Sample Exploration
Packages
SQL Database Connection
Tables in the DB:
| TABLE_CAT | TABLE_SCHEM | TABLE_NAME | TABLE_TYPE | REMARKS |
|---|---|---|---|---|
| NCP_DCEP | Dflt | TLC_AuditC | TABLE | NA |
| NCP_DCEP | Dflt | TLC_CohortKey | TABLE | NA |
| NCP_DCEP | Dflt | TLC_DX_PatSumByDXGroup | TABLE | NA |
| NCP_DCEP | Dflt | TLC_DX_PatSumByICD9 | TABLE | NA |
| NCP_DCEP | Dflt | TLC_EnrollDataBL | TABLE | NA |
| NCP_DCEP | Dflt | TLC_EnrollStatusBL | TABLE | NA |
| NCP_DCEP | Dflt | TLC_FilterLab | TABLE | NA |
| NCP_DCEP | Dflt | TLC_HERC_DISCHG | TABLE | NA |
| NCP_DCEP | Dflt | TLC_HERC_OP | TABLE | NA |
| NCP_DCEP | Dflt | TLC_HF_Smoke | TABLE | NA |
| NCP_DCEP | Dflt | TLC_ICDCode | TABLE | NA |
| NCP_DCEP | Dflt | TLC_LabA1C | TABLE | NA |
| NCP_DCEP | Dflt | TLC_NOSOS_NOPHA | TABLE | NA |
| NCP_DCEP | Dflt | TLC_NOSOS_PHA | TABLE | NA |
| NCP_DCEP | Dflt | TLC_Vital_BP | TABLE | NA |
| NCP_DCEP | Dflt | TLC_Vital_Weight | TABLE | NA |
| NCP_DCEP | Dflt | tmpRawImport_TLC_NCP_Cohort01 | TABLE | NA |
| NCP_DCEP | samp | BariatricSurgCodes | TABLE | NA |
| NCP_DCEP | samp | Distances2016 | TABLE | NA |
| NCP_DCEP | samp | heightSamples | TABLE | NA |
| NCP_DCEP | samp | MOVE2017_denom | TABLE | NA |
| NCP_DCEP | samp | MOVE2017_num_weights | TABLE | NA |
| NCP_DCEP | samp | MOVE2017_numerator | TABLE | NA |
| NCP_DCEP | samp | MOVEvisits2016 | TABLE | NA |
| NCP_DCEP | samp | NationalHeightSamples | TABLE | NA |
| NCP_DCEP | samp | NationalWeightSamples | TABLE | NA |
| NCP_DCEP | samp | PregnancyCodes | TABLE | NA |
| NCP_DCEP | samp | weightSamples | TABLE | NA |
| NCP_DCEP | sys | trace_xe_action_map | TABLE | NA |
| NCP_DCEP | sys | trace_xe_event_map | TABLE | NA |
Weight samples are held in Samp.NationalWeightSamples
Height samples are held in Samp.NationalHeightSamples
Pre-Process Data
We will be omitting the MOVE! cohort from the following analyses.
Pre-process data,
diab <- c(
"Non-Diabetic",
"Diabetes After",
"Diabetes Before",
"Diabetes Before and After"
)
race_eth_facs <- c("Non-Hispanic White",
"Hispanic White",
"Non-Hispanic Black",
"Hispanic Black",
"Other",
"Unknown")
weightSamples <- weightSamples %>%
filter(is.na(Pregnant)) %>%
select(-Pregnant) %>%
mutate_at(vars(PatientICN:PatientSID), as.character) %>%
mutate(
Sta3n = factor(Sta3n),
SampleYear = factor(SampleYear),
VisitDate = as.Date(VisitDateTime, tz = "UTC", '%Y-%m-%d'),
Bariatric = factor(Bariatric,
levels = 0:1,
labels = c("No", "Yes")),
InptWeight = ifelse(is.na(InptWeight), 0, InptWeight),
InptWeight = factor(InptWeight,
levels = 0:1,
labels = c("Outpatient", "Inpatient")),
Diabetic = ifelse(is.na(DiabetesTiming), 0, 1),
Diabetic = factor(Diabetic,
0:1,
c("Non-Diabetic", "Diabetic")),
DiabetesTiming = as.character(DiabetesTiming),
DiabetesTiming = ifelse(is.na(DiabetesTiming),
"Non-Diabetic",
DiabetesTiming),
DiabetesTiming = factor(DiabetesTiming, diab, diab),
race_eth = case_when(
Race == "WHITE" & Ethnicity == "NOT HISPANIC OR LATINO" ~ 0,
Race == "WHITE" & Ethnicity == "HISPANIC OR LATINO" ~ 1,
Race == "BLACK OR AFRICAN AMERICAN"
& Ethnicity == "NOT HISPANIC OR LATINO" ~ 2,
Race == "BLACK OR AFRICAN AMERICAN"
& Ethnicity == "HISPANIC OR LATINO" ~ 3,
(is.na(Race) | Race == "Missing")
& (is.na(Ethnicity) | Ethnicity == "UNKNOWN BY PATIENT") ~ 5,
TRUE ~ 4
),
race_eth = factor(race_eth, 0:5, race_eth_facs)
) %>%
filter(!is.na(Weight))
heightSamples <- heightSamples %>%
mutate_at(vars(PatientICN:PatientSID), as.character) %>%
inner_join(weightSamples %>%
distinct(PatientICN, VisitDateTime),
by = c("PatientICN", "VisitDateTime")) %>%
mutate(
SampleYear = case_when(
lubridate::year(VisitDateTime) %in% c(2007, 2008) ~ '2008',
lubridate::year(VisitDateTime) %in% c(2015, 2016) ~ '2016'
)
) %>%
filter(!is.na(Height))Overall Summaries
Number of people in each cohort, by sample year
weightSamples %>%
distinct(PatientICN, SampleYear) %>%
group_by(SampleYear) %>%
count() %>%
tableStyle()| SampleYear | n |
|---|---|
| 2008 | 98786 |
| 2016 | 98958 |
How many weight measurements per person, on average?
weightSamples %>%
group_by(SampleYear) %>%
count(PatientICN) %>%
summarise(
mean = mean(n),
SD = sd(n),
median = median(n),
min = min(n),
max = max(n)
) %>%
tableStyle()| SampleYear | mean | SD | median | min | max |
|---|---|---|---|---|---|
| 2008 | 12.29 | 15.97 | 9 | 1 | 1479 |
| 2016 | 12.21 | 24.85 | 8 | 1 | 4981 |
How many height measurements per person, on average?
heightSamples %>%
group_by(SampleYear) %>%
count(PatientICN) %>%
summarise(
mean = mean(n),
SD = sd(n),
median = median(n),
min = min(n),
max = max(n)
) %>%
tableStyle()| SampleYear | mean | SD | median | min | max |
|---|---|---|---|---|---|
| 2008 | 5.60 | 5.33 | 4 | 1 | 131 |
| 2016 | 5.53 | 5.00 | 4 | 1 | 105 |
Weight measurements per person
weightSamples %>%
count(PatientICN) %>%
ggplot(aes(x = n)) %>%
add(geom_histogram(
binwidth = 1,
color = "black",
fill = RColorBrewer::brewer.pal(3, "Set1")[1])) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(labs(
x = 'Weight Samples per Person',
y = 'Frequency'
))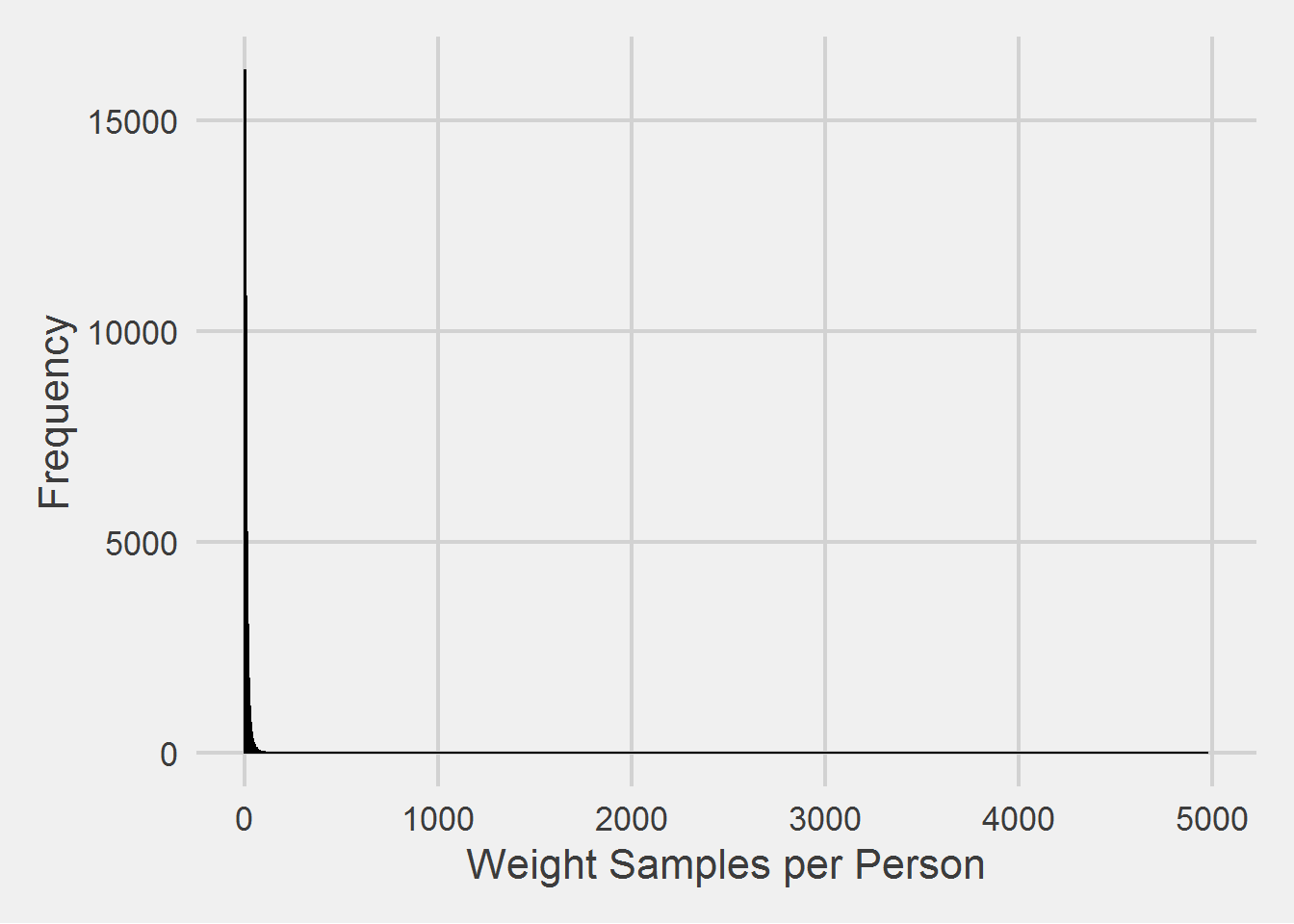
It may look like a lot of people are lacking weight samples (Weight Samples per Person = 0), but there are in fact, no people in the cohort without a weight sample; must pay attention to the scale!
