -16.75 | 9.70 | -245.37 | -19.00 | -14.00 | -11.20 | -7.20 |
| Chan 2017 | Obese | -23.10 | 15.63 | -509.50 | -26.61 | -19.00 | -14.70 | -9.00 |
| Jackson 2015 | Underweight | -13.80 | 8.75 | -55.75 | -16.10 | -11.80 | -8.15 | -5.00 |
| Jackson 2015 | Normal | -13.87 | 7.78 | -156.50 | -15.77 | -11.95 | -9.50 | -5.90 |
| Jackson 2015 | Overweight | -16.36 | 8.94 | -245.59 | -18.77 | -13.90 | -11.30 | -6.73 |
| Jackson 2015 | Obese | -22.55 | 12.82 | -126.70 | -25.59 | -18.60 | -14.50 | -8.35 |
| Buta 2018 | Underweight | -13.64 | 8.86 | -57.00 | -16.00 | -11.05 | -7.75 | -5.00 |
| Buta 2018 | Normal | -14.25 | 7.62 | -118.60 | -16.40 | -12.10 | -9.60 | -6.00 |
| Buta 2018 | Overweight | -16.85 | 9.81 | -245.37 | -19.10 | -14.10 | -11.30 | -7.20 |
| Buta 2018 | Obese | -23.17 | 13.60 | -159.20 | -27.00 | -19.00 | -14.70 | -9.00 |
| Kazerooni 2016 | Underweight | -12.37 | 5.70 | -28.70 | -16.00 | -10.85 | -8.00 | -6.00 |
| Kazerooni 2016 | Normal | -13.54 | 7.65 | -103.30 | -15.20 | -11.12 | -9.10 | -6.00 |
| Kazerooni 2016 | Overweight | -16.73 | 18.99 | -534.00 | -18.40 | -13.60 | -11.10 | -7.30 |
| Kazerooni 2016 | Obese | -22.79 | 13.07 | -165.97 | -26.20 | -19.00 | -14.74 | -8.00 |
| Noel 2012 | Underweight | -13.13 | 8.43 | -57.00 | -16.00 | -11.00 | -8.03 | -5.00 |
| Noel 2012 | Normal | -13.86 | 7.12 | -128.05 | -16.00 | -12.00 | -9.50 | -6.00 |
| Noel 2012 | Overweight | -16.66 | 9.39 | -245.59 | -19.17 | -14.00 | -11.32 | -6.95 |
| Noel 2012 | Obese | -22.74 | 12.80 | -119.60 | -26.01 | -18.80 | -14.50 | -7.40 |
