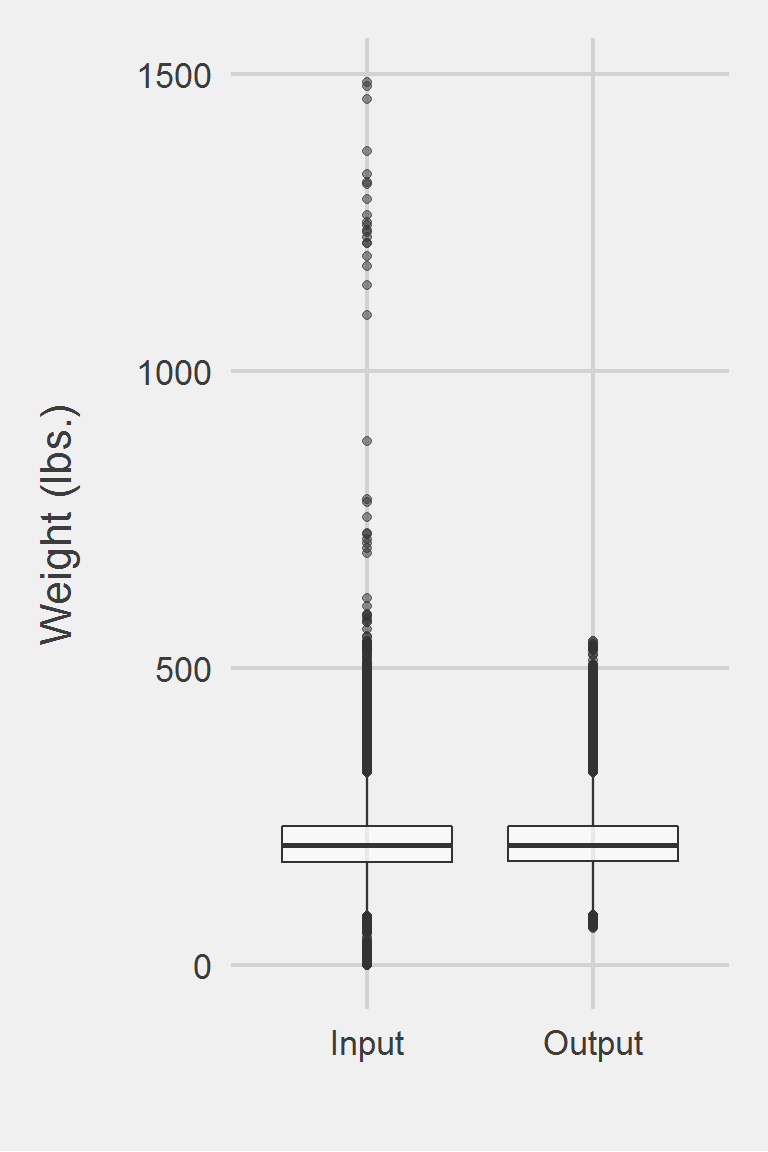Maciejewski 2016
Excerpt from published methods
Among the [cases], approximately 3.5% of weights were measured on the same day. If the standard deviation of the same-day weights was less than or equal to 2 lb, then the mean was taken. Otherwise, the standard deviation of each same-day weight with prior/post weight measurements was calculated and the same-day weight leading to the smallest standard deviation was retained. After sorting weight measures by date for each individual, rolling standard deviations were calculated using consecutive groups of three weight measures for each individual. The first group consisted of weight measures 1-3, the second 2-4, and so forth. The first and last groups were evaluated separately because the first (and last) weight measure could only be included in one group and the second (and next to last) could only be included in two groups. If the first (or last) two groups’ standard deviations were greater than 35 lb, the second (or next to last) weight measure was deleted. If the first (or last) standard deviation was greater than 35 lb, then paired standard deviations were calculated for each pair within the first (or last) group. If two of the paired standard deviations were greater than 45 lb and the remaining was less than 10 lb, the offending weight measure was deleted. After these deletions, the weight measures were reassembled in date order for each individual and rolling standard deviations were recalculated and assigned to the central weight measure of the group. Clusters of high standard deviations, indicating a potential outlier, were identified by flagging consecutive standard deviations greater than 10 lb. For each cluster of high standard deviations of three or more the interior weight measures were deleted keeping only the first and last measures of the cluster. Approximately 1.2% of weight measurements were identified as outliers and were deleted. Standard deviation cutoffs were determined via iterative trial and error driven by clinical plausibility of the specific measure rather than a standard rule and with guidance from clinical practitioners familiar with context of surgery and expected outcomes. Before the same-day and outlier cleaning, the cohort … had 89,757 measurements; after these cleaning steps, 85,556 (95.3%) weight measurements remained.
There is an extra step not mentioned in the methods but detailed elsewhere. All weights must first be between 50 and 700 lbs. (inclusive), then different weights recorded on the same day are dealt with as described above.