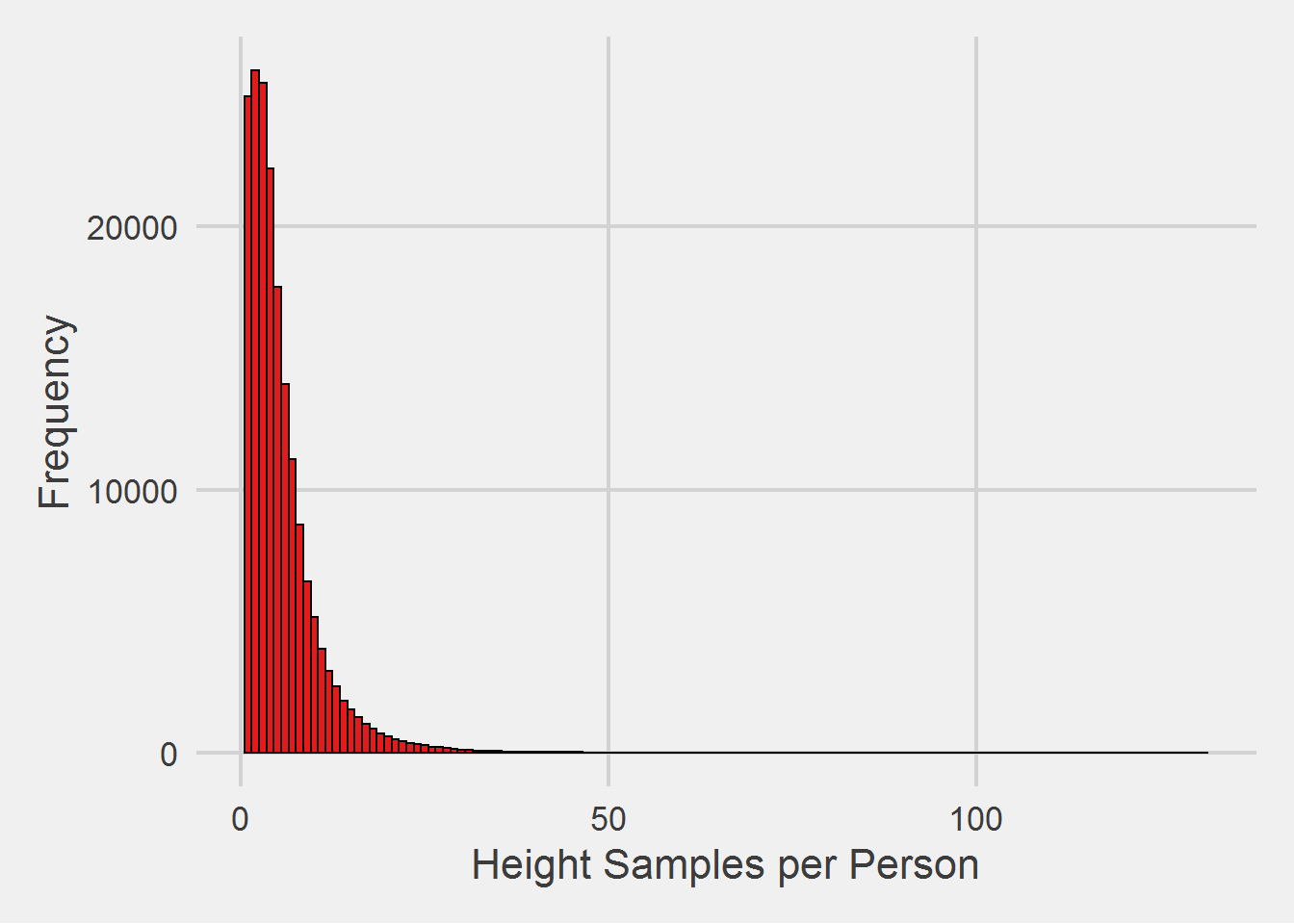
Height measurements per person
heightSamples %>%
count(PatientICN) %>%
ggplot(aes(x = n)) %>%
add(geom_histogram(
binwidth = 1,
color = "black",
fill = RColorBrewer::brewer.pal(3, "Set1")[1])) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(labs(
x = 'Height Samples per Person',
y = 'Frequency'
))
Age Distribution
At time of PCP visit
weightSamples %>%
distinct(PatientICN, AgeAtVisit, SampleYear) %>%
group_by(SampleYear) %>%
summarise(
mean = mean(AgeAtVisit),
sd = sd(AgeAtVisit),
median = median(AgeAtVisit),
min = min(AgeAtVisit),
max = max(AgeAtVisit)
) %>%
tableStyle()| SampleYear | mean | sd | median | min | max |
|---|---|---|---|---|---|
| 2008 | 64.04 | 14.8 | 63.77 | 18.18 | 101.09 |
| 2016 | 62.95 | 15.8 | 66.19 | 19.05 | 104.15 |
Race/Ethnicity
Without consideration of sample year,
weightSamples %>%
janitor::tabyl(Race) %>%
arrange(desc(n)) %>%
adorn_pct_formatting() %>%
tableStyle()| Race | n | percent | valid_percent |
|---|---|---|---|
| WHITE | 1738504 | 71.7% | 73.9% |
| BLACK OR AFRICAN AMERICAN | 445484 | 18.4% | 18.9% |
| Missing | 96399 | 4.0% | 4.1% |
| NA | 69886 | 2.9% |
|
| NATIVE HAWAIIAN OR OTHER PACIFIC ISLANDER | 22099 | 0.9% | 0.9% |
| AMERICAN INDIAN OR ALASKA NATIVE | 18299 | 0.8% | 0.8% |
| ASIAN | 17800 | 0.7% | 0.8% |
| MULTI-RACIAL | 14656 | 0.6% | 0.6% |
For our purposes, I’ll add any NA entries as Missing,
weightSamples <- weightSamples %>%
mutate(
Race = fct_explicit_na(Race, na_level = "Missing"),
Race = fct_infreq(Race)
)
weightSamples %>%
tabyl(Race, SampleYear) %>%
adorn_percentages("col") %>%
adorn_pct_formatting() %>%
adorn_ns(position = "front") %>%
tableStyle()| Race | 2008 | 2016 |
|---|---|---|
| WHITE | 870225 (71.7%) | 868279 (71.8%) |
| BLACK OR AFRICAN AMERICAN | 207869 (17.1%) | 237615 (19.7%) |
| Missing | 102163 (8.4%) | 64122 (5.3%) |
| NATIVE HAWAIIAN OR OTHER PACIFIC ISLANDER | 11344 (0.9%) | 10755 (0.9%) |
| AMERICAN INDIAN OR ALASKA NATIVE | 8299 (0.7%) | 10000 (0.8%) |
| ASIAN | 6473 (0.5%) | 11327 (0.9%) |
| MULTI-RACIAL | 8020 (0.7%) | 6636 (0.5%) |
“Missing” is the 3rd largest grouping, interesting …
These frequencies were by weight samples, now we’ll look at individual samples.
weightSamples %>%
distinct(SampleYear, PatientICN, Race) %>%
tabyl(Race, SampleYear) %>%
adorn_percentages("col") %>%
adorn_pct_formatting() %>%
adorn_ns(position = "front") %>%
tableStyle()| Race | 2008 | 2016 |
|---|---|---|
| WHITE | 71269 (72.1%) | 73200 (74.0%) |
| BLACK OR AFRICAN AMERICAN | 13501 (13.7%) | 16178 (16.3%) |
| Missing | 11401 (11.5%) | 6401 (6.5%) |
| NATIVE HAWAIIAN OR OTHER PACIFIC ISLANDER | 860 (0.9%) | 897 (0.9%) |
| AMERICAN INDIAN OR ALASKA NATIVE | 610 (0.6%) | 856 (0.9%) |
| ASIAN | 601 (0.6%) | 947 (1.0%) |
| MULTI-RACIAL | 544 (0.6%) | 479 (0.5%) |
“Missing” is still the 3rd largest group.
Let’s see if there are any Sta3n-differences, or maybe Sta6a differences, by frequency of “Missing”,
sta3n_race_miss <- weightSamples %>%
filter(SampleYear == "2016") %>%
distinct(PatientICN, Sta3n, Race) %>%
tabyl(Sta3n, Race) %>%
adorn_percentages() %>%
adorn_ns() %>%
select(Sta3n, Missing) %>%
separate(col = Missing, into = c("p", "N"), sep = "\\s+") %>%
mutate(
p = round(as.numeric(p), 4),
N = as.integer(str_remove_all(N, "\\(|\\)")),
q = cut_interval(p, n = 10),
qth = as.integer(q)
)Top 10% of Sta3n with “Missing” race, and bottom 10%
sta3n_race_miss %>%
filter(qth %in% c(1, 10)) %>%
arrange(desc(qth)) %>%
select(-qth) %>%
scrollTable()| Sta3n | p | N | q |
|---|---|---|---|
| 438 | 0.21 | 95 | (0.205,0.227] |
| 501 | 0.21 | 173 | (0.205,0.227] |
| 504 | 0.22 | 81 | (0.205,0.227] |
| 570 | 0.22 | 101 | (0.205,0.227] |
| 756 | 0.23 | 102 | (0.205,0.227] |
| 442 | 0.02 | 8 | [0.0058,0.0279] |
| 503 | 0.01 | 7 | [0.0058,0.0279] |
| 509 | 0.01 | 5 | [0.0058,0.0279] |
| 517 | 0.02 | 6 | [0.0058,0.0279] |
| 523 | 0.03 | 14 | [0.0058,0.0279] |
| 529 | 0.01 | 2 | [0.0058,0.0279] |
| 540 | 0.02 | 7 | [0.0058,0.0279] |
| 552 | 0.01 | 6 | [0.0058,0.0279] |
| 556 | 0.03 | 9 | [0.0058,0.0279] |
| 558 | 0.02 | 19 | [0.0058,0.0279] |
| 580 | 0.02 | 36 | [0.0058,0.0279] |
| 581 | 0.02 | 9 | [0.0058,0.0279] |
| 585 | 0.02 | 8 | [0.0058,0.0279] |
| 586 | 0.02 | 10 | [0.0058,0.0279] |
| 590 | 0.02 | 13 | [0.0058,0.0279] |
| 595 | 0.02 | 11 | [0.0058,0.0279] |
| 603 | 0.01 | 11 | [0.0058,0.0279] |
| 629 | 0.02 | 10 | [0.0058,0.0279] |
| 632 | 0.03 | 13 | [0.0058,0.0279] |
| 637 | 0.02 | 15 | [0.0058,0.0279] |
| 646 | 0.03 | 24 | [0.0058,0.0279] |
| 652 | 0.02 | 14 | [0.0058,0.0279] |
| 660 | 0.01 | 7 | [0.0058,0.0279] |
| 667 | 0.02 | 12 | [0.0058,0.0279] |
| 676 | 0.01 | 3 | [0.0058,0.0279] |
| 692 | 0.02 | 4 | [0.0058,0.0279] |
Overall distribution,
tmp <- sta3n_race_miss %>%
arrange(p) %>%
mutate(Sta3n_deident = row_number())
tmp %>%
ggplot(aes(x = Sta3n_deident, y = p)) %>%
add(geom_point(aes(size = N, fill = q), pch = 21)) %>%
add(gghighlight::gghighlight(
p > 0.15,
label_key = Sta3n
)) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(
axis.title = element_text(),
legend.position = "none"
)) %>%
add(labs(
x = 'Sta3n Deidentified',
y = 'Proportion Missing Race',
caption = "Point size relative to Sta3n numerator size;
Color Indicates Decile"
))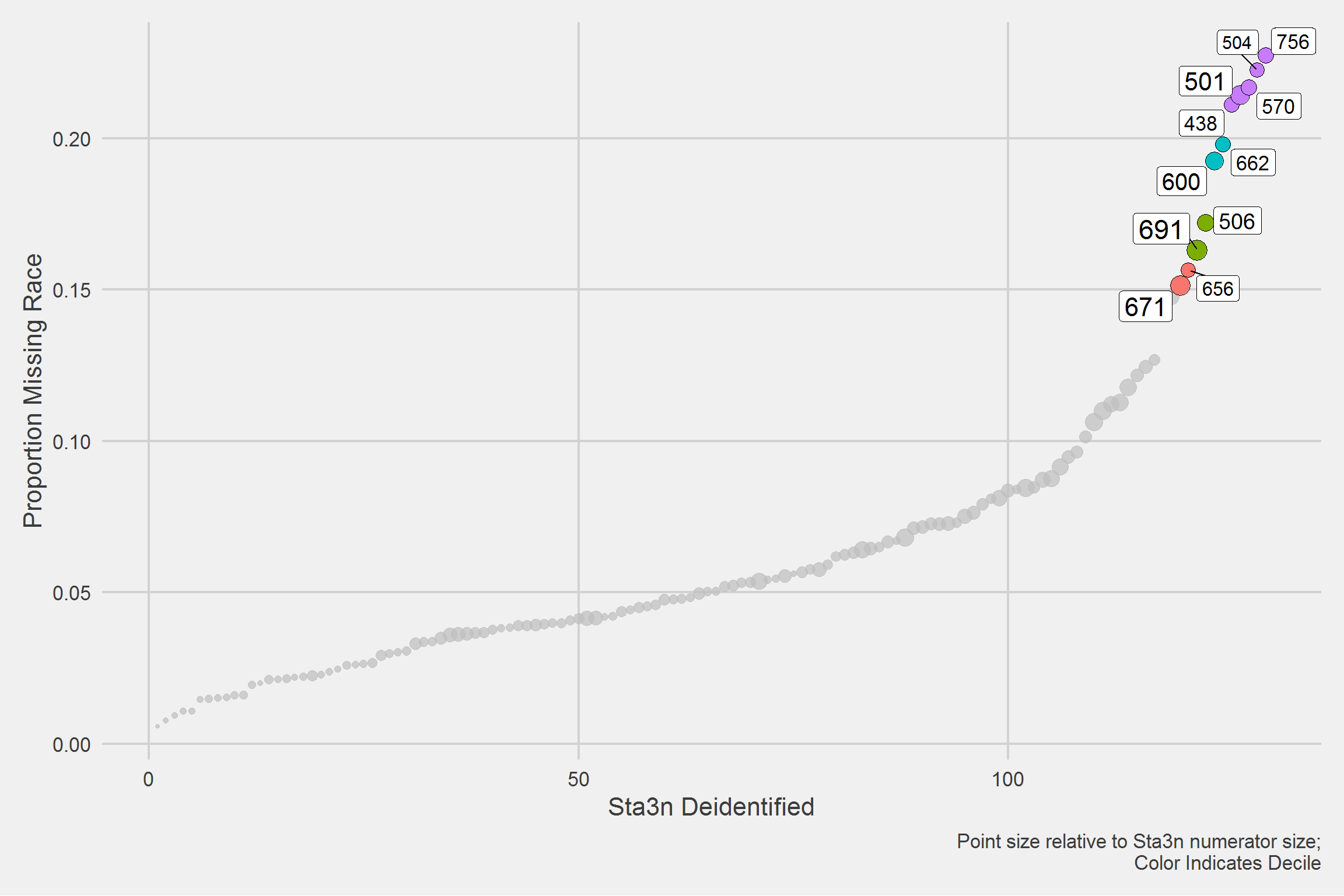
I admit this figure is bizarre, because I have the x-axis as de-identified Sta3n’s but I’m highlighting the actual Sta3n number, for those in excess of >15% missing Race.