| Rosenberger 2011 | Underweight | -13.20 | 9.27 | -57.00 | -14.09 | -10.70 | -8.00 | -5.00 |
| Rosenberger 2011 | Normal | -14.31 | 8.38 | -141.00 | -16.20 | -12.10 | -9.69 | -6.00 |
| Rosenberger 2011 | Overweight | -17.60 | 16.97 | -538.00 | -19.83 | -14.30 | -11.40 | -7.50 |
| Rosenberger 2011 | Obese | -23.98 | 18.99 | -509.50 | -27.20 | -19.00 | -14.70 | -9.00 |
Average Weight Change for those gaining \(\ge 5\)% weight loss:
DF %>%
filter(!is.na(diff) & gt5perc_weightgain == 1) %>%
select(Algorithm, BMI_cat, diff) %>%
group_by(Algorithm, BMI_cat) %>%
summarize(
mean = mean(diff),
SD = sd(diff),
min = min(diff),
Q1 = fivenum(diff)[2],
median = median(diff),
Q3 = fivenum(diff)[4],
max = max(diff)
) %>%
tableStyle()| Algorithm | BMI_cat | mean | SD | min | Q1 | median | Q3 | max |
|---|---|---|---|---|---|---|---|---|
| Raw Weights | Underweight | 28.14 | 45.27 | 5.50 | 8.00 | 11.30 | 21.61 | 195.80 |
| Raw Weights | Normal | 15.07 | 10.89 | 5.00 | 9.80 | 12.05 | 16.80 | 185.20 |
| Raw Weights | Overweight | 16.54 | 12.92 | 7.20 | 11.00 | 13.50 | 18.00 | 281.97 |
| Raw Weights | Obese | 21.49 | 17.19 | 8.20 | 14.00 | 17.60 | 23.30 | 296.10 |
| Janney 2016 | Underweight | 18.20 | 26.68 | 5.50 | 7.95 | 11.00 | 18.10 | 195.80 |
| Janney 2016 | Normal | 14.82 | 9.83 | 5.00 | 9.80 | 12.05 | 16.65 | 185.20 |
| Janney 2016 | Overweight | 16.48 | 12.67 | 7.20 | 11.00 | 13.50 | 18.00 | 281.97 |
| Janney 2016 | Obese | 20.56 | 11.49 | 8.20 | 14.00 | 17.50 | 23.00 | 145.50 |
| Littman 2012 | Underweight | 12.32 | 6.33 | 5.50 | 7.90 | 10.30 | 14.85 | 31.80 |
