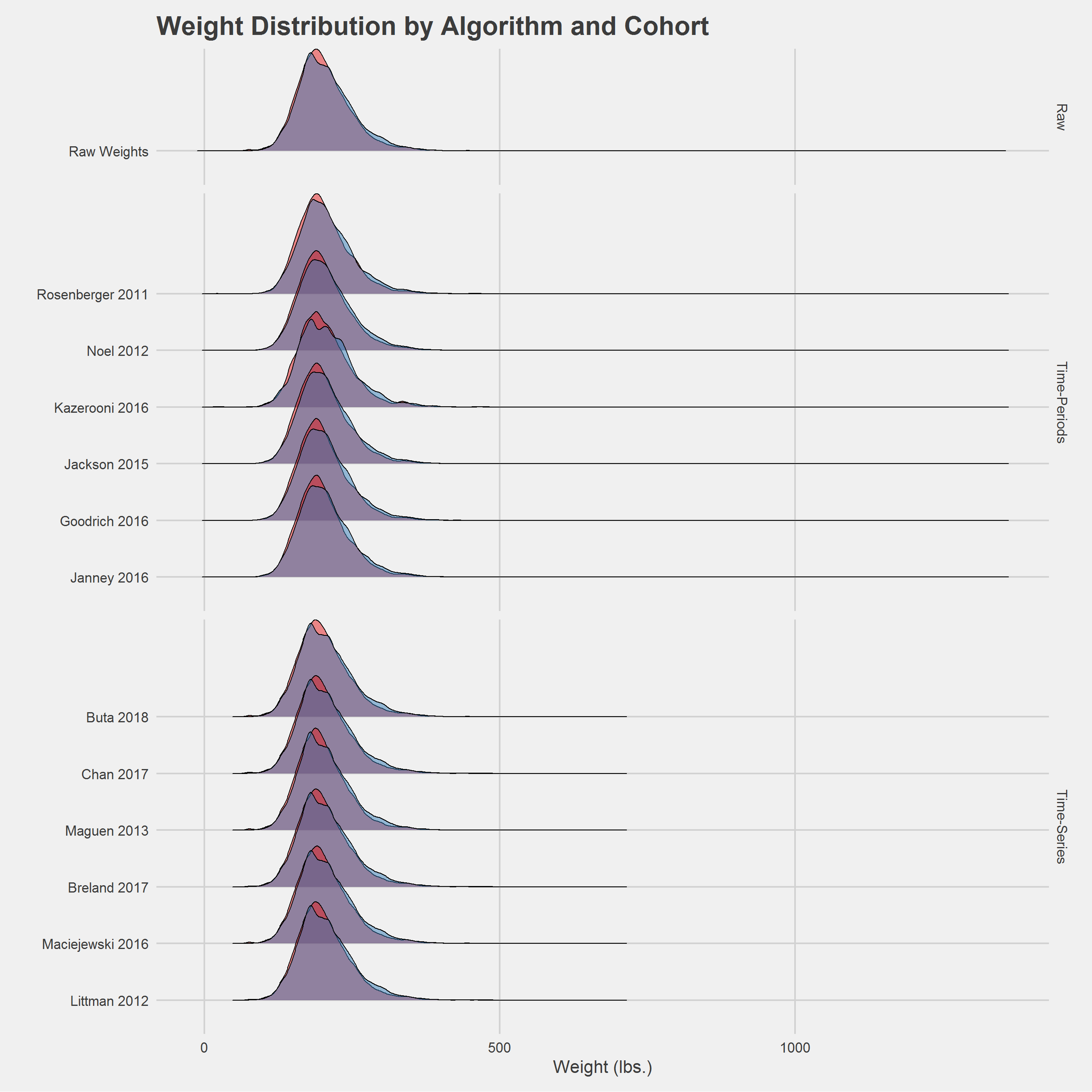
Overall Sample Comparison
weightSamples <- weightSamples %>%
left_join(
ModeOfHeight %>%
select(-freq),
by = "PatientICN"
) %>%
mutate(BMI = 703 * Weight / (Height ^ 2)) %>%
select(-Height) %>%
distinct(PatientICN, WeightDate, Weight, .keep_all = TRUE)
weight.ls <- vector("list", 2)
weight.ls[[1]] <- weightSamples %>%
filter(SampleYear == "2008" & !is.na(Weight))
weight.ls[[2]] <- weightSamples %>%
filter(SampleYear == "2016" & !is.na(Weight))
lsnames <- c("PCP2008", "PCP2016")
names(weight.ls) <- lsnames
rm(weightSamples)Aggregating over each year, the mean and standard deviation of weight is 205.2 (48.2).
| mean | min | max |
|---|---|---|
| 12.05 | 1 | 1525 |
On average, each veteran has 12 weights recorded over a 4-year collection period, and at least 1 person has 1,525 measurements collected. 5291 veterans have only 1 weight measurement recorded.
| SampleYear | mean | sd |
|---|---|---|
| 2008 | 202.71 | 47.65 |
| 2016 | 207.82 | 48.60 |
Between 2008 and 2016, the average weight increased by about 5 lbs. (202.7 to 207.8), and a 1 point increase in standard deviation.
| SampleYear | mean | min | max |
|---|---|---|---|
| 2008 | 12.09 | 1 | 1457 |
| 2016 | 11.88 | 1 | 1525 |
The number of weights recorded does not differ between the 2008 and 2016 cohorts, the average and range of each being nearly identical to the overall distribution.