What I’m seeing is that “missingness” is proportional to Sta3n size.
Let’s do this by Sta6a,
sta6a_race_miss <- weightSamples %>%
filter(SampleYear == "2016") %>%
distinct(PatientICN, Sta6a, Race) %>%
tabyl(Sta6a, Race) %>%
adorn_percentages() %>%
adorn_ns() %>%
select(Sta6a, Missing) %>%
separate(col = Missing, into = c("p", "N"), sep = "\\s+") %>%
mutate(
p = round(as.numeric(p), 4),
N = as.integer(str_remove_all(N, "\\(|\\)")),
q = cut_interval(p, n = 10),
qth = as.integer(q)
)Top 10% of Sta6a with “Missing” race, and bottom 10%
sta6a_race_miss %>%
filter(qth %in% c(1, 10)) %>%
arrange(desc(qth)) %>%
select(-qth) %>%
scrollTable()| Sta6a | p | N | q |
|---|---|---|---|
| 501G2 | 0.63 | 12 | (0.568,0.632] |
| 358 | 0.06 | 5 | [0,0.0632] |
| 402 | 0.04 | 10 | [0,0.0632] |
| 402GB | 0.00 | 0 | [0,0.0632] |
| 402GC | 0.05 | 1 | [0,0.0632] |
| 402GE | 0.00 | 0 | [0,0.0632] |
| 402GF | 0.00 | 0 | [0,0.0632] |
| 402HB | 0.02 | 2 | [0,0.0632] |
| 402HC | 0.03 | 2 | [0,0.0632] |
| 402HL | 0.00 | 0 | [0,0.0632] |
| 402QA | 0.00 | 0 | [0,0.0632] |
| 402QB | 0.00 | 0 | [0,0.0632] |
| 405 | 0.04 | 7 | [0,0.0632] |
| 405GA | 0.00 | 0 | [0,0.0632] |
| 405GC | 0.00 | 0 | [0,0.0632] |
| 405HA | 0.01 | 1 | [0,0.0632] |
| 405HC | 0.05 | 2 | [0,0.0632] |
| 436 | 0.04 | 5 | [0,0.0632] |
| 436GA | 0.00 | 0 | [0,0.0632] |
| 436GK | 0.00 | 0 | [0,0.0632] |
| 436GM | 0.00 | 0 | [0,0.0632] |
| 437GD | 0.06 | 3 | [0,0.0632] |
| 437GE | 0.02 | 1 | [0,0.0632] |
| 442 | 0.01 | 1 | [0,0.0632] |
| 442BU | 0.00 | 0 | [0,0.0632] |
| 442GB | 0.00 | 0 | [0,0.0632] |
| 442GC | 0.04 | 4 | [0,0.0632] |
| 442GD | 0.04 | 3 | [0,0.0632] |
| 442HK | 0.00 | 0 | [0,0.0632] |
| 442QA | 0.00 | 0 | [0,0.0632] |
| 442QB | 0.00 | 0 | [0,0.0632] |
| 459 | 0.05 | 12 | [0,0.0632] |
| 459GB | 0.05 | 2 | [0,0.0632] |
| 459GC | 0.04 | 1 | [0,0.0632] |
| 459GD | 0.00 | 0 | [0,0.0632] |
| 459GF | 0.00 | 0 | [0,0.0632] |
| 459GG | 0.02 | 1 | [0,0.0632] |
| 460 | 0.05 | 10 | [0,0.0632] |
| 460GA | 0.03 | 2 | [0,0.0632] |
| 460HG | 0.03 | 1 | [0,0.0632] |
| 460HK | 0.00 | 0 | [0,0.0632] |
| 463 | 0.04 | 7 | [0,0.0632] |
| 463GB | 0.03 | 1 | [0,0.0632] |
| 501GB | 0.03 | 1 | [0,0.0632] |
| 502 | 0.04 | 8 | [0,0.0632] |
| 502GA | 0.06 | 3 | [0,0.0632] |
| 502GB | 0.04 | 5 | [0,0.0632] |
| 502GE | 0.03 | 1 | [0,0.0632] |
| 502GF | 0.02 | 1 | [0,0.0632] |
| 503 | 0.01 | 1 | [0,0.0632] |
| 503GA | 0.01 | 1 | [0,0.0632] |
| 503GB | 0.01 | 1 | [0,0.0632] |
| 503GC | 0.02 | 2 | [0,0.0632] |
| 503GD | 0.04 | 1 | [0,0.0632] |
| 503GE | 0.04 | 1 | [0,0.0632] |
| 504HB | 0.00 | 0 | [0,0.0632] |
| 508 | 0.04 | 13 | [0,0.0632] |
| 508GA | 0.01 | 2 | [0,0.0632] |
| 508GE | 0.06 | 6 | [0,0.0632] |
| 508GF | 0.01 | 1 | [0,0.0632] |
| 508GG | 0.03 | 4 | [0,0.0632] |
| 508GH | 0.04 | 5 | [0,0.0632] |
| 508GI | 0.04 | 4 | [0,0.0632] |
| 508GK | 0.03 | 2 | [0,0.0632] |
| 508QF | 0.05 | 11 | [0,0.0632] |
| 509 | 0.00 | 0 | [0,0.0632] |
| 509A0 | 0.01 | 4 | [0,0.0632] |
| 509GA | 0.00 | 0 | [0,0.0632] |
| 509GB | 0.02 | 1 | [0,0.0632] |
| 512 | 0.02 | 5 | [0,0.0632] |
| 512A5 | 0.01 | 1 | [0,0.0632] |
| 512GC | 0.02 | 2 | [0,0.0632] |
| 512GD | 0.02 | 2 | [0,0.0632] |
| 512GE | 0.00 | 0 | [0,0.0632] |
| 512GF | 0.00 | 0 | [0,0.0632] |
| 512GG | 0.05 | 3 | [0,0.0632] |
| 515 | 0.06 | 12 | [0,0.0632] |
| 515BY | 0.05 | 14 | [0,0.0632] |
| 515GA | 0.04 | 3 | [0,0.0632] |
| 516 | 0.04 | 22 | [0,0.0632] |
| 516GA | 0.05 | 9 | [0,0.0632] |
| 516GF | 0.04 | 5 | [0,0.0632] |
| 517 | 0.03 | 6 | [0,0.0632] |
| 517GB | 0.00 | 0 | [0,0.0632] |
| 517QA | 0.00 | 0 | [0,0.0632] |
| 518GE | 0.04 | 1 | [0,0.0632] |
| 519GD | 0.00 | 0 | [0,0.0632] |
| 519HD | 0.00 | 0 | [0,0.0632] |
| 520 | 0.04 | 10 | [0,0.0632] |
| 520BZ | 0.05 | 15 | [0,0.0632] |
| 521 | 0.06 | 19 | [0,0.0632] |
| 521GA | 0.06 | 11 | [0,0.0632] |
| 521GE | 0.04 | 2 | [0,0.0632] |
| 521GH | 0.05 | 2 | [0,0.0632] |
| 521GI | 0.03 | 2 | [0,0.0632] |
| 521GJ | 0.06 | 1 | [0,0.0632] |
| 523 | 0.03 | 4 | [0,0.0632] |
| 523A4 | 0.01 | 1 | [0,0.0632] |
| 523A5 | 0.01 | 1 | [0,0.0632] |
| 523BY | 0.04 | 2 | [0,0.0632] |
| 523BZ | 0.05 | 3 | [0,0.0632] |
| 523GC | 0.05 | 1 | [0,0.0632] |
| 523GD | 0.00 | 0 | [0,0.0632] |
| 526GB | 0.00 | 0 | [0,0.0632] |
| 528 | 0.01 | 3 | [0,0.0632] |
| 528A4 | 0.00 | 0 | [0,0.0632] |
| 528A5 | 0.02 | 2 | [0,0.0632] |
| 528A6 | 0.03 | 3 | [0,0.0632] |
| 528A7 | 0.04 | 9 | [0,0.0632] |
| 528A8 | 0.01 | 2 | [0,0.0632] |
| 528G3 | 0.00 | 0 | [0,0.0632] |
| 528G4 | 0.00 | 0 | [0,0.0632] |
| 528G6 | 0.00 | 0 | [0,0.0632] |
| 528G7 | 0.04 | 1 | [0,0.0632] |
| 528G8 | 0.00 | 0 | [0,0.0632] |
| 528GB | 0.05 | 2 | [0,0.0632] |
| 528GC | 0.00 | 0 | [0,0.0632] |
| 528GD | 0.00 | 0 | [0,0.0632] |
| 528GE | 0.02 | 3 | [0,0.0632] |
| 528GL | 0.03 | 1 | [0,0.0632] |
| 528GO | 0.02 | 1 | [0,0.0632] |
| 528GP | 0.06 | 2 | [0,0.0632] |
| 528GQ | 0.02 | 1 | [0,0.0632] |
| 528GR | 0.04 | 1 | [0,0.0632] |
| 528GV | 0.00 | 0 | [0,0.0632] |
| 528GX | 0.00 | 0 | [0,0.0632] |
| 528GY | 0.03 | 1 | [0,0.0632] |
| 528GZ | 0.00 | 0 | [0,0.0632] |
| 529 | 0.01 | 1 | [0,0.0632] |
| 529GA | 0.00 | 0 | [0,0.0632] |
| 529GB | 0.00 | 0 | [0,0.0632] |
| 529GC | 0.03 | 1 | [0,0.0632] |
| 529GD | 0.00 | 0 | [0,0.0632] |
| 529GF | 0.00 | 0 | [0,0.0632] |
| 534GD | 0.06 | 8 | [0,0.0632] |
| 534QA | 0.06 | 2 | [0,0.0632] |
| 537 | 0.05 | 14 | [0,0.0632] |
| 537BY | 0.04 | 8 | [0,0.0632] |
| 537GA | 0.03 | 1 | [0,0.0632] |
| 537GD | 0.04 | 3 | [0,0.0632] |
| 537HA | 0.06 | 2 | [0,0.0632] |
| 538 | 0.05 | 8 | [0,0.0632] |
| 538GA | 0.03 | 1 | [0,0.0632] |