| Littman 2012 | Normal | 13.85 | 6.43 | 5.20 | 9.70 | 12.00 | 16.00 | 54.70 |
| Littman 2012 | Overweight | 15.66 | 7.11 | 7.20 | 11.00 | 13.40 | 18.00 | 70.50 |
| Littman 2012 | Obese | 19.90 | 9.48 | 8.20 | 14.00 | 17.40 | 22.90 | 108.00 |
| Maciejewski 2016 | Underweight | 20.58 | 32.48 | 5.45 | 7.95 | 11.00 | 18.65 | 195.80 |
| Maciejewski 2016 | Normal | 14.40 | 7.63 | 5.20 | 9.80 | 12.00 | 16.60 | 70.30 |
| Maciejewski 2016 | Overweight | 15.77 | 7.35 | 7.20 | 11.00 | 13.50 | 18.00 | 90.00 |
| Maciejewski 2016 | Obese | 19.70 | 8.83 | 8.20 | 13.90 | 17.30 | 22.60 | 99.80 |
| Breland 2017 | Underweight | 18.05 | 26.48 | 5.50 | 7.95 | 11.00 | 18.10 | 195.80 |
| Breland 2017 | Normal | 14.61 | 8.12 | 5.00 | 9.80 | 12.00 | 16.60 | 78.80 |
| Breland 2017 | Overweight | 16.09 | 9.12 | 7.20 | 11.00 | 13.50 | 18.00 | 207.20 |
| Breland 2017 | Obese | 20.32 | 10.59 | 8.20 | 14.00 | 17.50 | 23.00 | 152.60 |
| Maguen 2013 | Underweight | 11.11 | 5.65 | 5.50 | 7.55 | 9.45 | 11.75 | 30.70 |
| Maguen 2013 | Normal | 12.18 | 5.24 | 5.20 | 9.00 | 11.00 | 13.60 | 57.00 |
| Maguen 2013 | Overweight | 13.12 | 4.03 | 7.20 | 10.38 | 12.00 | 14.65 | 45.00 |
| Maguen 2013 | Obese | 15.84 | 5.15 | 8.20 | 12.80 | 14.80 | 17.80 | 98.40 |
| Goodrich 2016 | Underweight | 15.14 | 12.70 | 5.50 | 7.90 | 11.00 | 18.00 | 79.00 |
| Goodrich 2016 | Normal | 14.67 | 8.40 | 5.00 | 9.80 | 12.00 | 16.60 | 78.80 |
| Goodrich 2016 | Overweight | 16.14 | 9.25 | 7.20 | 11.00 | 13.50 | 18.00 | 205.20 |
| Goodrich 2016 | Obese | 20.42 | 10.95 | 8.20 | 14.00 | 17.40 | 23.00 | 145.50 |