By Algorithm
raw <- bind_rows(weight.ls, .id = "Cohort") %>%
select(Cohort, PatientICN, Weight, Sta3n)
#---- apply Janney et al. 2016 ----#
Janney2016.ls <- lapply(
weight.ls,
FUN = function(x){
Janney2016.f(
DF = x,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime",
startPoint = "VisitDateTime"
)
}
)
# post-process for joining with other algorithm output
Janney2016.df <- bind_rows(Janney2016.ls, .id = "Cohort") %>%
select(Cohort, PatientICN, Weight_OR, Sta3n) %>%
rename(Weight = Weight_OR) %>%
na.omit()
Janney2016.PCP16 <- Janney2016.ls[["PCP2016"]]
rm(Janney2016.ls)
#---- apply Littman et al. 2012 ----#
Littman2012.ls <- lapply(
weight.ls,
FUN = function(x) {
Littman2012.f(
DF = x,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime"
)
}
)
Littman2012.df <- bind_rows(Littman2012.ls, .id = "Cohort") %>%
select(Cohort, PatientICN, OutputMeasurement, Sta3n) %>%
rename(Weight = OutputMeasurement) %>%
na.omit()
Littman2012.PCP16 <- Littman2012.ls[["PCP2016"]]
rm(Littman2012.ls)
#---- Coerce Maciejewski et al. 2016 to a workable format ----#
maciejewski.df <- maciejewski %>%
filter(IO == "Output") %>%
mutate(
Cohort = paste0("PCP", SampleYear),
PatientICN = as.character(PatientICN)
) %>%
left_join(
raw %>%
distinct(PatientICN, Sta3n),
by = "PatientICN"
)
Maciejewski2016.df <- maciejewski.df %>%
select(Cohort, PatientICN, Weight, Sta3n) %>%
na.omit()
Maciejewski2016.PCP16 <- maciejewski.df %>% filter(Cohort == "PCP2016")
rm(maciejewski.df)
#---- Apply Breland et al. 2017 ----#
Breland2017.ls <- lapply(
weight.ls,
FUN = function(x) {
Breland2017.f(
DF = x,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime"
)
}
)
Breland2017.df <- bind_rows(Breland2017.ls, .id = "Cohort") %>%
select(Cohort, PatientICN, measures_aug_, Sta3n) %>%
rename(Weight = measures_aug_) %>%
na.omit()
Breland2017.PCP16 <- Breland2017.ls[["PCP2016"]]
rm(Breland2017.ls)
#---- Apply Maguen et al. 2013 ----#
Maguen2013.ls <- lapply(
weight.ls,
FUN = function(x) {
Maguen2013.f(
DF = x,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime",
variables = c("AgeAtVisit", "Gender")
)
}
)
Maguen2013.df <- bind_rows(Maguen2013.ls, .id = "Cohort") %>%
select(Cohort, PatientICN, Output, Sta3n) %>%
rename(Weight = Output) %>%
na.omit()
Maguen2013.PCP16 <- Maguen2013.ls[["PCP2016"]]
rm(Maguen2013.ls)
#---- Apply Goodrich 2016 ----#
Goodrich2016.ls <- lapply(
weight.ls,
FUN = function(x) {
Goodrich2016.f(
DF = x,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime",
startPoint = "VisitDateTime"
)
}
)
Goodrich2016.df <- bind_rows(Goodrich2016.ls, .id = "Cohort") %>%
select(Cohort, PatientICN, output, Sta3n) %>%
rename(Weight = output) %>%
na.omit()
Goodrich2016.PCP16 <- Goodrich2016.ls[["PCP2016"]]
rm(Goodrich2016.ls)
#---- Apply Chan & Raffa 2017 ----#
Chan2017.ls <- lapply(
weight.ls,
FUN = function(x) {
Chan2017.f(
DF = x,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime"
)
}
)
Chan2017.df <- bind_rows(Chan2017.ls, .id = "Cohort") %>%
select(Cohort, PatientICN, measures_aug_, Sta3n) %>%
rename(Weight = measures_aug_) %>%
na.omit()
Chan2017.PCP16 <- Chan2017.ls[["PCP2016"]]
rm(Chan2017.ls)
#---- Apply Jackson et al. 2015 ----#
Jackson2015.ls <- lapply(
weight.ls,
FUN = function(x) {
Jackson2015.f(
DF = x,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime",
startPoint = "VisitDateTime"
)
}
)
# re-insert Sta3n data
for (i in 1:length(Jackson2015.ls)) {
Jackson2015.ls[[i]] <- Jackson2015.ls[[i]] %>%
left_join(weight.ls[[i]] %>%
distinct(PatientICN, Sta3n),