| 538GB | 0.05 | 2 | [0,0.0632] |
| 538GD | 0.02 | 1 | [0,0.0632] |
| 538GE | 0.00 | 0 | [0,0.0632] |
| 539 | 0.04 | 8 | [0,0.0632] |
| 539GA | 0.02 | 1 | [0,0.0632] |
| 539GB | 0.04 | 4 | [0,0.0632] |
| 539GC | 0.01 | 1 | [0,0.0632] |
| 539GD | 0.02 | 1 | [0,0.0632] |
| 539GF | 0.00 | 0 | [0,0.0632] |
| 539QA | 0.00 | 0 | [0,0.0632] |
| 540 | 0.01 | 2 | [0,0.0632] |
| 540GA | 0.00 | 0 | [0,0.0632] |
| 540GB | 0.04 | 2 | [0,0.0632] |
| 540GC | 0.05 | 1 | [0,0.0632] |
| 540GD | 0.05 | 2 | [0,0.0632] |
| 540HK | 0.00 | 0 | [0,0.0632] |
| 541 | 0.05 | 15 | [0,0.0632] |
| 541BY | 0.00 | 0 | [0,0.0632] |
| 541BZ | 0.01 | 1 | [0,0.0632] |
| 541GB | 0.00 | 0 | [0,0.0632] |
| 541GC | 0.04 | 3 | [0,0.0632] |
| 541GD | 0.00 | 0 | [0,0.0632] |
| 541GE | 0.03 | 1 | [0,0.0632] |
| 541GG | 0.03 | 7 | [0,0.0632] |
| 541GH | 0.00 | 0 | [0,0.0632] |
| 541GI | 0.00 | 0 | [0,0.0632] |
| 541GJ | 0.00 | 0 | [0,0.0632] |
| 541GK | 0.05 | 3 | [0,0.0632] |
| 542 | 0.04 | 5 | [0,0.0632] |
| 542GE | 0.00 | 0 | [0,0.0632] |
| 544 | 0.02 | 11 | [0,0.0632] |
| 544BZ | 0.04 | 10 | [0,0.0632] |
| 544GB | 0.05 | 7 | [0,0.0632] |
| 544GC | 0.04 | 6 | [0,0.0632] |
| 544GD | 0.02 | 2 | [0,0.0632] |
| 544GE | 0.00 | 0 | [0,0.0632] |
| 544GF | 0.02 | 2 | [0,0.0632] |
| 544GG | 0.05 | 6 | [0,0.0632] |
| 546 | 0.04 | 10 | [0,0.0632] |
| 546BZ | 0.04 | 10 | [0,0.0632] |
| 546GA | 0.00 | 0 | [0,0.0632] |
| 546GE | 0.00 | 0 | [0,0.0632] |
| 546GH | 0.00 | 0 | [0,0.0632] |
| 548GA | 0.00 | 0 | [0,0.0632] |
| 548GB | 0.02 | 3 | [0,0.0632] |
| 548GC | 0.03 | 4 | [0,0.0632] |
| 548GE | 0.03 | 2 | [0,0.0632] |
| 548GF | 0.05 | 2 | [0,0.0632] |
| 549 | 0.05 | 39 | [0,0.0632] |
| 549BY | 0.05 | 28 | [0,0.0632] |
| 549GA | 0.00 | 0 | [0,0.0632] |
| 549GF | 0.03 | 1 | [0,0.0632] |
| 549GH | 0.05 | 2 | [0,0.0632] |
| 549QC | 0.04 | 4 | [0,0.0632] |
| 550 | 0.03 | 5 | [0,0.0632] |
| 550GA | 0.00 | 0 | [0,0.0632] |
| 550GC | 0.00 | 0 | [0,0.0632] |
| 550GD | 0.01 | 1 | [0,0.0632] |
| 552 | 0.02 | 6 | [0,0.0632] |
| 552GA | 0.00 | 0 | [0,0.0632] |
| 552GB | 0.00 | 0 | [0,0.0632] |
| 552GC | 0.00 | 0 | [0,0.0632] |
| 552GD | 0.00 | 0 | [0,0.0632] |
| 553 | 0.03 | 13 | [0,0.0632] |
| 553GB | 0.03 | 2 | [0,0.0632] |
| 554GI | 0.00 | 0 | [0,0.0632] |
| 556 | 0.01 | 2 | [0,0.0632] |
| 556GA | 0.06 | 2 | [0,0.0632] |
| 556GC | 0.03 | 2 | [0,0.0632] |
| 557 | 0.01 | 2 | [0,0.0632] |
| 557GA | 0.05 | 5 | [0,0.0632] |
| 557GC | 0.00 | 0 | [0,0.0632] |
| 557GE | 0.04 | 2 | [0,0.0632] |
| 557HA | 0.05 | 2 | [0,0.0632] |
| 558 | 0.02 | 8 | [0,0.0632] |
| 558GA | 0.02 | 4 | [0,0.0632] |
| 558GB | 0.02 | 4 | [0,0.0632] |
| 558GC | 0.04 | 3 | [0,0.0632] |
| 561A4 | 0.06 | 8 | [0,0.0632] |
| 561BZ | 0.01 | 2 | [0,0.0632] |
| 561GF | 0.04 | 1 | [0,0.0632] |
| 561GH | 0.00 | 0 | [0,0.0632] |
| 561GI | 0.03 | 2 | [0,0.0632] |
| 562 | 0.04 | 8 | [0,0.0632] |
| 562GA | 0.02 | 1 | [0,0.0632] |
| 562GB | 0.04 | 2 | [0,0.0632] |
| 562GC | 0.00 | 0 | [0,0.0632] |
| 562GD | 0.00 | 0 | [0,0.0632] |
| 562GE | 0.03 | 1 | [0,0.0632] |
| 564 | 0.04 | 11 | [0,0.0632] |
| 564BY | 0.05 | 13 | [0,0.0632] |
| 564GB | 0.01 | 2 | [0,0.0632] |
| 564GC | 0.01 | 1 | [0,0.0632] |
| 564GE | 0.03 | 1 | [0,0.0632] |
| 565 | 0.00 | 0 | [0,0.0632] |
| 565GA | 0.05 | 9 | [0,0.0632] |
| 565GC | 0.03 | 7 | [0,0.0632] |
| 565GE | 0.04 | 2 | [0,0.0632] |
| 565GG | 0.04 | 1 | [0,0.0632] |
| 565GL | 0.03 | 15 | [0,0.0632] |
| 568 | 0.01 | 1 | [0,0.0632] |
| 568A4 | 0.00 | 0 | [0,0.0632] |
| 568GA | 0.05 | 4 | [0,0.0632] |
| 568GB | 0.04 | 1 | [0,0.0632] |
| 568HA | 0.00 | 0 | [0,0.0632] |
| 568HB | 0.00 | 0 | [0,0.0632] |
| 568HF | 0.00 | 0 | [0,0.0632] |
| 568HJ | 0.00 | 0 | [0,0.0632] |
| 568HM | 0.00 | 0 | [0,0.0632] |
| 573A4 | 0.03 | 6 | [0,0.0632] |
| 573GD | 0.03 | 7 | [0,0.0632] |
| 573GE | 0.03 | 3 | [0,0.0632] |
| 573GF | 0.05 | 11 | [0,0.0632] |
| 573GI | 0.06 | 15 | [0,0.0632] |
| 573GJ | 0.00 | 0 | [0,0.0632] |
| 573GK | 0.00 | 0 | [0,0.0632] |
| 573GL | 0.05 | 2 | [0,0.0632] |
| 573GM | 0.02 | 1 | [0,0.0632] |
| 573QJ | 0.05 | 3 | [0,0.0632] |
| 575GB | 0.00 | 0 | [0,0.0632] |
| 578 | 0.04 | 15 | [0,0.0632] |
| 578GA | 0.03 | 3 | [0,0.0632] |
| 578GD | 0.00 | 0 | [0,0.0632] |
| 578GF | 0.04 | 2 | [0,0.0632] |
| 580 | 0.03 | 20 | [0,0.0632] |
| 580BY | 0.00 | 0 | [0,0.0632] |
| 580BZ | 0.00 | 0 | [0,0.0632] |
| 580GC | 0.01 | 2 | [0,0.0632] |
| 580GD | 0.02 | 4 | [0,0.0632] |
| 580GE | 0.04 | 5 | [0,0.0632] |
| 580GF | 0.05 | 2 | [0,0.0632] |
| 580GG | 0.01 | 1 | [0,0.0632] |
| 580GH | 0.01 | 2 | [0,0.0632] |
| 581 | 0.03 | 8 | [0,0.0632] |
| 581GA | 0.00 | 0 | [0,0.0632] |
| 581GB | 0.01 | 1 | [0,0.0632] |
| 583 | 0.05 | 31 | [0,0.0632] |
| 583GC | 0.03 | 1 | [0,0.0632] |
| 583GF | 0.06 | 1 | [0,0.0632] |
| 585 | 0.03 | 3 | [0,0.0632] |
| 585GA | 0.00 | 0 | [0,0.0632] |
| 585GB | 0.00 | 0 | [0,0.0632] |
| 585GC | 0.00 | 0 | [0,0.0632] |
| 585HA | 0.03 | 1 | [0,0.0632] |
| 586 | 0.00 | 0 | [0,0.0632] |
| 586GA | 0.00 | 0 | [0,0.0632] |
| 586GB | 0.02 | 1 | [0,0.0632] |
| 586GC | 0.00 | 0 | [0,0.0632] |
| 586GD | 0.00 | 0 | [0,0.0632] |
| 586GF | 0.04 | 1 | [0,0.0632] |
| 586GG | 0.05 | 2 | [0,0.0632] |
| 589A4 | 0.01 | 2 | [0,0.0632] |
| 589A5 | 0.03 | 8 | [0,0.0632] |
| 589A6 | 0.00 | 0 | [0,0.0632] |
| 589A7 | 0.03 | 7 | [0,0.0632] |
| 589G2 | 0.00 | 0 | [0,0.0632] |
| 589G3 | 0.00 | 0 | [0,0.0632] |
| 589G4 | 0.00 | 0 | [0,0.0632] |
| 589G7 | 0.02 | 1 | [0,0.0632] |
| 589G8 | 0.03 | 2 | [0,0.0632] |
| 589GC | 0.00 | 0 | [0,0.0632] |
| 589GD | 0.03 | 1 | [0,0.0632] |