| Chan 2017 | Underweight | 24.53 | 39.89 | 5.50 | 8.00 | 11.20 | 20.25 | 195.80 |
| Chan 2017 | Normal | 14.70 | 8.83 | 5.00 | 9.80 | 12.00 | 16.60 | 98.12 |
| Chan 2017 | Overweight | 16.17 | 10.83 | 7.20 | 11.00 | 13.50 | 18.00 | 278.27 |
| Chan 2017 | Obese | 20.38 | 11.50 | 8.20 | 14.00 | 17.40 | 23.00 | 182.15 |
| Jackson 2015 | Underweight | 19.07 | 28.57 | 5.80 | 7.89 | 10.37 | 19.17 | 195.80 |
| Jackson 2015 | Normal | 14.74 | 9.59 | 5.74 | 9.80 | 12.28 | 16.60 | 199.97 |
| Jackson 2015 | Overweight | 16.02 | 9.60 | 6.50 | 11.00 | 13.43 | 18.00 | 230.39 |
| Jackson 2015 | Obese | 20.00 | 10.39 | 8.00 | 13.78 | 17.00 | 22.80 | 122.60 |
| Buta 2018 | Underweight | 18.05 | 26.48 | 5.50 | 7.95 | 11.00 | 18.10 | 195.80 |
| Buta 2018 | Normal | 14.84 | 9.78 | 5.00 | 9.80 | 12.00 | 16.70 | 185.20 |
| Buta 2018 | Overweight | 16.32 | 11.12 | 7.20 | 11.00 | 13.50 | 18.00 | 278.27 |
| Buta 2018 | Obese | 20.69 | 12.12 | 8.20 | 14.00 | 17.56 | 23.08 | 152.60 |
| Kazerooni 2016 | Underweight | 18.73 | 27.46 | 6.50 | 7.55 | 11.45 | 17.75 | 142.37 |
| Kazerooni 2016 | Normal | 14.66 | 8.71 | 5.80 | 9.70 | 12.00 | 16.15 | 86.20 |
| Kazerooni 2016 | Overweight | 16.21 | 10.17 | 7.00 | 10.90 | 13.60 | 18.10 | 154.30 |
| Kazerooni 2016 | Obese | 21.14 | 18.80 | 9.00 | 13.50 | 16.60 | 22.60 | 299.30 |
| Noel 2012 | Underweight | 23.55 | 34.45 | 5.45 | 8.70 | 11.58 | 21.70 | 195.80 |
| Noel 2012 | Normal | 14.49 | 8.07 | 5.65 | 9.70 | 12.10 | 16.60 | 84.85 |
| Noel 2012 | Overweight | 15.94 | 8.35 | 6.95 | 11.00 | 13.50 | 18.00 | 142.13 |