Batch et al. (2018) used this same algorithm in their paper Outcome by Gender in the Veterans Health Administration Motivating Overweight/Obese Veterans Everywhere Weight Management Program.
Translation in pseudocode
Algorithm in R Code
#' @title Maciejewski 2016 Weight Measurment Algorithm
#' @param DF object of class data.frame, containing id and weights
#' @param id string corresponding to the name of the column of patient IDs in df
#' @param weights string corresponding to the name of the column of weights in df
Maciejewski2016.f <- function(DF, id, weights, timepoints) {
# TODO: skip this? Why reinvent the wheel.
}Algorithm in SAS Code
In an e-mail dated 9/17/2018 12:43 PM
…We came up with this idea of looking at rolling standard deviations when needing to clean weight data from bariatric surgery patients that had huge amounts of variability over time. For the macros, you need to run the code that defines both the %dolag and the %prepost macros but you will only ever need to explicitly use the %prepost macro. (The %prepost macro calls the %dolag macro.) [Redacted] modified the weight cleaning macros from their original versions to accept a few parameters instead of relying on the input data set having variables with certain names. And, note that the fixed numerical values were what we have found to work well in our bariatric surgery cohorts, they may need to be refined for other types of cohorts. In the LMHO study ([Redacted]’s current bariatric surgery study), [Redacted] executed the weight cleaning algorithm macros by running the following SAS code (after first defining the macros in my SAS session): %prepost(DSN = matches, IDVar = PERSON_ID, DateVar = WeightRecordedDate, AnalysisVar = Weight); The code to plot the weight trajectories was run in the same SAS session as (but after the successful execution of) the weight cleaning macros. This is because the DATA Steps that create the plotting data set rely on some data sets that the weight cleaning macros create internally…
/********************** pre-processing ***************************************
two steps before applying the below algorithm:
1. weights must be between 50 and 700 lbs.
2. Same day weights must be processed according to the published methods:
"...If the standard deviation of the same-day weights was less than or
equal to 2 lb, then the mean was taken. Otherwise, the standard
deviation of each same-day weight with prior/post weight measurements
was calculated and the same-day weight leading to the smallest standard
deviation was retained."
*****************************************************************************/
/***************************** SameDayMeasures() *****************************
* `&DSN.` is just the name of the data set that contains the data to be
cleaned, in "long" format (i.e., one record per date per patient).
* `&IDVar.` is the name of the variable that uniquely identifies each
patient in the data set (e.g., ScrSSN, PatientICN, PERSON_ID, StudyID).
* `&DateVar.` is the name of the variable that contains the date on which
the value of &ANALYSISVAR. was recorded, i.e., the date of the weight
measurement record. NB: All of the following code was used with data
sets where there was only ever one weight measurement record per
calendar date per patient.
* `&AnalysisVar.` is the name of the numeric variable in &DSN. that these
macros are supposed to "clean". In this case, it should be the name of
the variable that contains weight measurement values.
* `&clearSpace.` if value is "TRUE" then all intermediate datasets are
removed from the workspace. If literally any other valid string "" that
does not evaluate to "TRUE" is passed as the parameter, then all
intermediate datasets are retained. Default is TRUE.
*****************************************************************************/
%MACRO SameDayMeasures(DSN = ,
IDVar = ,
DateVar = ,
AnalysisVar = ,
clearSpace = "TRUE");
/* Remove outliers and missing values first */
DATA df_step1;
SET &DSN;
IF &AnalysisVar < 50 OR &AnalysisVar > 700 THEN &AnalysisVar = .;
IF &AnalysisVar = . THEN DELETE;
KEEP &IDVar &DateVar &AnalysisVar;
RUN;
/* Identify same day weights */
PROC SQL;
CREATE TABLE df_step2 AS
SELECT DISTINCT *
FROM df_step1;
CREATE TABLE SameDayAnalysisVar AS
SELECT a.&IDVar,
a.&DateVar,
a.&AnalysisVar,
b.Cnt
FROM df_step2 AS a
LEFT JOIN (
SELECT &IDVar, &DateVar, COUNT(*) AS Cnt
FROM df_step2
GROUP BY &IDVar, &DateVar
HAVING COUNT(*) > 1
) AS b
ON a.&IDVar = b.&IDVar
AND
a.&DateVar = b.&DateVar
WHERE Cnt ne .