by = "PatientICN")
}
Jackson2015.df <- bind_rows(Jackson2015.ls, .id = "Cohort") %>%
select(Cohort, PatientICN, Sta3n, output) %>%
rename(Weight = output) %>%
na.omit()
Jackson2015.PCP16 <- Jackson2015.ls[["PCP2016"]]
rm(Jackson2015.ls)
#---- Apply Buta et al., 2018 ----#
Buta2018.ls <- lapply(
weight.ls,
FUN = function(x) {
Buta2018.f(
DF = x,
id = "PatientICN",
measures = "BMI",
tmeasures = "WeightDateTime"
)
}
)
Buta2018.df <- bind_rows(Buta2018.ls, .id = "Cohort") %>%
select(Cohort, PatientICN, Sta3n, Weight) %>%
na.omit()
Buta2018.PCP16 <- Buta2018.ls[["PCP2016"]]
rm(Buta2018.ls)
#---- Apply Kazerooni & Lim, 2016 ----#
Kazerooni2016.ls <- lapply(
weight.ls,
FUN = function(x) {
Kazerooni2016.f(
DF = x,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime",
startPoint = "VisitDateTime"
)
}
)
Kazerooni2016.df <- bind_rows(Kazerooni2016.ls, .id = "Cohort") %>%
select(Cohort, PatientICN, Sta3n, Weight) %>%
na.omit()
Kazerooni2016.PCP16 <- Kazerooni2016.ls[["PCP2016"]]
rm(Kazerooni2016.ls)
#---- Apply Noel et al., 2012 ----#
Noel2012.ls <- lapply(
weight.ls,
FUN = function(x) {
Noel2012.f(
DF = x,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime"
)
}
)
Noel2012.df <- bind_rows(Noel2012.ls, .id = "Cohort") %>%
distinct(Cohort, PatientICN, Sta3n, FYQ, Qmedian) %>%
select(-FYQ) %>%
rename(Weight = Qmedian) %>%
na.omit()
Noel2012.PCP16 <- Noel2012.ls[["PCP2016"]]
rm(Noel2012.ls)
#---- Apply Rosenberger et al., 2011 ----#
Rosenberger2011.ls <- lapply(
weight.ls,
FUN = function(x) {
Rosenberger2011.f(
DF = x,
id = "PatientICN",
tmeasures = "WeightDateTime",
startPoint = "VisitDateTime",
pad = 1
)
}
)
Rosenberger2011.df <- bind_rows(Rosenberger2011.ls, .id = "Cohort") %>%
select(Cohort, PatientICN, Sta3n, Weight) %>%
na.omit()
Rosenberger2011.PCP16 <- Rosenberger2011.ls[["PCP2016"]]
rm(Rosenberger2011.ls)
# stack original (raw), janney 2016, littman 2012, maciejewski 2016,
# Breland 2017, Maguen 2013, Goodrich 2016, Chan & Raffa 2017,
# Jackson 2015, Buta 2018, Kazerooni & Lim 2016, Noel 2012,
# and Rosenberger 2011 processed weights
cohort.fac <- c("PCP2008", "PCP2016")
cohort.labels <- c("PCP 2008", "PCP 2016")
algos.fac <- c("Raw Weights",
"Janney 2016",
"Littman 2012",
"Maciejewski 2016",
"Breland 2017",
"Maguen 2013",
"Goodrich 2016",
"Chan 2017",
"Jackson 2015",
"Buta 2018",
"Kazerooni 2016",
"Noel 2012",
"Rosenberger 2011")
DF <- bind_rows(
list(`Raw Weights` = raw,
`Janney 2016` = Janney2016.df,
`Littman 2012` = Littman2012.df,
`Maciejewski 2016` = Maciejewski2016.df,
`Breland 2017` = Breland2017.df,
`Maguen 2013` = Maguen2013.df,
`Goodrich 2016` = Goodrich2016.df,
`Chan 2017` = Chan2017.df,
`Jackson 2015` = Jackson2015.df,
`Buta 2018` = Buta2018.df,
`Kazerooni 2016` = Kazerooni2016.df,
`Noel 2012` = Noel2012.df,
`Rosenberger 2011` = Rosenberger2011.df),
.id = "Algorithm"
)
rm(
Buta2018.df,
Kazerooni2016.df,
Noel2012.df,
Rosenberger2011.df,
Breland2017.df,
Chan2017.df,
Goodrich2016.df,
Jackson2015.df,
Janney2016.df,
Littman2012.df,
Maciejewski2016.df,
Maguen2013.df,
raw
)
DF$Cohort <- factor(DF$Cohort, levels = cohort.fac, labels = cohort.labels)
DF$Algorithm <- factor(DF$Algorithm, levels = algos.fac, labels = algos.fac)
DF$Type <- case_when(
DF$Algorithm == "Raw Weights" ~ 1,
DF$Algorithm %in% algos.fac[c(2, 7, 9, 11:13)] ~ 2,
DF$Algorithm %in% algos.fac[c(3:6, 8, 10)] ~ 3
)
DF$Type <- factor(DF$Type, 1:3, c("Raw", "Time-Periods", "Time-Series"))Total weight measurements, by algorithm and cohort year.