| Noel 2012 | Obese | 20.13 | 10.75 | 8.20 | 13.90 | 17.20 | 22.87 | 145.00 |
| Rosenberger 2011 | Underweight | 18.29 | 18.77 | 5.80 | 7.90 | 11.50 | 21.40 | 102.80 |
| Rosenberger 2011 | Normal | 14.78 | 8.16 | 5.20 | 10.00 | 12.40 | 16.75 | 78.20 |
| Rosenberger 2011 | Overweight | 16.37 | 9.23 | 7.20 | 11.00 | 13.70 | 18.40 | 154.30 |
| Rosenberger 2011 | Obese | 20.89 | 15.08 | 7.91 | 14.00 | 17.60 | 23.20 | 298.70 |
Visual summaries
p <- DF %>%
filter(!is.na(diff)) %>%
ggplot(aes(x = diff, y = Algorithm, fill = Algorithm)) %>%
add(ggridges::geom_density_ridges(
jittered_points = TRUE,
point_shape = "|",
position = ggridges::position_points_jitter(height = 0),
scale = 3,
size = 0.25,
rel_min_height = 10^-5
)) %>%
add(facet_grid(Type ~ BMI_cat, scales = "free_y", space = "free_y")) %>%
add(scale_fill_viridis_d(option = "A")) %>%
add(theme(
legend.position = "none",
text = element_text(size = 20),
axis.text = element_text(color = "black"),
panel.grid = element_blank(),
strip.background = element_blank()
)) %>%
add(labs(
x = "Weight Change (Lbs.)",
y = "",
title = "Weight Change 1-year Post Baseline"
))
p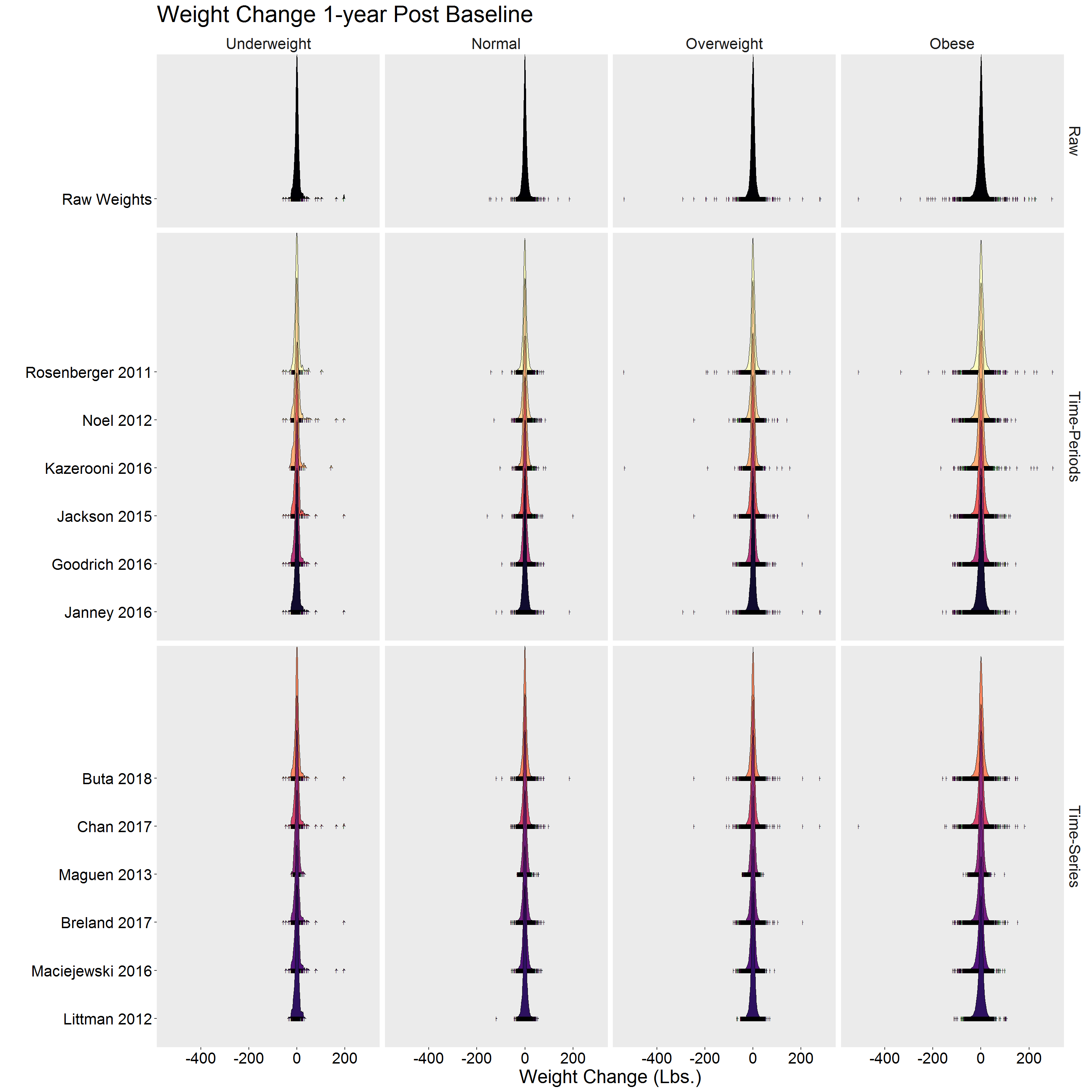
No discernible differences
Alternative view
p <- DF %>%
filter(!is.na(diff)) %>%
group_by(Algorithm) %>%
mutate(dens = approxfun(density(diff))(diff)) %>%
ggplot(aes(y = Algorithm, x = diff, color = dens)) %>%
add(geom_jitter(size = 2)) %>%
add(scale_color_viridis_c()) %>%
add(facet_grid(Type ~ BMI_cat, scales = "free_y", space = "free_y")) %>%
add(theme(