ORDER BY &IDVar, &DateVar;
QUIT;
* enumerate AnalysisVar by IDVar and DateVar;
DATA SameDayAnalysisVar;
SET SameDayAnalysisVar;
BY &IDVar &DateVar;
IF first.&IDVar OR first.&DateVar THEN Count = 0;
Count + 1;
DROP Cnt;
RUN;
PROC MEANS DATA = SameDayAnalysisVar NOPRINT NWAY;
CLASS &IDVar &DateVar;
VAR &AnalysisVar;
OUTPUT OUT = AnalysisVarStats (DROP = _TYPE_ _FREQ_)
MEAN = Mean_AnalysisVar
STD = SD_AnalysisVar;
RUN;
* AnalysisVar2SD: of the same-day measures,
those with SD < 2, we keep the mean;
* AnalysisVarFlagged: of the same-day measures,
those with SD > 2, must be processed further;
DATA AnalysisVar2SD (DROP = SD_AnalysisVar)
AnalysisVarFlagged;
SET AnalysisVarStats;
IF SD_AnalysisVar <= 2 THEN OUTPUT AnalysisVar2SD;
ELSE OUTPUT AnalysisVarFlagged;
RUN;
PROC SORT DATA = AnalysisVar2SD; BY &IDVar &DateVar; RUN;
PROC SORT DATA = df_step2; BY &IDVar &DateVar; RUN;
DATA df_step3;
MERGE df_step2 AnalysisVar2SD;
BY &IDVar &DateVar;
IF Mean_AnalysisVar ne . THEN &AnalysisVar = Mean_AnalysisVar;
DROP Mean_AnalysisVar;
RUN;
PROC SQL;
CREATE TABLE MeanFixed AS
SELECT DISTINCT *
FROM df_step3
ORDER BY &IDVar, &DateVar;
QUIT;
/*
Calculate the standard deviation of each same-day weight with prior/post
weight measurements, retain the same-day weight leading to the smallest
standard deviation
*/
PROC SQL;
CREATE TABLE AnalysisVarFlagged2 AS
SELECT a.*
FROM MeanFixed AS a
RIGHT JOIN (
SELECT DISTINCT &IDVar
FROM AnalysisVarFlagged
) AS b
ON a.&IDVar = b.&IDVar;
QUIT;
PROC SORT DATA = AnalysisVarFlagged2; BY &IDVar &DateVar; RUN;
DATA AnalysisVarFlagged2;
SET AnalysisVarFlagged2;
BY &IDVar &DateVar;
IF first.&IDVar OR first.&DateVar THEN Cnt = 0;
Cnt + 1;
PROC TRANSPOSE DATA = AnalysisVarFlagged2
OUT = AnalysisVarWide (KEEP = &IDVar &DateVar &AnalysisVar:)
PREFIX = &AnalysisVar;
BY &IDVar &DateVar;
ID Cnt;
VAR &AnalysisVar;
RUN;
PROC SQL noprint;
CREATE TABLE AnalysisVarFlagged3 AS
SELECT a.*,
(a.&DateVar - b.&DateVar) AS Diff
FROM AnalysisVarWide AS a
LEFT JOIN AnalysisVarFlagged AS b
ON a.&IDVar = b.&IDVar
ORDER BY a.&IDVar , a.&DateVar;
SELECT name
INTO :list SEPARATED BY ' '
FROM dictionary.columns
WHERE LIBNAME = 'WORK' AND MEMNAME = 'ANALYSISVARFLAGGED3';
CREATE TABLE backward AS
SELECT &IDVar,
%SCAN(&list, 3, ' ') AS Before,
Diff
FROM AnalysisVarFlagged3
WHERE Diff < 0
ORDER BY &IDVar, Diff DESC;
CREATE TABLE forward AS
SELECT &IDVar,
%SCAN(&list, 3, ' ') AS After,
Diff
FROM AnalysisVarFlagged3
WHERE Diff > 0
ORDER BY &IDVar, Diff;
CREATE TABLE MultipleAnalysisVars AS
SELECT &IDVar,
&DateVar,
%SCAN(&list, 3, ' '),
%SCAN(&list, 4, ' '),
%SCAN(&list, 5, ' '),
%SCAN(&list, 6, ' ')
FROM AnalysisVarFlagged3
where Diff = 0;
QUIT;
DATA backward;
SET backward;
BY &IDVar;
IF first.&IDVar;
RUN;
DATA forward;
SET forward;
BY &IDVar;
IF first.&IDVar;
RUN;
PROC SQL;
CREATE TABLE SameDayAnalysisVars AS
SELECT a.*,
b.before,
c.after
FROM MultipleAnalysisVars AS a
LEFT JOIN backward AS b
ON a.&IDVar = b.&IDVar
LEFT JOIN forward AS c
ON a.&IDVar = c.&IDVar;
QUIT;