| Algorithm | PCP 2008 | PCP 2016 |
|---|---|---|
| Raw Weights | 1193976 | 1175995 |
| Janney 2016 | 209591 | 199830 |
| Littman 2012 | 1176644 | 1161661 |
| Maciejewski 2016 | 1168701 | 1146995 |
| Breland 2017 | 1191360 | 1175177 |
| Maguen 2013 | 1050974 | 1037293 |
| Goodrich 2016 | 209447 | 199803 |
| Chan 2017 | 1186602 | 1170114 |
| Jackson 2015 | 264829 | 251501 |
| Buta 2018 | 1145268 | 1131996 |
| Kazerooni 2016 | 87489 | 71961 |
| Noel 2012 | 721118 | 683008 |
| Rosenberger 2011 | 246594 | 227215 |
Aggregating over Year
Distribution of weight measurements, by algorithm, aggregating over 2008 and 2016,
DF %>%
filter(!is.na(Weight)) %>%
group_by(Algorithm) %>%
dplyr::summarise(
weight_n = n(),
mean = mean(Weight),
SD = sd(Weight),
median = median(Weight),
IQR = IQR(Weight),
min = min(Weight),
max = max(Weight)
) %>%
tableStyle()| Algorithm | weight_n | mean | SD | median | IQR | min | max |
|---|---|---|---|---|---|---|---|
| Raw Weights | 2369971 | 205.24 | 48.19 | 199.6 | 58.60 | 0.0 | 2423.35 |
| Janney 2016 | 409421 | 203.61 | 44.72 | 198.0 | 55.20 | 72.0 | 595.25 |
| Littman 2012 | 2338305 | 205.43 | 47.31 | 199.9 | 58.60 | 75.0 | 595.00 |
| Maciejewski 2016 | 2315696 | 205.47 | 47.30 | 200.0 | 58.60 | 53.9 | 632.50 |
| Breland 2017 | 2366537 | 205.31 | 47.44 | 199.7 | 58.50 | 75.0 | 694.46 |
| Maguen 2013 | 2088267 | 203.06 | 45.41 | 198.0 | 56.62 | 70.1 | 586.00 |
| Goodrich 2016 | 409250 | 203.57 | 44.59 | 198.0 | 55.10 | 80.0 | 500.00 |
| Chan 2017 | 2356716 | 205.31 | 47.47 | 199.7 | 58.50 | 50.0 | 727.50 |
| Jackson 2015 | 516330 | 203.83 | 44.79 | 198.4 | 55.60 | 75.0 | 629.50 |
| Buta 2018 | 2277264 | 205.36 | 47.50 | 199.9 | 58.71 | 60.0 | 571.10 |
| Kazerooni 2016 | 159450 | 206.19 | 47.53 | 200.7 | 58.30 | 0.0 | 1490.00 |
| Noel 2012 | 1404126 | 204.69 | 45.37 | 199.0 | 56.15 | 70.0 | 630.00 |
| Rosenberger 2011 | 473809 | 205.14 | 45.73 | 200.0 | 55.96 | 0.0 | 1478.30 |
Number of Patients/Veterans Retained, by algorithm, aggregating over 2008 and 2016,
DF %>%
filter(!is.na(Weight)) %>%