legend.position = "none",
text = element_text(size = 20),
axis.text = element_text(color = "black"),
panel.grid = element_blank(),
strip.background = element_blank()
)) %>%
add(labs(
x = "Weight Change (Lbs.)",
y = "",
title = ""
))
p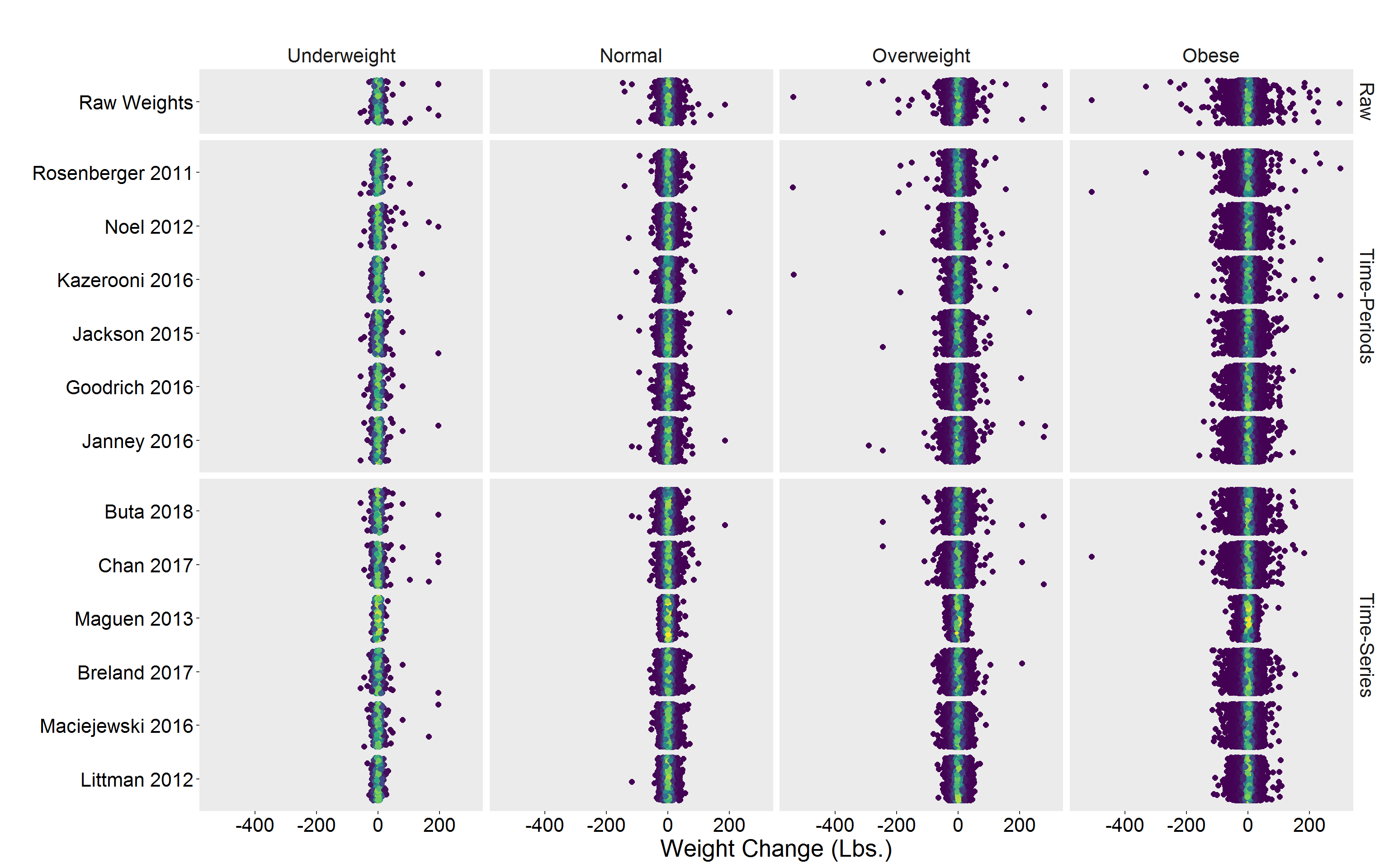
Site/Facility Level Analyses
Sta3n_wtdx_diff <- DF %>%
select(Algorithm, BMI_cat, Sta3n, diff, diff_perc) %>%
reshape2::melt(id.vars = c("BMI_cat", "Algorithm", "Sta3n")) %>%
group_by(BMI_cat, Algorithm, Sta3n, variable) %>%
na.omit() %>%
summarize(
n = n(),
mean = mean(value),
median = median(value)
) %>%
pivot_wider(
names_from = variable,
values_from = c("n", "mean", "median")
)
Sta3n_wtpct <- DF %>%
select(BMI_cat, Algorithm, Sta3n, contains("weight")) %>%
reshape2::melt(id.vars = c("BMI_cat", "Algorithm", "Sta3n")) %>%
group_by(BMI_cat, Algorithm, Sta3n, variable) %>%
summarize(prop = sum(value == 1) / n()) %>%
pivot_wider(names_from = variable, values_from = "prop")
Sta3n_wtdx <- Sta3n_wtdx_diff %>%
left_join(Sta3n_wtpct, by = c("BMI_cat", "Algorithm", "Sta3n")) %>%
left_join(
DF %>%
distinct(Algorithm, Type),
by = "Algorithm"
) %>%
select(
Type,
BMI_cat,
Sta3n,
n_diff,
mean_diff,
median_diff,
mean_diff_perc,
median_diff_perc,
gt5perc_weightloss,
gt5perc_weightgain
) %>%
ungroup()
rm(Sta3n_wtdx_diff, Sta3n_wtpct)
COLS <- c("#2E364F", "#2D5D7C", "#F3F0E2", "#EF5939")
p <- Sta3n_wtdx %>%
ggplot(aes(mean_diff, group = BMI_cat, fill = BMI_cat)) %>%
add(geom_histogram(bins = 100, alpha = 0.7, color = "black")) %>%
add(theme_minimal(16)) %>%
add(theme(legend.position = "none")) %>%