* find min(SD) of AnalysisVar{i} vs. Before and After measures;
DATA AnalysisVar1 (KEEP = &IDVar &DateVar %SCAN(&list, 3, ' '))
AnalysisVar2 (KEEP = &IDVar &DateVar %SCAN(&list, 4, ' '))
AnalysisVar3 (KEEP = &IDVar &DateVar %SCAN(&list, 5, ' '))
AnalysisVar4 (KEEP = &IDVar &DateVar %SCAN(&list, 6, ' '))
dump;
SET SameDayAnalysisVars;
SD1 = STD(of %SCAN(&list, 3, ' '), Before, After);
SD2 = STD(of %SCAN(&list, 4, ' '), Before, After);
IF %SCAN(&list, 5, ' ') ne . THEN
DO;
SD3 = STD(of %SCAN(&list, 5, ' '), Before, After);
END;
IF %SCAN(&list, 6, ' ') ne . THEN
DO;
SD4 = STD(of %SCAN(&list, 6, ' '), Before, After);
END;
MinAnalysisVar = MIN(of SD1-SD4);
IF SD1 = MinAnalysisVar THEN OUTPUT AnalysisVar1;
ELSE IF SD2 = MinAnalysisVar THEN OUTPUT AnalysisVar2;
ELSE IF SD3 = MinAnalysisVar THEN OUTPUT AnalysisVar3;
ELSE IF SD4 = MinAnalysisVar THEN OUTPUT AnalysisVar4;
ELSE OUTPUT dump;
RUN;
* join back to phase 1 AnalysisVar;
PROC SQL;
CREATE TABLE Phase1Output AS
SELECT DISTINCT a.&IDVar,
a.&Datevar,
CASE WHEN b.%SCAN(&list, 3, ' ') NE . THEN b.%SCAN(&list, 3, ' ')
WHEN c.%SCAN(&list, 4, ' ') NE . THEN c.%SCAN(&list, 4, ' ')
WHEN d.%SCAN(&list, 5, ' ') NE . THEN d.%SCAN(&list, 5, ' ')
WHEN e.%SCAN(&list, 6, ' ') NE . THEN e.%SCAN(&list, 6, ' ')
ELSE a.&AnalysisVar
END AS AnalysisVarFixed
FROM MeanFixed AS a
LEFT JOIN AnalysisVar1 AS b
ON a.&IDVar = b.&IDVar
AND
a.&DateVar = b.&DateVar
LEFT JOIN AnalysisVar2 AS c
ON a.&IDVar = c.&IDVar
AND
a.&DateVar = c.&DateVar
LEFT JOIN AnalysisVar3 AS d
ON a.&IDVar = d.&IDVar
AND
a.&DateVar = d.&DateVar
LEFT JOIN AnalysisVar4 AS e
ON a.&IDVar = e.&IDVar
AND
a.&DateVar = e.&DateVar;
/****** clear workspace if clearSpace = 'TRUE' ******/
DATA sets;
SET sashelp.class;
RUN;
PROC SQL noprint;
SELECT CASE WHEN &clearSpace = 'TRUE'
THEN 'PROC DELETE
DATA = MeanFixed
df_step1
df_step2
df_step3
AnalysisVar2SD
AnalysisVarFlagged
AnalysisVarFlagged2
AnalysisVarFlagged3
AnalysisVarStats
AnalysisVarWide
SameDayAnalysisVar
Backward
Forward
MultipleAnalysisVars
SameDayAnalysisVars
AnalysisVar1
AnalysisVar2
AnalysisVar3
AnalysisVar4
Dump;
RUN;'
ELSE ' ' END
INTO :deleteStatement
FROM sets;
QUIT;
&deleteStatement;
%MEND;
/********************************** dolag() **********************************
* `&DSN.` is just the name of the data set that contains the data to be
cleaned, in "long" format (i.e., one record per date per patient).
* `&IDVAR.` is the name of the variable that uniquely identifies each
patient in the data set (e.g., ScrSSN, PatientICN, PERSON_ID, StudyID).
* `&DATEVAR.` is the name of the variable that contains the date on which
the value of &ANALYSISVAR. was recorded, i.e., the date of the weight
measurement record. NB: All of the following code was used with data
sets where there was only ever one weight measurement record per
calendar date per patient.
* `&ANALYSISVAR.` is the name of the numeric variable in &DSN. that these
macros are supposed to "clean". In this case, it should be the name of
the variable that contains weight measurement values.