group_by(Algorithm) %>%
distinct(PatientICN) %>%
count() %>%
tableStyle()| Algorithm | n |
|---|---|
| Raw Weights | 196625 |
| Janney 2016 | 191104 |
| Littman 2012 | 191323 |
| Maciejewski 2016 | 196622 |
| Breland 2017 | 196621 |
| Maguen 2013 | 195492 |
| Goodrich 2016 | 191109 |
| Chan 2017 | 191327 |
| Jackson 2015 | 192453 |
| Buta 2018 | 179015 |
| Kazerooni 2016 | 53015 |
| Noel 2012 | 196622 |
| Rosenberger 2011 | 131012 |
Number of weights retained, per person, by algorithm, aggregating over 2008 and 2016 samples,
DF %>%
filter(!is.na(Weight)) %>%
group_by(Algorithm, PatientICN) %>%
count() %>%
ungroup() %>%
group_by(Algorithm) %>%
summarize(
mean = mean(n),
SD = sd(n),
min = min(n),
Q1 = fivenum(n)[2],
median = median(n),
Q3 = fivenum(n)[4],
max = max(n)
) %>%
tableStyle()| Algorithm | mean | SD | min | Q1 | median | Q3 | max |
|---|---|---|---|---|---|---|---|
| Raw Weights | 12.05 | 17.27 | 1 | 5 | 8 | 15 | 1525 |
| Janney 2016 | 2.14 | 0.79 | 1 | 2 | 2 | 3 | 6 |
| Littman 2012 | 12.22 | 17.15 | 2 | 5 | 8 | 15 | 1524 |
| Maciejewski 2016 | 11.78 | 13.78 | 1 | 5 | 8 | 14 | 784 |
| Breland 2017 | 12.04 | 17.22 | 1 | 5 | 8 | 14 | 1524 |
| Maguen 2013 | 10.68 | 14.97 | 1 | 4 | 7 | 13 | 1516 |
| Goodrich 2016 | 2.14 | 0.79 | 1 | 2 | 2 | 3 | 6 |
| Chan 2017 | 12.32 | 17.27 | 2 | 5 | 8 | 15 | 1517 |
| Jackson 2015 | 2.68 | 1.01 | 1 | 2 | 3 | 3 | 8 |
| Buta 2018 | 12.72 | 17.83 | 2 | 5 | 9 | 15 | 1524 |
| Kazerooni 2016 | 3.01 | 0.15 | 3 | 3 | 3 | 3 | 6 |
| Noel 2012 | 7.14 | 3.76 | 1 | 4 | 7 | 10 | 34 |
| Rosenberger 2011 | 3.62 | 0.55 | 3 | 3 | 4 | 4 | 8 |
By Cohort
DF %>%
filter(!is.na(Weight)) %>%
group_by(Algorithm, Cohort) %>%
dplyr::summarise(
weight_n = n(),
mean = mean(Weight),
SD = sd(Weight),
median = median(Weight),
IQR = IQR(Weight),
min = min(Weight),
max = max(Weight)
) %>%
ungroup() %>%
tableStyle()| Algorithm | Cohort | weight_n | mean | SD | median | IQR | min | max |
|---|---|---|---|---|---|---|---|---|
| Raw Weights | PCP 2008 | 1193976 | 202.71 | 47.65 | 197.00 | 57.00 | 0.00 | 2423.35 |