| 589GE | 0.02 | 1 | [0,0.0632] |
| 589GF | 0.02 | 1 | [0,0.0632] |
| 589GH | 0.00 | 0 | [0,0.0632] |
| 589GI | 0.00 | 0 | [0,0.0632] |
| 589GJ | 0.00 | 0 | [0,0.0632] |
| 589GM | 0.00 | 0 | [0,0.0632] |
| 589GN | 0.00 | 0 | [0,0.0632] |
| 589GP | 0.00 | 0 | [0,0.0632] |
| 589GR | 0.03 | 1 | [0,0.0632] |
| 589GU | 0.06 | 1 | [0,0.0632] |
| 589GV | 0.00 | 0 | [0,0.0632] |
| 589GW | 0.04 | 2 | [0,0.0632] |
| 589GX | 0.02 | 1 | [0,0.0632] |
| 589GY | 0.06 | 2 | [0,0.0632] |
| 589HK | 0.00 | 0 | [0,0.0632] |
| 589JA | 0.00 | 0 | [0,0.0632] |
| 589JE | 0.00 | 0 | [0,0.0632] |
| 589JF | 0.00 | 0 | [0,0.0632] |
| 590 | 0.03 | 12 | [0,0.0632] |
| 590GB | 0.01 | 1 | [0,0.0632] |
| 590GC | 0.00 | 0 | [0,0.0632] |
| 590GD | 0.00 | 0 | [0,0.0632] |
| 593 | 0.00 | 0 | [0,0.0632] |
| 593GE | 0.04 | 6 | [0,0.0632] |
| 593GH | 0.04 | 1 | [0,0.0632] |
| 595 | 0.00 | 0 | [0,0.0632] |
| 595GA | 0.02 | 3 | [0,0.0632] |
| 595GC | 0.01 | 1 | [0,0.0632] |
| 595GD | 0.03 | 3 | [0,0.0632] |
| 595GE | 0.00 | 0 | [0,0.0632] |
| 596 | 0.03 | 11 | [0,0.0632] |
| 596GA | 0.06 | 5 | [0,0.0632] |
| 596GC | 0.00 | 0 | [0,0.0632] |
| 596GD | 0.06 | 4 | [0,0.0632] |
| 598 | 0.06 | 1 | [0,0.0632] |
| 598A0 | 0.01 | 5 | [0,0.0632] |
| 598GE | 0.00 | 0 | [0,0.0632] |
| 598GF | 0.02 | 1 | [0,0.0632] |
| 598GG | 0.03 | 2 | [0,0.0632] |
| 603 | 0.00 | 0 | [0,0.0632] |
| 603GA | 0.04 | 4 | [0,0.0632] |
| 603GB | 0.01 | 1 | [0,0.0632] |
| 603GC | 0.01 | 1 | [0,0.0632] |
| 603GD | 0.00 | 0 | [0,0.0632] |
| 603GE | 0.02 | 4 | [0,0.0632] |
| 603GF | 0.02 | 1 | [0,0.0632] |
| 603GG | 0.00 | 0 | [0,0.0632] |
| 603GH | 0.00 | 0 | [0,0.0632] |
| 607 | 0.02 | 4 | [0,0.0632] |
| 607GC | 0.00 | 0 | [0,0.0632] |
| 607GE | 0.04 | 2 | [0,0.0632] |
| 607HA | 0.06 | 7 | [0,0.0632] |
| 608GA | 0.02 | 1 | [0,0.0632] |
| 608GC | 0.05 | 3 | [0,0.0632] |
| 608HA | 0.04 | 1 | [0,0.0632] |
| 610A4 | 0.03 | 6 | [0,0.0632] |
| 612B4 | 0.05 | 9 | [0,0.0632] |
| 612GF | 0.03 | 5 | [0,0.0632] |
| 613 | 0.05 | 9 | [0,0.0632] |
| 613GA | 0.01 | 1 | [0,0.0632] |
| 613GB | 0.05 | 3 | [0,0.0632] |
| 613GC | 0.01 | 1 | [0,0.0632] |
| 613GE | 0.00 | 0 | [0,0.0632] |
| 613GG | 0.04 | 2 | [0,0.0632] |
| 614 | 0.04 | 8 | [0,0.0632] |
| 614GE | 0.02 | 2 | [0,0.0632] |
| 614GF | 0.02 | 2 | [0,0.0632] |
| 614GG | 0.03 | 2 | [0,0.0632] |
| 618GD | 0.05 | 5 | [0,0.0632] |
| 618GJ | 0.05 | 2 | [0,0.0632] |
| 618QA | 0.00 | 0 | [0,0.0632] |
| 619A4 | 0.02 | 5 | [0,0.0632] |
| 619GA | 0.06 | 2 | [0,0.0632] |
| 619GB | 0.05 | 5 | [0,0.0632] |
| 619GD | 0.02 | 1 | [0,0.0632] |
| 619GE | 0.00 | 0 | [0,0.0632] |
| 619GF | 0.03 | 6 | [0,0.0632] |
| 619QB | 0.03 | 4 | [0,0.0632] |
| 620 | 0.02 | 1 | [0,0.0632] |
| 620A4 | 0.05 | 5 | [0,0.0632] |
| 620GA | 0.05 | 2 | [0,0.0632] |
| 620GB | 0.00 | 0 | [0,0.0632] |
| 620GD | 0.03 | 1 | [0,0.0632] |
| 620GF | 0.00 | 0 | [0,0.0632] |
| 620GG | 0.00 | 0 | [0,0.0632] |
| 620GH | 0.00 | 0 | [0,0.0632] |
| 621 | 0.04 | 19 | [0,0.0632] |
| 621BU | 0.00 | 0 | [0,0.0632] |
| 621BY | 0.01 | 3 | [0,0.0632] |
| 621GA | 0.06 | 2 | [0,0.0632] |
| 621GG | 0.05 | 2 | [0,0.0632] |
| 621GI | 0.04 | 2 | [0,0.0632] |
| 621GK | 0.00 | 0 | [0,0.0632] |
| 623 | 0.04 | 8 | [0,0.0632] |
| 623GA | 0.00 | 0 | [0,0.0632] |
| 623GB | 0.00 | 0 | [0,0.0632] |
| 626 | 0.06 | 21 | [0,0.0632] |
| 626GE | 0.05 | 7 | [0,0.0632] |
| 626GF | 0.01 | 3 | [0,0.0632] |
| 626GN | 0.05 | 1 | [0,0.0632] |
| 626GO | 0.00 | 0 | [0,0.0632] |
| 626QB | 0.00 | 0 | [0,0.0632] |
| 629 | 0.02 | 5 | [0,0.0632] |
| 629BY | 0.00 | 0 | [0,0.0632] |
| 629GA | 0.02 | 1 | [0,0.0632] |
| 629GB | 0.02 | 1 | [0,0.0632] |
| 629GC | 0.03 | 2 | [0,0.0632] |
| 629GD | 0.00 | 0 | [0,0.0632] |
| 629GE | 0.00 | 0 | [0,0.0632] |
| 629GF | 0.04 | 1 | [0,0.0632] |
| 630GC | 0.00 | 0 | [0,0.0632] |
| 631 | 0.03 | 3 | [0,0.0632] |
| 631GC | 0.00 | 0 | [0,0.0632] |
| 631GD | 0.03 | 1 | [0,0.0632] |
| 631GE | 0.01 | 1 | [0,0.0632] |
| 632 | 0.03 | 7 | [0,0.0632] |
| 632GA | 0.02 | 2 | [0,0.0632] |
| 632HA | 0.06 | 1 | [0,0.0632] |
| 632HB | 0.03 | 1 | [0,0.0632] |
| 632HC | 0.00 | 0 | [0,0.0632] |
| 632HD | 0.04 | 2 | [0,0.0632] |
| 635 | 0.04 | 14 | [0,0.0632] |
| 635GB | 0.05 | 4 | [0,0.0632] |
| 635GD | 0.06 | 2 | [0,0.0632] |
| 635GE | 0.05 | 1 | [0,0.0632] |
| 635GG | 0.00 | 0 | [0,0.0632] |
| 635HB | 0.00 | 0 | [0,0.0632] |
| 635QB | 0.01 | 2 | [0,0.0632] |
| 636 | 0.02 | 5 | [0,0.0632] |
| 636A4 | 0.05 | 7 | [0,0.0632] |
| 636A5 | 0.02 | 3 | [0,0.0632] |
| 636A6 | 0.02 | 4 | [0,0.0632] |
| 636A8 | 0.01 | 1 | [0,0.0632] |
| 636BU | 0.00 | 0 | [0,0.0632] |
| 636GA | 0.03 | 1 | [0,0.0632] |
| 636GC | 0.00 | 0 | [0,0.0632] |
| 636GD | 0.04 | 1 | [0,0.0632] |
| 636GF | 0.06 | 7 | [0,0.0632] |
| 636GG | 0.02 | 1 | [0,0.0632] |
| 636GK | 0.02 | 1 | [0,0.0632] |
| 636GL | 0.00 | 0 | [0,0.0632] |
| 636GM | 0.00 | 0 | [0,0.0632] |
| 636GP | 0.06 | 1 | [0,0.0632] |
| 636GQ | 0.00 | 0 | [0,0.0632] |
| 636GR | 0.00 | 0 | [0,0.0632] |
| 636GS | 0.00 | 0 | [0,0.0632] |
| 637 | 0.01 | 5 | [0,0.0632] |
| 637GA | 0.00 | 0 | [0,0.0632] |
| 637GB | 0.03 | 2 | [0,0.0632] |
| 637GC | 0.05 | 8 | [0,0.0632] |
| 640BY | 0.06 | 8 | [0,0.0632] |
| 642 | 0.05 | 17 | [0,0.0632] |
| 642GD | 0.04 | 3 | [0,0.0632] |
| 644BY | 0.05 | 15 | [0,0.0632] |
| 644GD | 0.00 | 0 | [0,0.0632] |
| 646 | 0.02 | 3 | [0,0.0632] |
| 646A4 | 0.00 | 1 | [0,0.0632] |
| 646GA | 0.06 | 6 | [0,0.0632] |
| 646GB | 0.01 | 1 | [0,0.0632] |
| 646GC | 0.00 | 0 | [0,0.0632] |
| 646GE | 0.02 | 1 | [0,0.0632] |
| 648GD | 0.06 | 2 | [0,0.0632] |
| 649GB | 0.00 | 0 | [0,0.0632] |