*****************************************************************************/
%macro dolag(DSN = , IDVar = , DateVar = , AnalysisVar = );
%LET DSN = &DSN.;
%LET IDVar = &IDVAR.;
%LET DateVar = &DATEVAR.;
%LET AnalysisVar = &ANALYSISVAR.;
* get lagged values, calculate standard deviations, flag SD > 20;
DATA &DSN.;
SYSECHO "DATA Step: Get lagged values, calculate SDs, flag SD > 20";
SET &DSN.;
BY &IDVAR. &DATEVAR.;
before = lag2(&ANALYSISVAR.);
center = lag(&ANALYSISVAR.);
after = &ANALYSISVAR.;
lagid1 = lag(&IDVAR.);
lagid2 = lag2(&IDVAR.);
lagdt = lag(&DATEVAR.);
IF (lagid2 = &IDVAR.) THEN stdev = std(before, center, after);
IF (stdev > 20) THEN highsd = 1;
KEEP &IDVAR. &DATEVAR. before center after lag: stdev highsd;
RUN;
/* save last observation for each individual */
DATA &DSN.l(rename = (a = after b = before c = center));
SYSECHO "DATA Step: Save last observation for each &IDVAR.";
SET &DSN.;
BY &IDVAR. &DATEVAR.;
IF last.&IDVAR.;
b = center;
c = after;
a = .;
stdev = .;
KEEP &IDVAR. &DATEVAR. b c a stdev;
RUN;
/* reset values when person_id changes */
DATA &DSN.;
SYSECHO "DATA Step: Reset values for each new &IDVAR.";
SET &DSN.;
BY &IDVAR. &DATEVAR.;
DROP &DATEVAR.;
IF (lagid2 NE &IDVAR.) THEN before = .;
IF (lagid1 NE &IDVAR.) THEN center = .;
RUN;
/* move each SD to center of measures */
DATA &DSN.;
SYSECHO "DATA Step: Move each SD to center of measures";
SET &DSN.;
IF (lagid1 = &IDVAR.) THEN &DATEVAR. = lagdt;
DROP lag:;
* IF (NOT missing(&DATEVAR.));
RUN;
DATA &DSN.;
SYSECHO "DATA Step: Keep only non-missing &DATEVAR.";
SET &DSN.;
IF (&DATEVAR. NE .);
RUN;
PROC SORT DATA = &DSN.;
SYSECHO "PROC SORT: By &IDVAR. &DATEVAR.";
BY &IDVAR. &DATEVAR.;
RUN;
/* tack on last observation for each individual and flag first and last
observations for each individual */
DATA &DSN.;
SYSECHO "DATA Step: Add last obs, flag first and last obs per &IDVAR.";
SET &DSN. &DSN.l;
BY &IDVAR. &DATEVAR.;
RUN;
%mend dolag;
/********************************* prepost() *********************************
* `&DSN.` is just the name of the data set that contains the data to be
cleaned, in "long" format (i.e., one record per date per patient).
* `&IDVAR.` is the name of the variable that uniquely identifies each
patient in the data set (e.g., ScrSSN, PatientICN, PERSON_ID, StudyID).
* `&DATEVAR.` is the name of the variable that contains the date on which
the value of &ANALYSISVAR. was recorded, i.e., the date of the weight
measurement record. NB: All of the following code was used with data
sets where there was only ever one weight measurement record per
calendar date per patient.
* `&ANALYSISVAR.` is the name of the numeric variable in &DSN. that these
macros are supposed to "clean". In this case, it should be the name of
the variable that contains weight measurement values.
******************************************************************************/
%macro prepost(DSN = , IDVar = , DateVar = , AnalysisVar = );
%LET DSN = &DSN.;
%LET IDVar = &IDVAR.;
%LET DateVar = &DATEVAR.;
%LET AnalysisVar = &ANALYSISVAR.;
PROC SORT DATA = &DSN.;
SYSECHO "PROC SORT: By &IDVAR. &DATEVAR.";
BY &IDVAR. &DATEVAR.;
RUN;
%dolag(DSN = &DSN.,
IDVar = &IDVAR.,
DateVar = &DATEVAR.,
AnalysisVar = &ANALYSISVAR.);
/* number position within individuals measures from beginning */
DATA &DSN.;
SYSECHO "DATA Step: Determine chronological order of records";
SET &DSN.;
BY &IDVAR. &DATEVAR.;
RETAIN fctr 0;
IF (first.&IDVAR.) THEN fctr = 0;
fctr + 1;
RUN;
/* number position within individuals measures from end */
PROC SORT data = &DSN.;
SYSECHO "PROC SORT: By &IDVAR. DESCENDING &DATEVAR.";
BY &IDVAR. descending &DATEVAR.;
RUN;
DATA &DSN.;
SYSECHO "DATA Step: Determine reverse chronological order of records";
SET &DSN.;
BY &IDVAR. descending &DATEVAR.;
RETAIN lctr 0;
IF (first.&IDVAR.) THEN lctr = 0;
lctr + 1;
RUN;
*Re-sort with dates in ascending order;
PROC SORT DATA = &DSN.;
SYSECHO "PROC SORT: By &IDVAR. &DATEVAR.";
BY &IDVAR. &DATEVAR.;
RUN;
/* find those with highsd in first three observations */
PROC FREQ DATA = &DSN.;