| Raw Weights | PCP 2016 | 1175995 | 207.82 | 48.60 | 202.30 | 60.40 | 0.00 | 1486.20 |
| Janney 2016 | PCP 2008 | 209591 | 201.18 | 43.86 | 195.70 | 53.70 | 72.00 | 595.25 |
| Janney 2016 | PCP 2016 | 199830 | 206.15 | 45.46 | 201.00 | 56.70 | 78.40 | 546.00 |
| Littman 2012 | PCP 2008 | 1176644 | 202.91 | 46.34 | 197.00 | 56.80 | 75.00 | 595.00 |
| Littman 2012 | PCP 2016 | 1161661 | 207.97 | 48.14 | 202.60 | 60.00 | 75.20 | 546.00 |
| Maciejewski 2016 | PCP 2008 | 1168701 | 202.91 | 46.33 | 197.00 | 56.70 | 53.90 | 632.50 |
| Maciejewski 2016 | PCP 2016 | 1146995 | 208.08 | 48.13 | 202.70 | 60.00 | 62.00 | 546.00 |
| Breland 2017 | PCP 2008 | 1191360 | 202.81 | 46.49 | 197.00 | 57.00 | 75.00 | 632.50 |
| Breland 2017 | PCP 2016 | 1175177 | 207.85 | 48.25 | 202.40 | 60.30 | 75.00 | 694.46 |
| Maguen 2013 | PCP 2008 | 1050974 | 200.57 | 44.31 | 195.00 | 54.80 | 70.10 | 586.00 |
| Maguen 2013 | PCP 2016 | 1037293 | 205.58 | 46.37 | 200.70 | 58.28 | 70.30 | 541.10 |
| Goodrich 2016 | PCP 2008 | 209447 | 201.12 | 43.65 | 195.70 | 53.70 | 80.00 | 496.04 |
| Goodrich 2016 | PCP 2016 | 199803 | 206.13 | 45.42 | 201.00 | 56.70 | 80.00 | 500.00 |
| Chan 2017 | PCP 2008 | 1186602 | 202.79 | 46.53 | 197.00 | 57.00 | 50.00 | 632.50 |
| Chan 2017 | PCP 2016 | 1170114 | 207.86 | 48.27 | 202.40 | 60.30 | 54.00 | 727.50 |
| Jackson 2015 | PCP 2008 | 264829 | 201.48 | 43.96 | 196.00 | 53.70 | 75.00 | 629.50 |
| Jackson 2015 | PCP 2016 | 251501 | 206.31 | 45.52 | 201.05 | 56.96 | 75.63 | 553.00 |
| Buta 2018 | PCP 2008 | 1145268 | 202.84 | 46.57 | 197.00 | 57.00 | 60.50 | 571.10 |
| Buta 2018 | PCP 2016 | 1131996 | 207.91 | 48.29 | 202.60 | 60.20 | 60.00 | 540.00 |
| Kazerooni 2016 | PCP 2008 | 87489 | 203.87 | 47.06 | 198.00 | 56.80 | 0.00 | 1490.00 |
| Kazerooni 2016 | PCP 2016 | 71961 | 209.00 | 47.95 | 204.00 | 60.00 | 0.00 | 1233.70 |
| Noel 2012 | PCP 2008 | 721118 | 202.26 | 44.50 | 196.50 | 54.20 | 70.00 | 630.00 |
| Noel 2012 | PCP 2016 | 683008 | 207.25 | 46.13 | 202.00 | 57.60 | 70.00 | 588.90 |