| 649GD | 0.05 | 1 | [0,0.0632] |
| 649QC | 0.00 | 0 | [0,0.0632] |
| 650 | 0.02 | 8 | [0,0.0632] |
| 652 | 0.01 | 9 | [0,0.0632] |
| 652GA | 0.02 | 2 | [0,0.0632] |
| 652GB | 0.03 | 1 | [0,0.0632] |
| 652GE | 0.03 | 2 | [0,0.0632] |
| 652GF | 0.00 | 0 | [0,0.0632] |
| 653 | 0.04 | 5 | [0,0.0632] |
| 653BY | 0.06 | 12 | [0,0.0632] |
| 654 | 0.04 | 15 | [0,0.0632] |
| 654GB | 0.00 | 0 | [0,0.0632] |
| 654GC | 0.00 | 0 | [0,0.0632] |
| 654GD | 0.04 | 1 | [0,0.0632] |
| 655GA | 0.05 | 3 | [0,0.0632] |
| 655GB | 0.05 | 4 | [0,0.0632] |
| 655GE | 0.00 | 0 | [0,0.0632] |
| 655GF | 0.00 | 0 | [0,0.0632] |
| 655GG | 0.00 | 0 | [0,0.0632] |
| 655GH | 0.00 | 0 | [0,0.0632] |
| 655GI | 0.06 | 1 | [0,0.0632] |
| 657 | 0.01 | 3 | [0,0.0632] |
| 657A0 | 0.03 | 5 | [0,0.0632] |
| 657A4 | 0.01 | 1 | [0,0.0632] |
| 657A5 | 0.05 | 7 | [0,0.0632] |
| 657GA | 0.00 | 0 | [0,0.0632] |
| 657GB | 0.01 | 1 | [0,0.0632] |
| 657GD | 0.00 | 0 | [0,0.0632] |
| 657GG | 0.00 | 0 | [0,0.0632] |
| 657GH | 0.03 | 2 | [0,0.0632] |
| 657GI | 0.00 | 0 | [0,0.0632] |
| 657GJ | 0.06 | 12 | [0,0.0632] |
| 657GL | 0.05 | 4 | [0,0.0632] |
| 657GN | 0.00 | 0 | [0,0.0632] |
| 657GQ | 0.03 | 1 | [0,0.0632] |
| 657GT | 0.00 | 0 | [0,0.0632] |
| 657GV | 0.00 | 0 | [0,0.0632] |
| 658GC | 0.05 | 3 | [0,0.0632] |
| 659 | 0.03 | 10 | [0,0.0632] |
| 659BY | 0.03 | 9 | [0,0.0632] |
| 659BZ | 0.02 | 4 | [0,0.0632] |
| 659GA | 0.03 | 9 | [0,0.0632] |
| 660 | 0.01 | 2 | [0,0.0632] |
| 660GA | 0.00 | 0 | [0,0.0632] |
| 660GB | 0.03 | 3 | [0,0.0632] |
| 660GC | 0.00 | 0 | [0,0.0632] |
| 660GD | 0.00 | 0 | [0,0.0632] |
| 660GE | 0.00 | 0 | [0,0.0632] |
| 660GG | 0.02 | 1 | [0,0.0632] |
| 660GJ | 0.01 | 1 | [0,0.0632] |
| 660GK | 0.00 | 0 | [0,0.0632] |
| 663 | 0.05 | 15 | [0,0.0632] |
| 663GC | 0.03 | 3 | [0,0.0632] |
| 664GB | 0.05 | 10 | [0,0.0632] |
| 664GD | 0.05 | 5 | [0,0.0632] |
| 666 | 0.03 | 2 | [0,0.0632] |
| 666GB | 0.03 | 1 | [0,0.0632] |
| 666GC | 0.00 | 0 | [0,0.0632] |
| 666GF | 0.00 | 0 | [0,0.0632] |
| 667 | 0.03 | 7 | [0,0.0632] |
| 667GA | 0.01 | 1 | [0,0.0632] |
| 667GB | 0.02 | 2 | [0,0.0632] |
| 667GC | 0.02 | 2 | [0,0.0632] |
| 667QA | 0.00 | 0 | [0,0.0632] |
| 671A4 | 0.05 | 6 | [0,0.0632] |
| 671GO | 0.06 | 8 | [0,0.0632] |
| 671GP | 0.00 | 0 | [0,0.0632] |
| 672 | 0.02 | 12 | [0,0.0632] |
| 672B0 | 0.06 | 11 | [0,0.0632] |
| 672GA | 0.05 | 1 | [0,0.0632] |
| 672GB | 0.00 | 0 | [0,0.0632] |
| 672GC | 0.00 | 0 | [0,0.0632] |
| 672GD | 0.02 | 1 | [0,0.0632] |
| 672QC | 0.00 | 0 | [0,0.0632] |
| 673 | 0.04 | 23 | [0,0.0632] |
| 673BZ | 0.05 | 11 | [0,0.0632] |
| 673GB | 0.04 | 8 | [0,0.0632] |
| 673GC | 0.03 | 3 | [0,0.0632] |
| 673GF | 0.04 | 2 | [0,0.0632] |
| 674A4 | 0.03 | 5 | [0,0.0632] |
| 674BY | 0.06 | 20 | [0,0.0632] |
| 674GA | 0.06 | 4 | [0,0.0632] |
| 674GB | 0.00 | 0 | [0,0.0632] |
| 674GD | 0.03 | 3 | [0,0.0632] |
| 675GA | 0.05 | 23 | [0,0.0632] |
| 675GE | 0.05 | 4 | [0,0.0632] |
| 676 | 0.02 | 2 | [0,0.0632] |
| 676GA | 0.00 | 0 | [0,0.0632] |
| 676GC | 0.00 | 0 | [0,0.0632] |
| 676GD | 0.01 | 1 | [0,0.0632] |
| 676GE | 0.00 | 0 | [0,0.0632] |
| 678GC | 0.02 | 1 | [0,0.0632] |
| 678GD | 0.00 | 0 | [0,0.0632] |
| 678GE | 0.05 | 2 | [0,0.0632] |
| 678GF | 0.02 | 2 | [0,0.0632] |
| 678GG | 0.03 | 2 | [0,0.0632] |
| 679 | 0.06 | 12 | [0,0.0632] |
| 679HK | 0.00 | 0 | [0,0.0632] |
| 687 | 0.01 | 1 | [0,0.0632] |
| 687GA | 0.04 | 3 | [0,0.0632] |
| 687GB | 0.02 | 1 | [0,0.0632] |
| 687HA | 0.05 | 3 | [0,0.0632] |
| 688GB | 0.00 | 0 | [0,0.0632] |
| 689 | 0.04 | 12 | [0,0.0632] |
| 689GC | 0.06 | 3 | [0,0.0632] |
| 689GD | 0.05 | 2 | [0,0.0632] |
| 689GE | 0.06 | 2 | [0,0.0632] |
| 689HC | 0.01 | 1 | [0,0.0632] |
| 689PA | 0.00 | 0 | [0,0.0632] |
| 692 | 0.01 | 1 | [0,0.0632] |
| 692GB | 0.00 | 0 | [0,0.0632] |
| 693 | 0.01 | 3 | [0,0.0632] |
| 693B4 | 0.02 | 3 | [0,0.0632] |
| 693GA | 0.00 | 0 | [0,0.0632] |
| 693GC | 0.00 | 0 | [0,0.0632] |
| 693GG | 0.00 | 0 | [0,0.0632] |
| 695 | 0.03 | 13 | [0,0.0632] |
| 695BY | 0.02 | 4 | [0,0.0632] |
| 695GD | 0.04 | 6 | [0,0.0632] |
| 740GD | 0.02 | 1 | [0,0.0632] |
| 757GA | 0.03 | 2 | [0,0.0632] |
| 757GB | 0.00 | 0 | [0,0.0632] |
| 757GC | 0.04 | 2 | [0,0.0632] |
| 757GD | 0.00 | 0 | [0,0.0632] |
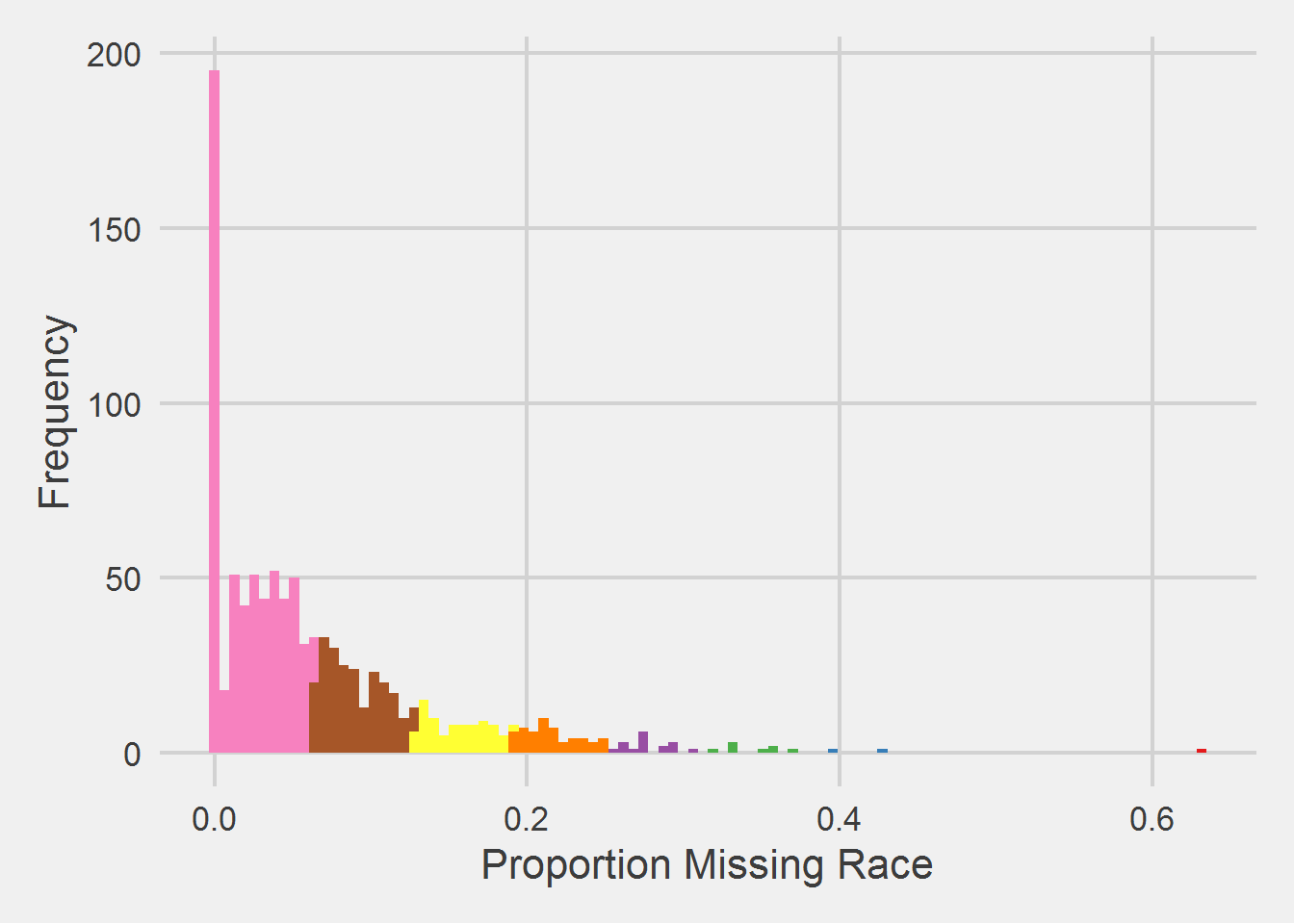
Looks like there is only one in the top 10% (0.568,0.632], and that’s 501G2. The distribution of missing proportion looks highly skewed, let’s examine:
sta6a_race_miss %>%
ggplot(aes(x = p)) %>%
add(geom_histogram(aes(fill = q), bins = 100)) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(
axis.title = element_text(),
legend.position = "none"
)) %>%
add(scale_fill_brewer(palette = "Set1", direction = -1)) %>%
add(labs(
x = "Proportion Missing Race",
y = "Frequency"
))
Let’s see if there any similarities for those missing race > 15%
sta6a_race_miss %>%
filter(p >= 0.15) %>%
left_join(
weightSamples %>%
distinct(Sta3n, Sta6a),
by = "Sta6a"
) %>%
arrange(p) %>%
mutate(Sta6a_deident = row_number()) %>%
ggplot(aes(x = Sta6a_deident, y = p)) %>%
add(geom_point(aes(fill = Sta3n, size = N), pch = 21)) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(
axis.title = element_text(),
legend.position = "none"
)) %>%
add(labs(
y = "Proportion Missing Race",
x = "Sta6a Deidentified",
caption = "Point color represents parent Sta3n"
))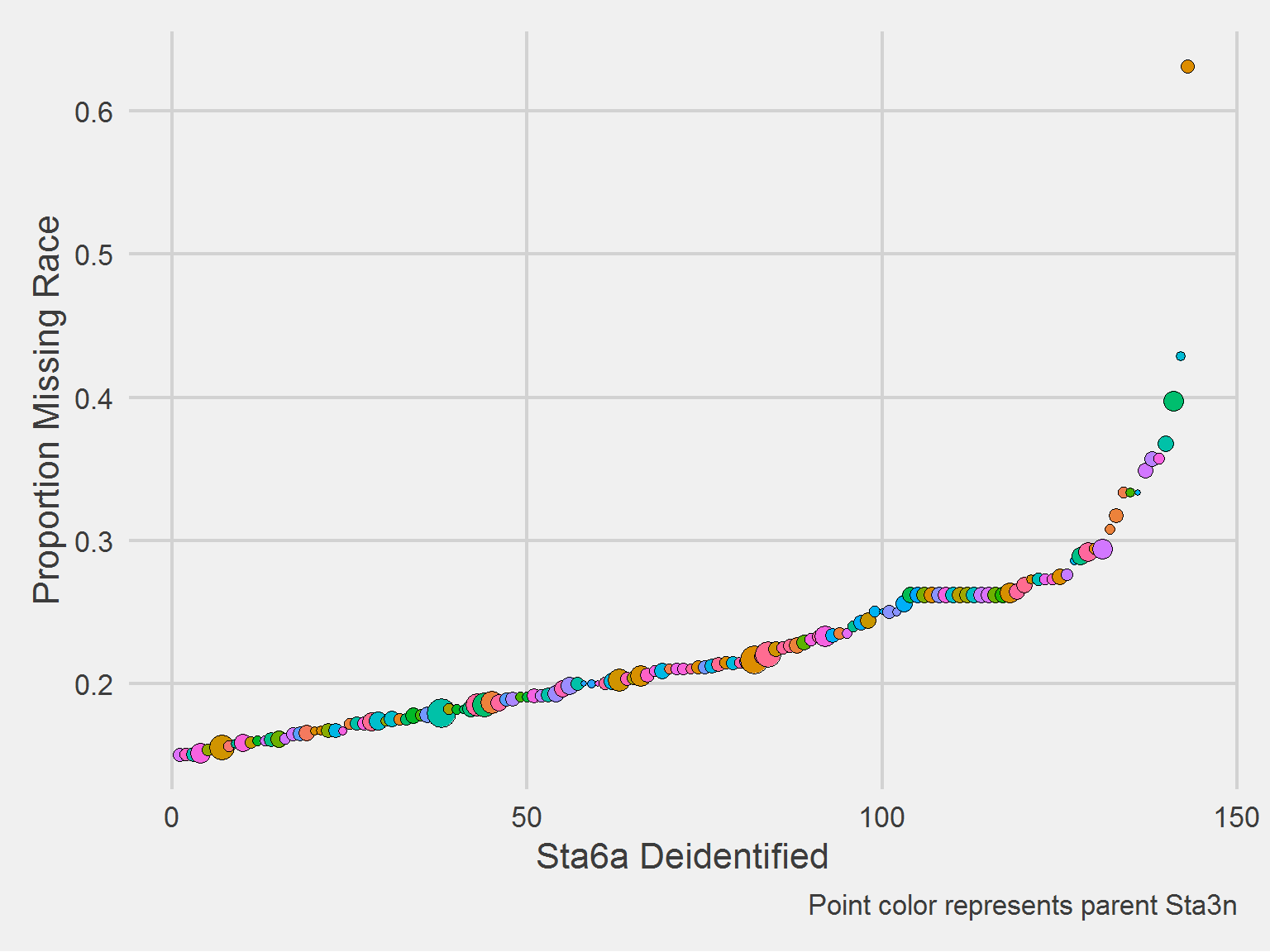
I see no patterns in terms of Sta3n dependence, the orange Sta3n may be fully represented here. There does seem to be a sharp inflection point at the 30% missingness point.