add(scale_fill_manual(values = COLS)) %>%
add(labs(
x = "Weight Change (lbs.)",
y = "Frequency",
fill = ""
))
q <- Sta3n_wtdx %>%
filter(!is.infinite(mean_diff_perc)) %>%
ggplot(aes(mean_diff_perc, group = BMI_cat, fill = BMI_cat)) %>%
add(geom_histogram(bins = 100, alpha = 0.7, color = "black")) %>%
add(theme_minimal(16)) %>%
add(theme(legend.position = c(0.8, 0.8))) %>%
add(scale_fill_manual(values = COLS)) %>%
add(labs(
x = "Weight Change %",
y = "Frequency",
fill = ""
))
r <- Sta3n_wtdx %>%
ggplot(aes(gt5perc_weightloss, group = BMI_cat, fill = BMI_cat)) %>%
add(geom_histogram(bins = 100, alpha = 0.7, color = "black")) %>%
add(theme_minimal(16)) %>%
add(theme(legend.position = "none")) %>%
add(scale_fill_manual(values = COLS)) %>%
add(labs(
x = ">= 5% Weight Loss",
y = "Frequency"
))
s <- Sta3n_wtdx %>%
filter(!is.na(gt5perc_weightgain)) %>%
ggplot(aes(gt5perc_weightgain, group = BMI_cat, fill = BMI_cat)) %>%
add(geom_histogram(bins = 100, alpha = 0.7, color = "black")) %>%
add(theme_minimal(16)) %>%
add(theme(legend.position = "none")) %>%
add(scale_fill_manual(values = COLS)) %>%
add(labs(
x = ">= 5% Weight Gain",
y = "Frequency"
))
library(patchwork)
p + q + r + s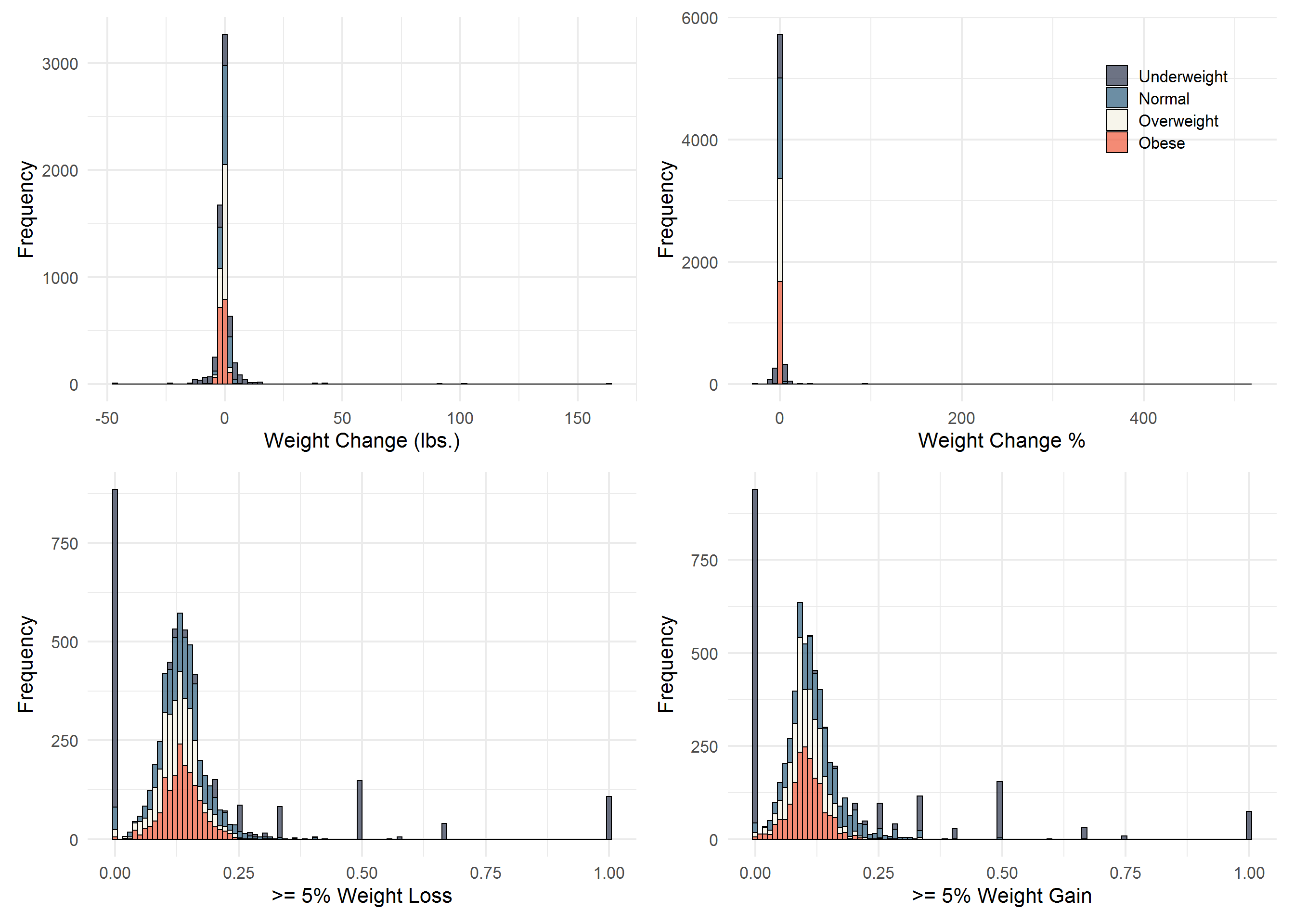
library(gridExtra)
tmp <- Sta3n_wtdx %>%
select(BMI_cat, Type, Algorithm, Sta3n, gt5perc_weightloss)
raw <- tmp %>%
filter(Algorithm == "Raw Weights") %>%
group_by(BMI_cat) %>%
arrange(gt5perc_weightloss) %>%
mutate(Sta3n_rank = row_number()) %>%
select(-Type, -Algorithm) %>%
rename(raw_wtloss = gt5perc_weightloss)