SYSECHO "PROC FREQ: Identify &IDVAR.s with high SD in first 3 obs";
TABLES &IDVAR. / NOPRINT OUT = skipfirst;
WHERE ((fctr IN (2 3 4)) AND (highsd = 1));
RUN;
DATA skipfirst;
SYSECHO "DATA Step: Keep &IDVAR.s with high SD in all first 3 obs";
SET skipfirst;
IF count = 3;
KEEP &IDVAR.;
RUN;
/* allow those with highsd in first three observations to skip first obs processing */
DATA first&DSN.;
SYSECHO "DATA Step: Exclude &IDVAR.s from first obs processing";
MERGE &DSN. skipfirst (IN = ins);
BY &IDVAR.;
IF NOT (ins);
RUN;
/* select sd > 35 when in first two sd calculations */
DATA first&DSN.;
SYSECHO "DATA Step: Find SD > 35 in first 2 SD calculations";
SET first&DSN.;
BY &IDVAR. &DATEVAR.;
IF ((fctr IN (2 3)) AND (stdev > 35));
RUN;
DATA first&DSN.;
SYSECHO "DATA Step: Flag ""first 3"" obs for deletion";
SET first&DSN.;
BY &IDVAR. &DATEVAR.;
KEEP &IDVAR. del;
/* if first two sds > 35 mark second obs for deletion */
IF not(first.&IDVAR. AND last.&IDVAR.) THEN DO;
del = 2;
IF (last.&IDVAR.) THEN output;
END;
ELSE DO;
sd_after = std(before, center);
sd_before = std(center, after);
sd_center = std(before, after);
/*
if only first sd > 35 check sds of each pair within group of
3 - flag value causing each of its sds to be greater than 45
when remaining sd < 10
*/
IF (MIN(sd_after, sd_before, sd_center) NE sd_after) THEN DO;
IF ((sd_before < sd_center) AND
(sd_before < 10) AND
(sd_center ge 45) AND
(sd_after ge 45))
THEN del = 1;
ELSE DO;
IF ((sd_center < 10) AND
(sd_before ge 45) AND
(sd_after ge 45))
THEN del = 2;
END;
END;
IF (del IN (1 2)) THEN OUTPUT;
END;
RUN;
/* find those with highsd in last three observations */
PROC FREQ DATA = &DSN.;
SYSECHO "PROC FREQ: Identify &IDVAR.s with high SD in last 3 obs";
TABLES &IDVAR. / NOPRINT OUT = skiplast;
WHERE ((lctr IN (2 3 4)) AND (highsd = 1));
RUN;
DATA skiplast;
SET skiplast;
IF count = 3;
KEEP &IDVAR.;
RUN;
/* allow those to skip first obs processing */
DATA last&DSN.;
MERGE &DSN. skiplast(in = ins);
BY &IDVAR.;
IF not(ins);
RUN;
DATA last&DSN.;
SET last&DSN.;
BY &IDVAR. &DATEVAR.;
IF ((lctr IN (2 3)) AND (stdev > 35));
RUN;
DATA last&DSN.;
SET last&DSN.;
BY &IDVAR. &DATEVAR.;
KEEP &IDVAR. del;
/* if last two sds > 35 mark second from last obs for deletion */
IF not(first.&IDVAR. AND last.&IDVAR.) THEN DO;
del = 2;
IF (last.&IDVAR.) THEN OUTPUT;
END;
ELSE DO;
sd_after = std(before, center);
sd_before = std(center, after);
sd_center = std(before, after);
/*
if only last sd > 35 check sds of each pair within group of
3 - flag value causing each of its sds to be greater than 45
when remaining sd < 10
*/
IF (min(sd_after, sd_before, sd_center) NE sd_before) THEN DO;
IF ((sd_after < sd_center) AND
(sd_after < 10) AND
(sd_center ge 45) AND
(sd_before ge 45))
THEN del = 1;
ELSE DO;
IF ((sd_center < 10) AND
(sd_after ge 45) AND
(sd_before ge 45))
THEN del = 2;
END;
END;
IF (del IN (1 2)) THEN OUTPUT;
END;
RUN;
/* eliminate measures flagged for deletion */
DATA &DSN.;
MERGE &DSN. first&DSN.(in = inf) last&DSN.(in = inl);
BY &IDVAR.;
KEEP &IDVAR. &DATEVAR. center;
IF ((inf) AND (del = fctr)) THEN DELETE;
IF ((inl) AND (del = lctr)) THEN DELETE;
RENAME center = &ANALYSISVAR.;
RUN;
PROC SORT DATA = &DSN.;
BY &IDVAR. &DATEVAR.;
RUN;
%dolag(DSN = &DSN.,
IDVar = &IDVAR.,
DateVar = &DATEVAR.,
AnalysisVar = &ANALYSISVAR.);
/* number blocks of consecutive high sd */
DATA &DSN.;
SET &DSN.;
BY &IDVAR. &DATEVAR.;
RETAIN highctr 0;
IF (first.&IDVAR.) THEN highctr = 0;
IF highsd = 1 THEN highctr + 1;
ELSE highctr = 0;
RUN;
%MEND prepost;Example in SAS & R
/******************************* Set Library ********************************/
ods html close; ods html;
libname DCEP "X:\Damschroder-NCP PEI\6. Aim 2 Weight Algorithms\CDW Data";
/**************************** Import Sample Data ****************************/
* data stored in SQL DB;
LIBNAME samp OLEDB
INIT_STRING = "Provider = SQLNCLI11;
Integrated Security = SSPI;
Persist Security Info = True;
Initial Catalog = NCP_DCEP;
Data Source = vhacdwrb03.vha.med.va.gov"
Schema = samp;
/* examine data
%dim(Samp.heightSamples);