Next I would like to see if there is an association between number of weight samples collected, and race.
weightSamples %>%
filter(SampleYear == "2016") %>%
group_by(Race) %>%
count(PatientICN) %>%
summarise(
mean = mean(n),
SD = sd(n),
median = median(n),
min = min(n),
max = max(n)
) %>%
tableStyle()| Race | mean | SD | median | min | max |
|---|---|---|---|---|---|
| WHITE | 11.86 | 25.47 | 8 | 1 | 4981 |
| BLACK OR AFRICAN AMERICAN | 14.69 | 22.65 | 10 | 1 | 1262 |
| Missing | 10.02 | 19.06 | 6 | 1 | 848 |
| NATIVE HAWAIIAN OR OTHER PACIFIC ISLANDER | 11.99 | 11.58 | 8 | 1 | 94 |
| AMERICAN INDIAN OR ALASKA NATIVE | 11.68 | 11.49 | 8 | 1 | 105 |
| ASIAN | 11.96 | 52.13 | 7 | 1 | 1199 |
| MULTI-RACIAL | 13.85 | 12.29 | 10 | 1 | 102 |
Now, let’s do the same for the combination of race and ethnicity,
weightSamples %>%
filter(SampleYear == "2016") %>%
group_by(race_eth) %>%
count(PatientICN) %>%
summarise(
mean = mean(n),
SD = sd(n),
median = median(n),
min = min(n),
max = max(n)
) %>%
tableStyle()| race_eth | mean | SD | median | min | max |
|---|---|---|---|---|---|
| Non-Hispanic White | 11.76 | 25.42 | 8 | 1 | 4981 |
| Hispanic White | 13.65 | 28.04 | 9 | 1 | 1259 |
| Non-Hispanic Black | 14.71 | 22.88 | 10 | 1 | 1262 |
| Hispanic Black | 15.64 | 15.90 | 11 | 1 | 135 |
| Other | 11.15 | 23.44 | 7 | 1 | 1199 |
| Unknown | 7.44 | 9.56 | 4 | 1 | 134 |
Sex
Total number of weight samples by Sex and Sample Year
weightSamples %>%
janitor::tabyl(Gender, SampleYear) %>%
adorn_percentages("col") %>%
adorn_pct_formatting() %>%
adorn_ns() %>%
tableStyle()| Gender | 2008 | 2016 |
|---|---|---|
| F | 5.5% (67199) | 6.6% (79952) |
| M | 94.5% (1147194) | 93.4% (1128782) |
How many people per sex?
weightSamples %>%
distinct(SampleYear, PatientICN, Gender) %>%
tabyl(Gender, SampleYear) %>%
adorn_percentages("col") %>%
adorn_pct_formatting() %>%
adorn_ns(position = "front") %>%
tableStyle()| Gender | 2008 | 2016 |
|---|---|---|
| F | 4849 (4.9%) | 6200 (6.3%) |
| M | 93937 (95.1%) | 92758 (93.7%) |
Association between number of weight samples collected, and Sex/Gender, by Cohort Year,
weightSamples %>%
group_by(SampleYear, Gender) %>%
count(PatientICN) %>%
summarise(
mean = mean(n),
SD = sd(n),
median = median(n),
min = min(n),
max = max(n)
) %>%
tableStyle()| SampleYear | Gender | mean | SD | median | min | max |
|---|---|---|---|---|---|---|
| 2008 | F | 13.86 | 15.25 | 10 | 1 | 353 |
| 2008 | M | 12.21 | 16.00 | 9 | 1 | 1479 |
| 2016 | F | 12.90 | 12.78 | 9 | 1 | 282 |
| 2016 | M | 12.17 | 25.46 | 8 | 1 | 4981 |
weightSamples %>%
group_by(SampleYear, Gender) %>%
count(PatientICN) %>%
ggplot(aes(x = Gender, y = n)) %>%
add(geom_violin(fill = "maroon")) %>%
add(facet_wrap(vars(SampleYear), ncol = 1)) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(coord_flip())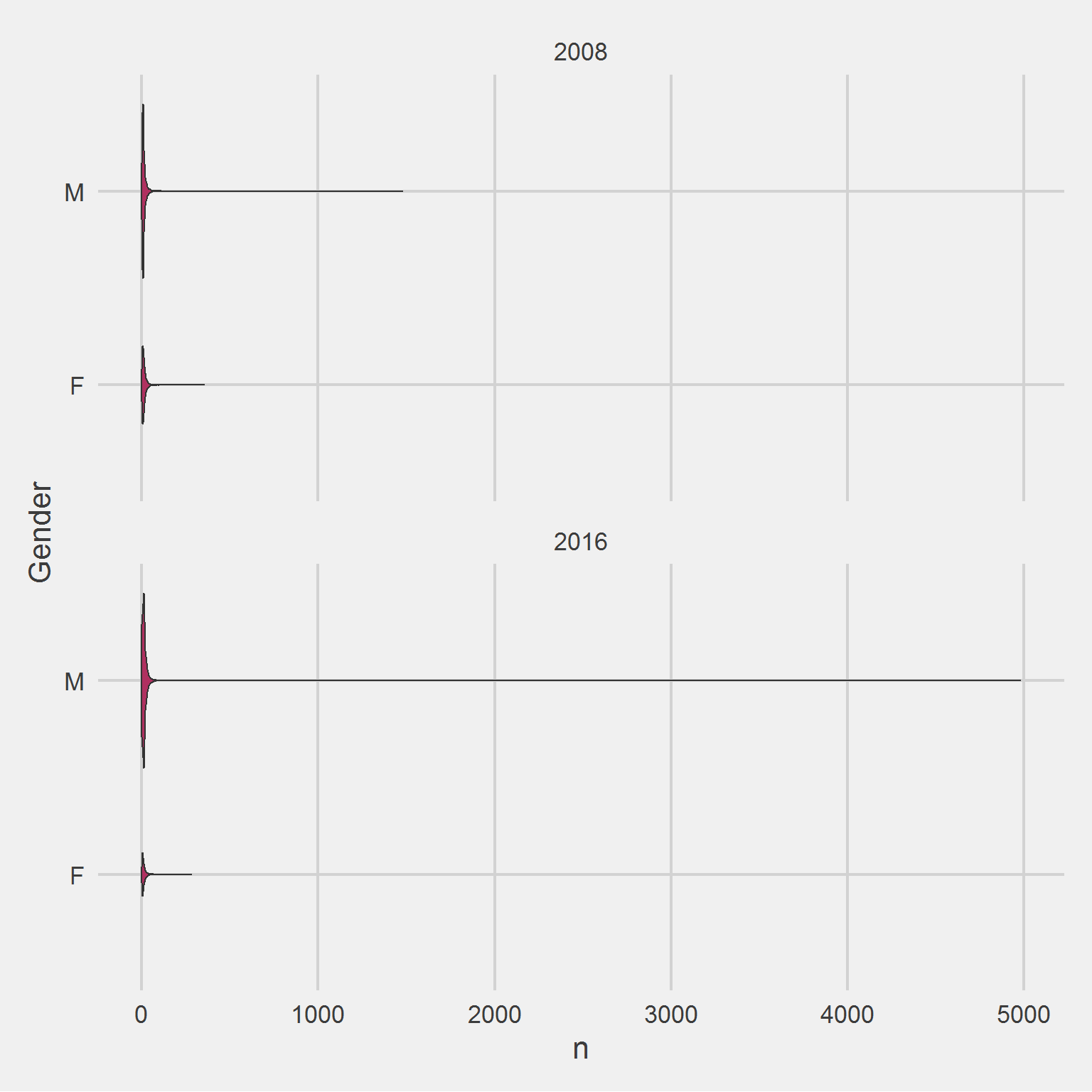
Removing the outlier,
weightSamples %>%
group_by(SampleYear, Gender) %>%
count(PatientICN) %>%
filter(n < 1000) %>%
ggplot(aes(x = Gender, y = n)) %>%
add(geom_boxplot(fill = "maroon")) %>%
add(facet_wrap(vars(SampleYear), ncol = 1)) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(coord_flip())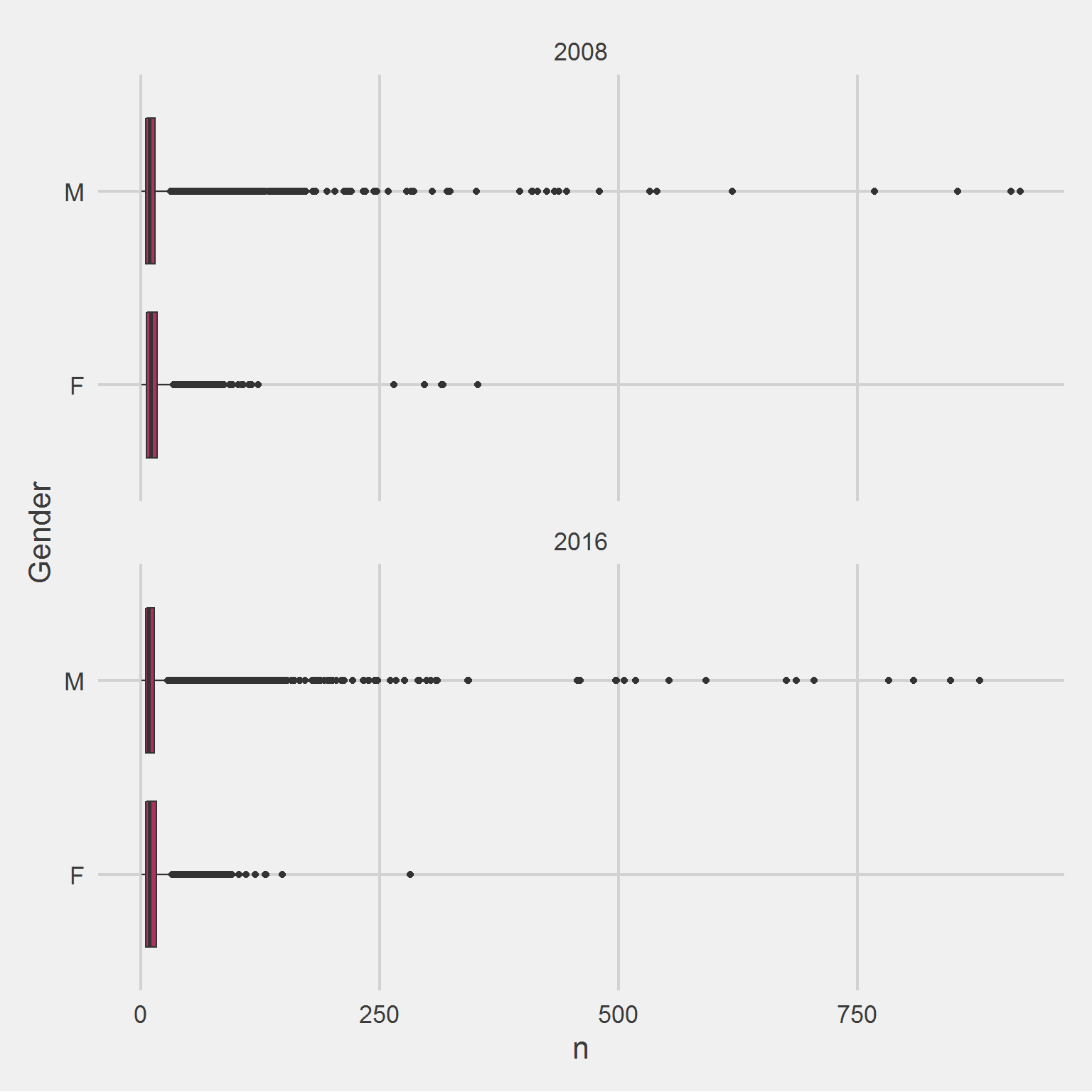
All weight in lbs. by sex and sample year
weightSamples %>%
group_by(SampleYear, Gender) %>%
summarise(
mean = mean(Weight),
SD = sd(Weight),
median = median(Weight),
min = min(Weight),
max = max(Weight)
) %>%
tableStyle()| SampleYear | Gender | mean | SD | median | min | max |
|---|---|---|---|---|---|---|
| 2008 | F | 181.85 | 47.15 | 176.9 | 0 | 1475.70 |
| 2008 | M | 203.88 | 47.40 | 198.0 | 0 | 2423.35 |
| 2016 | F | 184.65 | 44.50 | 180.7 | 0 | 1479.90 |
| 2016 | M | 209.19 | 48.41 | 203.6 | 0 | 1486.20 |
Both Males and Females have implausible minimum and maximum weight values.