tp <- tmp %>%
filter(Type == "Time-Periods") %>%
left_join(raw, by = c("Sta3n", "BMI_cat")) %>%
mutate(loss_diff = gt5perc_weightloss - raw_wtloss)
ts <- tmp %>%
filter(Type == "Time-Series") %>%
left_join(raw, by = c("Sta3n", "BMI_cat")) %>%
mutate(loss_diff = gt5perc_weightloss - raw_wtloss)
p1 <- tp %>%
ggplot(aes(x = Sta3n_rank, y = raw_wtloss)) %>%
add(facet_wrap(vars(Type, BMI_cat))) %>%
add(geom_line(size = 1)) %>%
add(geom_segment(
aes(
x = Sta3n_rank,
y = raw_wtloss,
xend = Sta3n_rank,
yend = gt5perc_weightloss
),
alpha = 0.3
)) %>%
add(geom_point(
aes(
x = Sta3n_rank,
y = gt5perc_weightloss,
color = Algorithm
),
size = 2,
alpha = 0.7
)) %>%
add(expand_limits(y = c(0.0, 0.3))) %>%
add(theme_minimal(16)) %>%
add(theme(
legend.position = "bottom",
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank()
)) %>%
add(guides(color = guide_legend(override.aes = list(size = 5)))) %>%
add(scale_color_brewer(palette = "Dark2")) %>%
add(labs(
x = "Sta3n",
y = "Proportion Achieving 5% Weight Loss",
color = ""
))
p2 <- p1 %+% ts
p2 <- p2 %>% add(labs(y = ""))
grid.arrange(p1, p2, nrow = 2)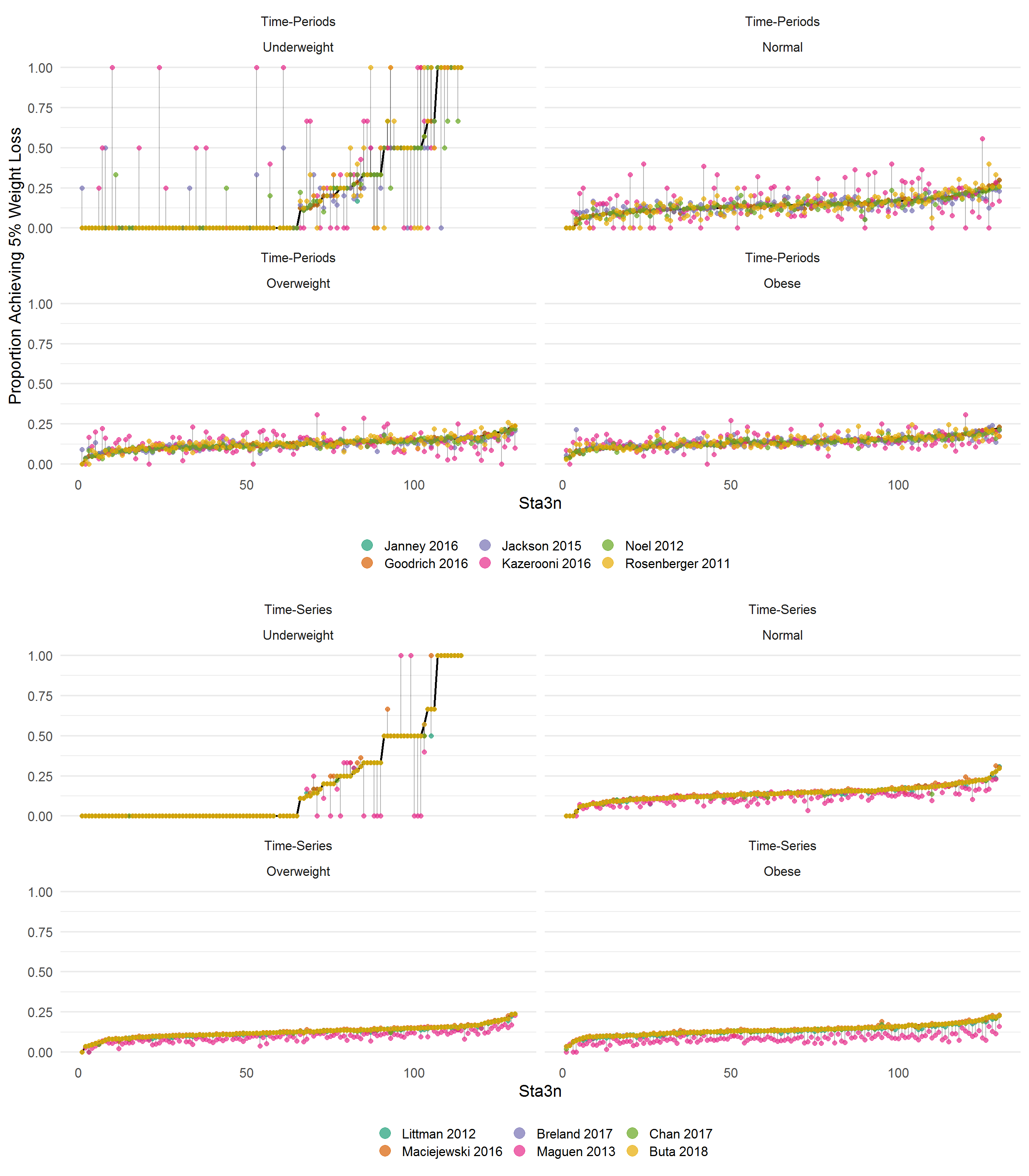
Sites are ordered/ranked along the x-axis using the percent of patients with at least 5% weight loss using the raw data. The left hand side shows the “time-period” algorithms, while the right displays the “time-series” algorithms. Results suggest that generally, time-series algorithms may result in more similar rates by site, save for Maguen 2013. Results are more mixed using the “time-period” algorithms.