* nrow = 94146, ncol = 5;
%dim(Samp.weightSamples);
* nrow = 279604, ncol = 8;
*/
/******************************* Examine Data *******************************/
PROC UNIVARIATE DATA = Samp.heightSamples;
VAR height;
RUN;
PROC UNIVARIATE DATA = Samp.weightSamples;
VAR weight;
RUN;
PROC FREQ DATA = Samp.weightSamples;
TABLES (Sta3n Sta6a DivisionName) * SampleYear
(Sta3n Sta6a DivisionName) * Source
/ nocum missing;
RUN;
/***************************** Macro invocation *****************************/
/*** PCP Source ***/
DATA PCP; SET Samp.weightSamples; WHERE Source = 'PCP'; RUN; * nrow = 225363;
%LET DSN = PCP;
%LET IDVar = PatientICN;
%LET DateVar = WeightDate;
%LET AnalysisVar = Weight;
/***** Step 1: Remove missing values and outliers, fix same day weights *****/
%SameDayMeasures(DSN = &DSN.,
IDVar = &IDVar.,
DateVar = &DateVar.,
AnalysisVar = &AnalysisVar.,
clearSpace = 'TRUE');
* nrow_before = 225363, ncol = 15;
* nrow_after = 219958, ncol = 3;
* reduction by 5405 (2.4%);
/************ Step 2: Rolling SD by cluster and person algorithm ************/
/*
* internal function for prepost();
* ran here as test;
%dolag(DSN = MOVEweights,
IDVar = PatientICN,
DateVar = WeightDateTime,
AnalysisVar = Weight);
*/
%LET DSN = Phase1Output;
%LET AnalysisVar = AnalysisVarFixed;
%prepost(DSN = &DSN.,
IDVar = &IDVar.,
DateVar = &DateVar.,
AnalysisVar = &AnalysisVar.);
* Augments &DSN. and creates &DSNl;
* nrow_before = 219958, ncol = 3;
* nrow_after = 219871, ncol = 8;
* reduction by 87 (.04%);
DATA PCPcleaned; SET Phase1Output; RUN; * nrow = 219871, ncol = 8;
/*** MOVE! Source ***/
DATA MOVE; SET Samp.weightSamples; WHERE Source = 'MOVE'; RUN; * nrow = 54241;
/***** Step 1: Remove missing values and outliers, fix same day weights *****/
%LET DSN = MOVE;
%LET AnalysisVar = Weight;
%SameDayMeasures(DSN = &DSN.,
IDVar = &IDVar.,
DateVar = &DateVar.,
AnalysisVar = &AnalysisVar.,
clearSpace = 'TRUE');
* nrow_before = 54241, ncol = 15;
* nrow_after = 52752, ncol = 3;
* reduction by 1489 (2.7%);
/************ Step 2: Rolling SD by cluster and person algorithm ************/
%LET DSN = Phase1Output;
%LET AnalysisVar = AnalysisVarFixed;
%prepost(DSN = &DSN.,
IDVar = &IDVar.,
DateVar = &DateVar.,
AnalysisVar = &AnalysisVar.);
* Augments &DSN. and creates &DSNl;
* nrow_before = 52752, ncol = 3;
* nrow_after = 52744, ncol = 8;
* reduction by 8 (.015%);
DATA MOVEcleaned; SET Phase1Output; RUN; * nrow = 52744, ncol = 8;
/*************************** Post-Macro Mess 'Round *************************/
/* enumerate clusters with high SD */
DATA MOVEcleaned;
SET MOVEcleaned;
BY &IDVAR. &DATEVAR.;
RETAIN cluster 0;
lagctr = lag(highctr);
IF (first.&IDVAR.) THEN cluster = 0;
ELSE DO;
IF ((lagctr = 0) AND (highctr = 1)) THEN cluster + 1;
END;
RUN;
* nrow = 219871, ncol = 10;
/* count number of high sd in each cluster */
PROC MEANS DATA = MOVEcleaned NOPRINT;
CLASS &IDVAR. cluster;
VAR highctr;
WAYS 2;
OUTPUT MAX = maxctr OUT = clusters (DROP = _TYPE_ _FREQ_);
RUN;
* nrow = 2352, ncol = 3;
/* sort for merging */
PROC SORT DATA = MOVEcleaned;
BY &IDVAR. cluster highctr;
RUN;
/* merge max number of each cluster to mark interior measures for deletion */
DATA MOVEcleaned;
MERGE MOVEcleaned clusters (IN = inc);
BY &IDVAR. cluster;
IF inc THEN DO;
IF 1 < highctr < maxctr THEN DELETE;
END;
RUN;
* nrow = 52692, ncol = 11;
/* save permanent
* whole sample;
DATA Dcep.weightSamples; SET Samp.weightSamples; RUN;
* PCP sample;
DATA Dcep.PCPcleaned; SET PCPcleaned; RUN;
* MOVE sample;
DATA Dcep.MOVEcleaned; SET MOVEcleaned; RUN;
*/The SAS macro (%prepost()) augments the input data by removing the “central” measurement when it is flagged for having a high standard deviation, i.e., the ‘middle’ weight in a cluster of three weights, organized by date/time + some other magical happenings I haven’t quite figured out yet…
Summary Statistics
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
0.0 1.7 3.0 4.0 5.0 259.4 387911 There were 15665 clusters with a high standard deviation identified. In total, -2311755 observations were removed.