| Rosenberger 2011 | PCP 2008 | 246594 | 202.68 | 45.13 | 197.00 | 54.30 | 0.00 | 1478.30 |
| Rosenberger 2011 | PCP 2016 | 227215 | 207.80 | 46.23 | 202.90 | 57.90 | 0.00 | 1314.50 |
Number of Patients/Veterans Retained
DF %>%
filter(!is.na(Weight)) %>%
group_by(Cohort, Algorithm) %>%
distinct(PatientICN) %>%
count() %>%
ungroup() %>%
tableStyle()| Cohort | Algorithm | n |
|---|---|---|
| PCP 2008 | Raw Weights | 98786 |
| PCP 2008 | Janney 2016 | 96420 |
| PCP 2008 | Littman 2012 | 96292 |
| PCP 2008 | Maciejewski 2016 | 98782 |
| PCP 2008 | Breland 2017 | 98782 |
| PCP 2008 | Maguen 2013 | 98246 |
| PCP 2008 | Goodrich 2016 | 96419 |
| PCP 2008 | Chan 2017 | 96294 |
| PCP 2008 | Jackson 2015 | 96984 |
| PCP 2008 | Buta 2018 | 89944 |
| PCP 2008 | Kazerooni 2016 | 29163 |
| PCP 2008 | Noel 2012 | 98783 |
| PCP 2008 | Rosenberger 2011 | 68232 |
| PCP 2016 | Raw Weights | 98958 |
| PCP 2016 | Janney 2016 | 95742 |
| PCP 2016 | Littman 2012 | 96130 |
| PCP 2016 | Maciejewski 2016 | 98958 |
| PCP 2016 | Breland 2017 | 98958 |
| PCP 2016 | Maguen 2013 | 98352 |
| PCP 2016 | Goodrich 2016 | 95748 |
| PCP 2016 | Chan 2017 | 96132 |
| PCP 2016 | Jackson 2015 | 96559 |
| PCP 2016 | Buta 2018 | 90159 |
| PCP 2016 | Kazerooni 2016 | 23987 |
| PCP 2016 | Noel 2012 | 98958 |
| PCP 2016 | Rosenberger 2011 | 63405 |
Number of weights retained, per person, by algorithm and Cohort,
DF %>%
filter(!is.na(Weight)) %>%
group_by(Cohort, Algorithm, PatientICN) %>%
count() %>%
ungroup() %>%
group_by(Cohort, Algorithm) %>%
summarize(
mean = mean(n),
SD = sd(n),
min = min(n),
Q1 = fivenum(n)[2],
median = median(n),
Q3 = fivenum(n)[4],
max = max(n)
) %>%
ungroup() %>%
tableStyle()| Cohort | Algorithm | mean | SD | min | Q1 | median | Q3 | max |
|---|---|---|---|---|---|---|---|---|
| PCP 2008 | Raw Weights | 12.09 | 15.47 | 1 | 5 | 8 | 15 | 1457 |
| PCP 2008 | Janney 2016 | 2.17 | 0.77 | 1 | 2 | 2 | 3 | 3 |
| PCP 2008 | Littman 2012 | 12.22 | 15.14 | 2 | 5 | 9 | 15 | 1448 |
| PCP 2008 | Maciejewski 2016 | 11.83 | 12.93 | 1 | 5 | 8 | 14 | 784 |
| PCP 2008 | Breland 2017 | 12.06 | 15.39 | 1 | 5 | 8 | 15 | 1447 |
| PCP 2008 | Maguen 2013 | 10.70 | 12.93 | 1 | 5 | 8 | 13 | 1421 |
| PCP 2008 | Goodrich 2016 | 2.17 | 0.77 | 1 | 2 | 2 | 3 | 3 |
| PCP 2008 | Chan 2017 | 12.32 | 15.42 | 2 | 5 | 9 | 15 | 1445 |
| PCP 2008 | Jackson 2015 | 2.73 | 0.98 | 1 | 2 | 3 | 3 | 4 |
| PCP 2008 | Buta 2018 | 12.73 | 15.91 | 2 | 5 | 9 | 15 | 1448 |
| PCP 2008 | Kazerooni 2016 | 3.00 | 0.00 | 3 | 3 | 3 | 3 | 3 |
| PCP 2008 | Noel 2012 | 7.30 | 3.66 | 1 | 4 | 7 | 10 | 17 |
| PCP 2008 | Rosenberger 2011 | 3.61 | 0.49 | 3 | 3 | 4 | 4 | 4 |
| PCP 2016 | Raw Weights | 11.88 | 18.71 | 1 | 4 | 8 | 14 | 1525 |
| PCP 2016 | Janney 2016 | 2.09 | 0.76 | 1 | 2 | 2 | 3 | 3 |
| PCP 2016 | Littman 2012 | 12.08 | 18.76 | 2 | 5 | 8 | 14 | 1524 |
| PCP 2016 | Maciejewski 2016 | 11.59 | 14.34 | 1 | 4 | 8 | 14 | 637 |
| PCP 2016 | Breland 2017 | 11.88 | 18.69 | 1 | 4 | 8 | 14 | 1524 |
| PCP 2016 | Maguen 2013 | 10.55 | 16.60 | 1 | 4 | 7 | 13 | 1516 |
| PCP 2016 | Goodrich 2016 | 2.09 | 0.76 | 1 | 2 | 2 | 3 | 3 |
| PCP 2016 | Chan 2017 | 12.17 | 18.76 | 2 | 5 | 8 | 15 | 1517 |
| PCP 2016 | Jackson 2015 | 2.60 | 0.98 | 1 | 2 | 3 | 3 | 4 |
| PCP 2016 | Buta 2018 | 12.56 | 19.36 | 2 | 5 | 8 | 15 | 1524 |
| PCP 2016 | Kazerooni 2016 | 3.00 | 0.00 | 3 | 3 | 3 | 3 | 3 |
| PCP 2016 | Noel 2012 | 6.90 | 3.72 | 1 | 4 | 6 | 9 | 17 |
| PCP 2016 | Rosenberger 2011 | 3.58 | 0.49 | 3 | 3 | 4 | 4 | 4 |
Bootstrapped Estimates
N <- 100
weight_samps <- vector("list", N)
key_cols <- c("Algorithm", "Cohort")
pb <- txtProgressBar(min = 0, max = N, initial = 0, style = 3)
for (i in 1:N) {
setTxtProgressBar(pb, i)
samp <- sample(pts, 1000, replace = TRUE)
weight_samps[[i]] <- DF %>% filter(PatientICN %in% samp)
setDT(weight_samps[[i]])
setkeyv(weight_samps[[i]], key_cols)
weight_samps[[i]] <-
weight_samps[[i]][,
list(mean = mean(Weight), SD = sd(Weight)),
by = key_cols
]
}Visualization
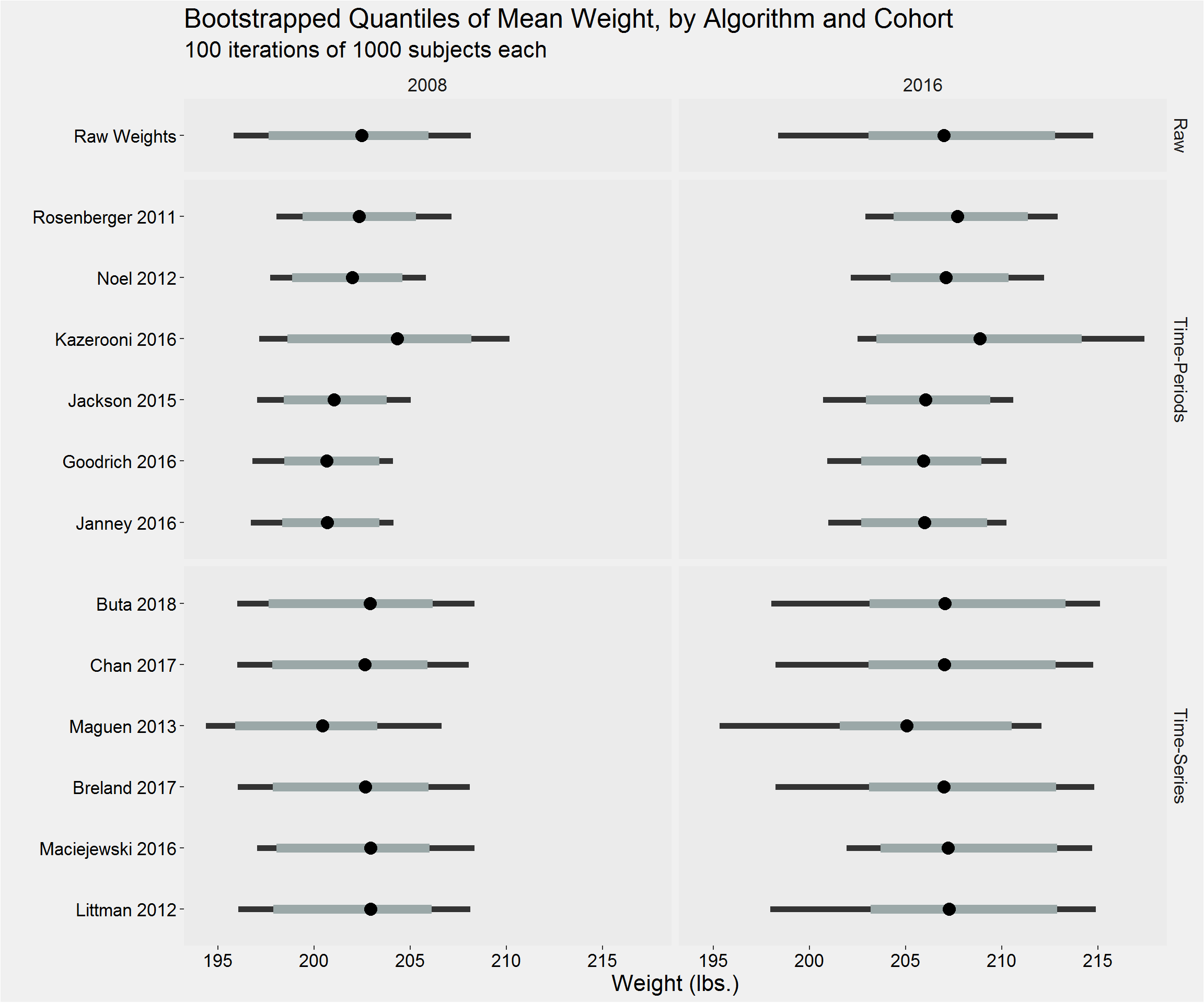
Distribution of the mean
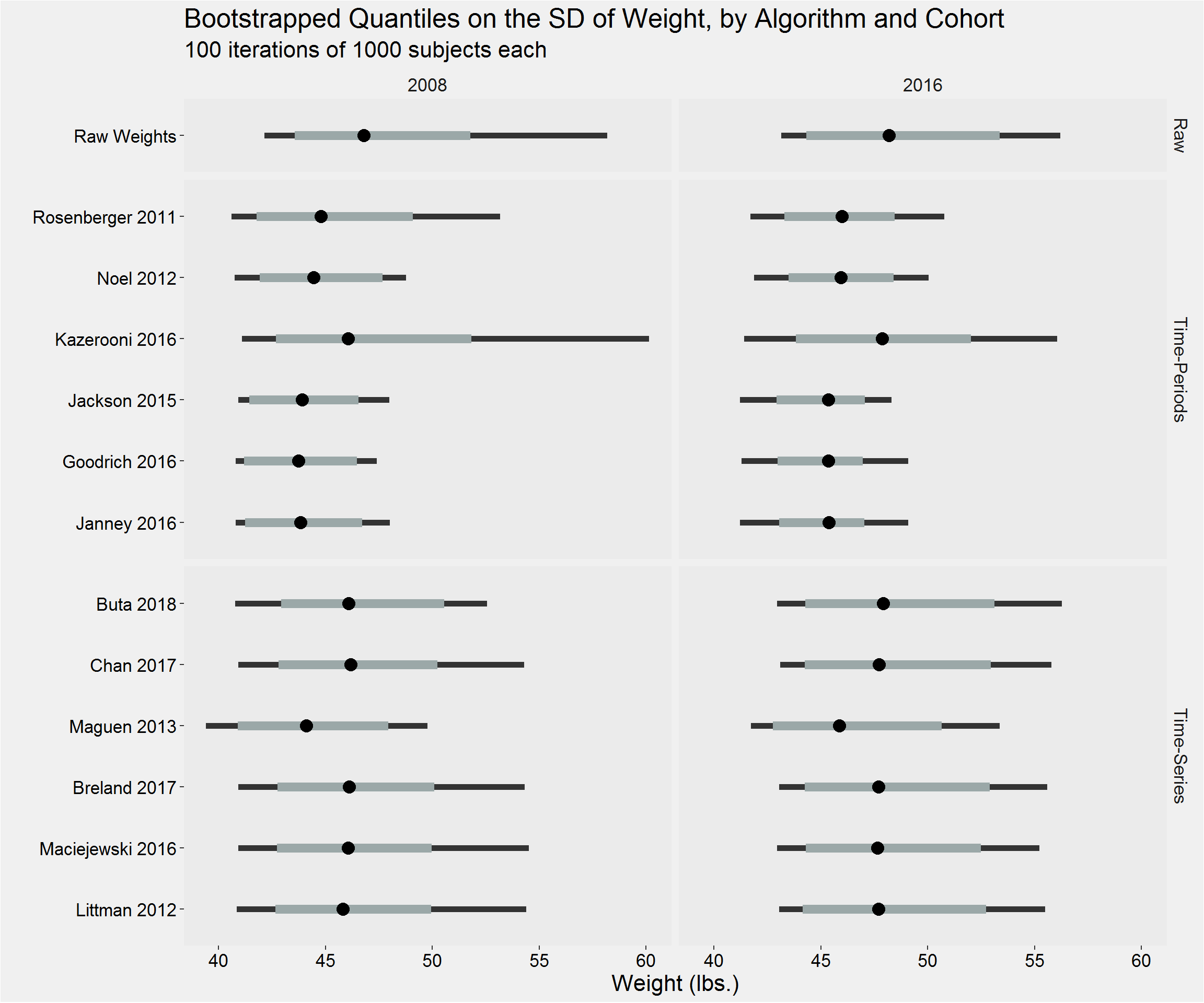
Distribution of the sample standard deviation
Alternative, ridgeline plot