Distribution of raw weight data versus algorithm processed data
Descriptive statistics by group
group: Input
vars n mean sd median trimmed mad min max range skew
X1 1 2430339 205.02 48.16 199.2 201.68 43.29 0 2423.35 2423.35 1.3
kurtosis se
X1 14.06 0.03
------------------------------------------------------------
group: Output
vars n mean sd median trimmed mad min max range skew kurtosis
X1 1 2311755 205.45 47.3 199.9 202.11 42.85 53.9 632.5 578.6 0.88 1.73
se
X1 0.03The Maciejewski algorithm processed weights removed 5.08% of the values. For the cohort described in Maciejewski et al. 2016, this algorithm reduces their sample by \(\approx 5\)%
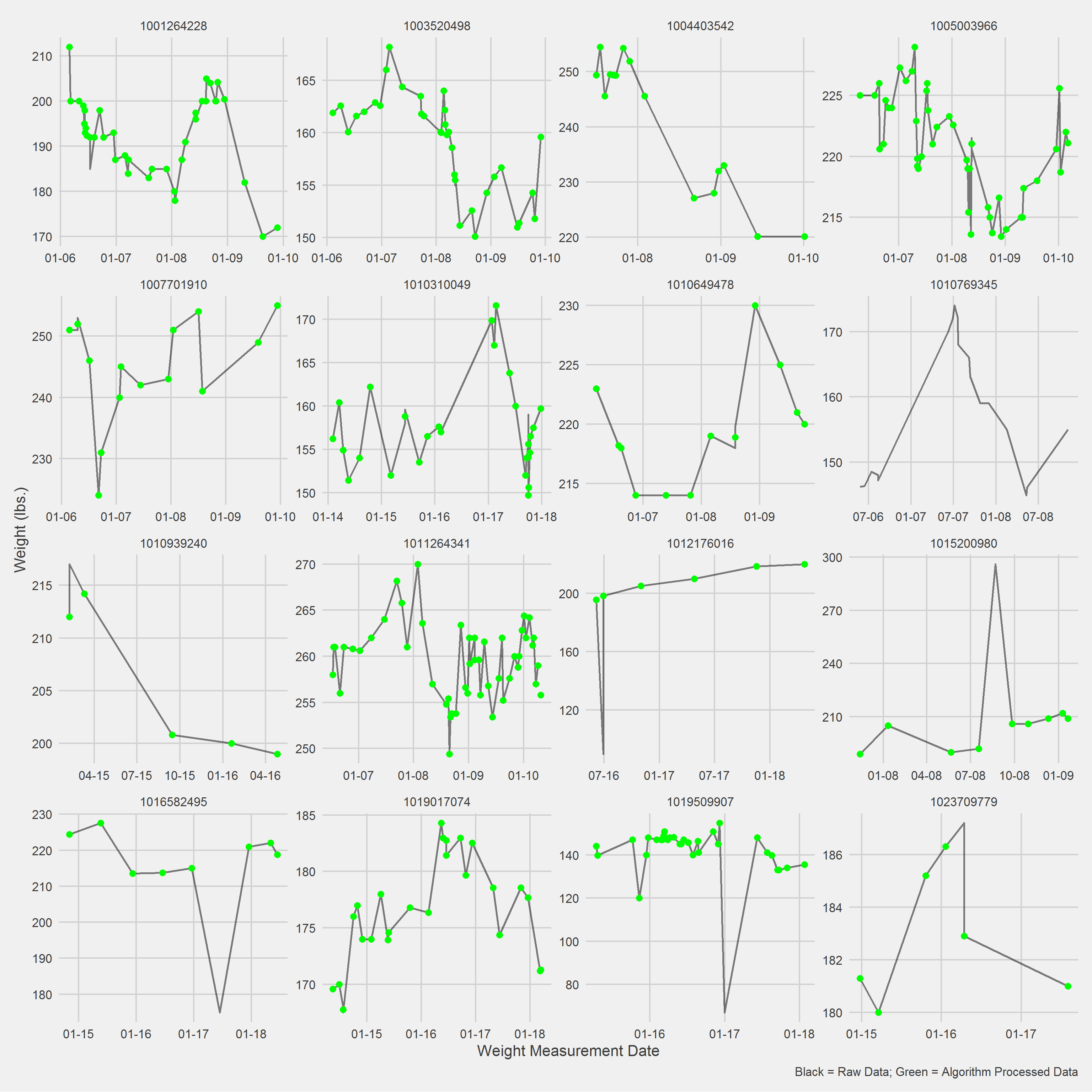
Displaying a Vignette of 16 selected patients with at least 1 weight observation removed.
Left boxplot is raw data from the sample of 2016, PCP visit subjects while the right boxplot describes the output from running %SameDayMeasures() and %Prepost() in SAS.