weightSamples %>%
ggplot(aes(x = Gender, y = Weight)) %>%
add(geom_boxplot(fill = "maroon")) %>%
add(facet_wrap(vars(SampleYear), ncol = 1)) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(coord_flip())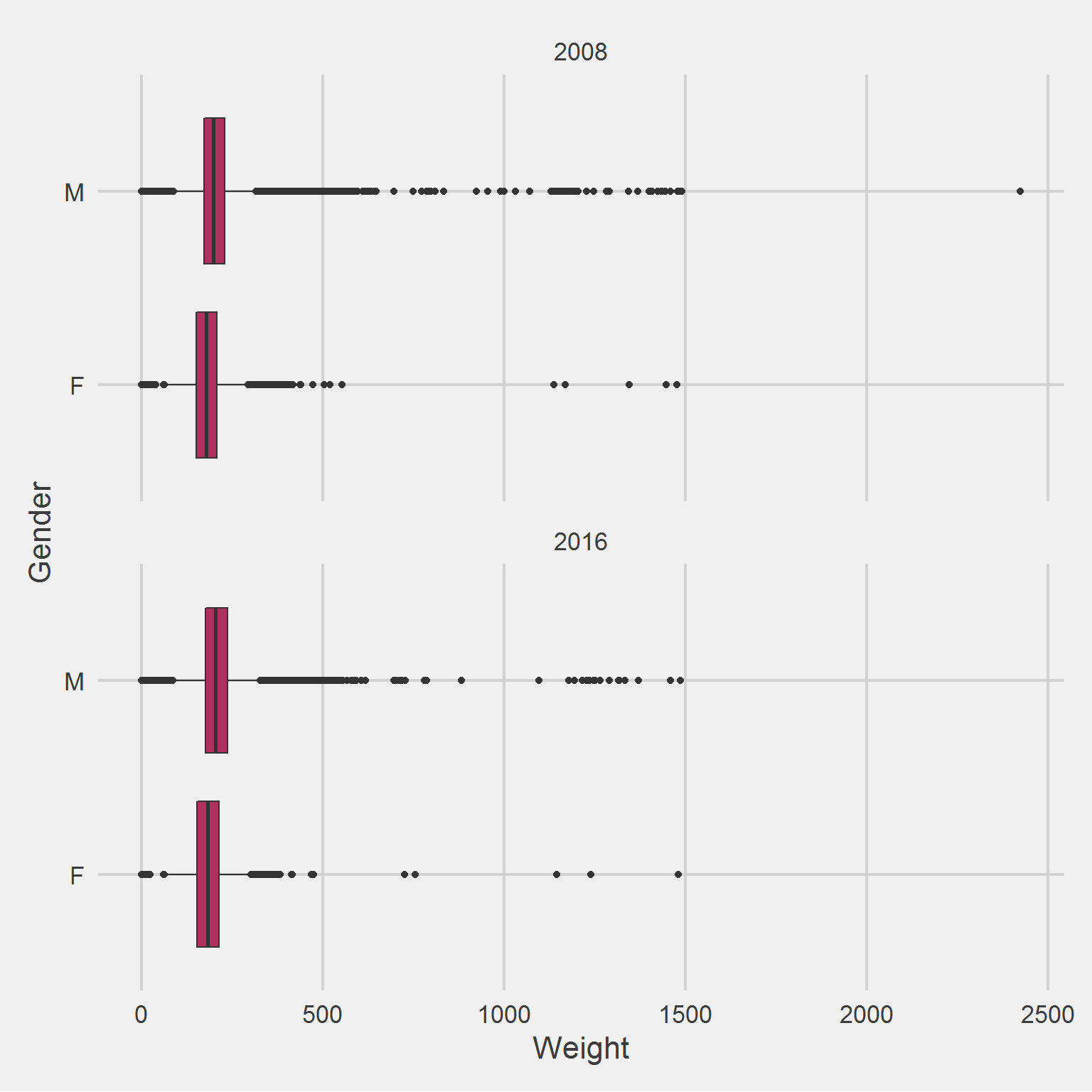
And similar distributions of “outlier” weights.
Sta3n
By Sta3n
weightSamples %>%
tabyl(Sta3n) %>%
adorn_pct_formatting() %>%
arrange(desc(percent)) %>%
scrollTable()| Sta3n | n | percent |
|---|---|---|
| 528 | 56881 | 2.3% |
| 541 | 53921 | 2.2% |
| 589 | 52707 | 2.2% |
| 516 | 50372 | 2.1% |
| 573 | 51013 | 2.1% |
| 549 | 45440 | 1.9% |
| 580 | 45190 | 1.9% |
| 636 | 47227 | 1.9% |
| 657 | 44990 | 1.9% |
| 672 | 38494 | 1.6% |
| 673 | 38711 | 1.6% |
| 671 | 35408 | 1.5% |
| 691 | 36344 | 1.5% |
| 508 | 34916 | 1.4% |
| 674 | 33385 | 1.4% |
| 612 | 30388 | 1.3% |
| 554 | 28104 | 1.2% |
| 644 | 29386 | 1.2% |
| 663 | 28535 | 1.2% |
| 695 | 28249 | 1.2% |
| 521 | 27573 | 1.1% |
| 537 | 25579 | 1.1% |
| 558 | 25933 | 1.1% |
| 593 | 27322 | 1.1% |
| 618 | 26092 | 1.1% |
| 626 | 27005 | 1.1% |
| 501 | 24199 | 1.0% |
| 546 | 23980 | 1.0% |
| 548 | 23035 | 1.0% |
| 561 | 24788 | 1.0% |
| 586 | 23629 | 1.0% |
| 652 | 23530 | 1.0% |
| 659 | 23088 | 1.0% |
| 678 | 24530 | 1.0% |
| 534 | 20881 | 0.9% |
| 564 | 22035 | 0.9% |
| 578 | 22697 | 0.9% |
| 605 | 21139 | 0.9% |
| 614 | 20809 | 0.9% |
| 630 | 20928 | 0.9% |
| 635 | 21014 | 0.9% |
| 640 | 21702 | 0.9% |
| 648 | 22815 | 0.9% |
| 689 | 22498 | 0.9% |
| 402 | 18290 | 0.8% |
| 512 | 19902 | 0.8% |
| 520 | 19286 | 0.8% |
| 523 | 19365 | 0.8% |
| 544 | 20143 | 0.8% |
| 565 | 20296 | 0.8% |
| 583 | 20377 | 0.8% |
| 598 | 20047 | 0.8% |
| 600 | 18812 | 0.8% |
| 603 | 18758 | 0.8% |
| 621 | 18688 | 0.8% |
| 629 | 19817 | 0.8% |
| 664 | 19776 | 0.8% |
| 675 | 19001 | 0.8% |
| 539 | 15870 | 0.7% |
| 552 | 17003 | 0.7% |
| 570 | 16611 | 0.7% |
| 595 | 17779 | 0.7% |
| 596 | 16656 | 0.7% |
| 619 | 17616 | 0.7% |
| 646 | 17584 | 0.7% |
| 658 | 16001 | 0.7% |
| 667 | 17058 | 0.7% |
| 688 | 16352 | 0.7% |
| 506 | 14142 | 0.6% |
| 553 | 14955 | 0.6% |
| 581 | 13357 | 0.6% |
| 590 | 13396 | 0.6% |
| 610 | 14076 | 0.6% |
| 632 | 14182 | 0.6% |
| 642 | 15087 | 0.6% |
| 662 | 15484 | 0.6% |
| 693 | 15502 | 0.6% |
| 436 | 12086 | 0.5% |
| 437 | 12091 | 0.5% |
| 438 | 11401 | 0.5% |
| 504 | 11365 | 0.5% |
| 509 | 13169 | 0.5% |
| 526 | 11339 | 0.5% |
| 550 | 11149 | 0.5% |
| 556 | 11384 | 0.5% |
| 557 | 12827 | 0.5% |
| 607 | 12426 | 0.5% |
| 613 | 12603 | 0.5% |
| 623 | 12703 | 0.5% |
| 637 | 13133 | 0.5% |
| 650 | 11432 | 0.5% |
| 654 | 12902 | 0.5% |
| 655 | 13110 | 0.5% |
| 656 | 11449 | 0.5% |
| 660 | 13285 | 0.5% |
| 668 | 10951 | 0.5% |
| 757 | 12660 | 0.5% |
| 405 | 9189 | 0.4% |
| 459 | 9244 | 0.4% |
| 502 | 10591 | 0.4% |
| 515 | 10853 | 0.4% |
| 531 | 9849 | 0.4% |
| 538 | 9867 | 0.4% |
| 540 | 8745 | 0.4% |
| 653 | 9249 | 0.4% |
| 756 | 8952 | 0.4% |
| 442 | 6101 | 0.3% |
| 460 | 7849 | 0.3% |
| 503 | 7858 | 0.3% |
| 517 | 7667 | 0.3% |
| 562 | 7968 | 0.3% |
| 568 | 7763 | 0.3% |
| 585 | 8069 | 0.3% |
| 608 | 7816 | 0.3% |
| 620 | 7697 | 0.3% |
| 631 | 6634 | 0.3% |
| 649 | 6811 | 0.3% |
| 676 | 8254 | 0.3% |
| 463 | 4559 | 0.2% |
| 518 | 4112 | 0.2% |
| 519 | 6035 | 0.2% |
| 529 | 5561 | 0.2% |
| 542 | 5623 | 0.2% |
| 575 | 4934 | 0.2% |
| 666 | 4625 | 0.2% |
| 679 | 4680 | 0.2% |
| 687 | 4998 | 0.2% |
| 740 | 6001 | 0.2% |
| 692 | 2688 | 0.1% |
| 358 | 1089 | 0.0% |
Sta3n by person
weightSamples %>%
distinct(PatientICN, Sta3n) %>%
tabyl(Sta3n) %>%
adorn_pct_formatting() %>%
arrange(desc(percent)) %>%
scrollTable()| Sta3n | n | percent |
|---|---|---|
| 528 | 4488 | 2.3% |
| 573 | 4556 | 2.3% |
| 589 | 4547 | 2.3% |
| 636 | 4334 | 2.2% |
| 549 | 3761 | 1.9% |
| 516 | 3546 | 1.8% |
| 541 | 3496 | 1.8% |
| 657 | 3640 | 1.8% |
| 580 | 3182 | 1.6% |
| 673 | 3058 | 1.6% |
| 508 | 2628 | 1.3% |
| 544 | 2524 | 1.3% |
| 626 | 2555 | 1.3% |
| 674 | 2578 | 1.3% |
| 691 | 2627 | 1.3% |
| 612 | 2417 | 1.2% |
| 618 | 2351 | 1.2% |
| 644 | 2286 | 1.2% |
| 671 | 2369 | 1.2% |
| 672 | 2331 | 1.2% |
| 548 | 2167 | 1.1% |
| 554 | 2247 | 1.1% |
| 605 | 2179 | 1.1% |
| 659 | 2177 | 1.1% |
| 663 | 2114 | 1.1% |
| 520 | 1895 | 1.0% |
| 565 | 1967 | 1.0% |
| 621 | 1870 | 1.0% |
| 648 | 2050 | 1.0% |
| 664 | 2054 | 1.0% |
| 695 | 1897 | 1.0% |
| 521 | 1752 | 0.9% |
| 534 | 1843 | 0.9% |
| 561 | 1834 | 0.9% |
| 564 | 1814 | 0.9% |
| 578 | 1754 | 0.9% |
| 583 | 1686 | 0.9% |
| 598 | 1694 | 0.9% |
| 635 | 1727 | 0.9% |
| 640 | 1830 | 0.9% |
| 646 | 1801 | 0.9% |
| 689 | 1838 | 0.9% |
| 501 | 1520 | 0.8% |
| 512 | 1579 | 0.8% |
| 546 | 1512 | 0.8% |
| 558 | 1648 | 0.8% |
| 586 | 1549 | 0.8% |
| 593 | 1500 | 0.8% |
| 595 | 1531 | 0.8% |
| 603 | 1505 | 0.8% |
| 610 | 1491 | 0.8% |
| 614 | 1476 | 0.8% |
| 652 | 1495 | 0.8% |
| 675 | 1619 | 0.8% |
| 678 | 1589 | 0.8% |
| 402 | 1400 | 0.7% |
| 506 | 1308 | 0.7% |
| 515 | 1355 | 0.7% |
| 523 | 1283 | 0.7% |
| 537 | 1345 | 0.7% |
| 600 | 1463 | 0.7% |
| 619 | 1392 | 0.7% |
| 642 | 1446 | 0.7% |
| 660 | 1309 | 0.7% |
| 688 | 1285 | 0.7% |
| 693 | 1383 | 0.7% |
| 436 | 1189 | 0.6% |
| 437 | 1085 | 0.6% |
| 509 | 1096 | 0.6% |
| 539 | 1169 | 0.6% |
| 550 | 1177 | 0.6% |
| 552 | 1130 | 0.6% |
| 553 | 1244 | 0.6% |
| 557 | 1162 | 0.6% |
| 590 | 1218 | 0.6% |
| 596 | 1203 | 0.6% |
| 613 | 1132 | 0.6% |
| 623 | 1215 | 0.6% |
| 629 | 1266 | 0.6% |
| 630 | 1235 | 0.6% |
| 632 | 1094 | 0.6% |
| 637 | 1253 | 0.6% |
| 656 | 1142 | 0.6% |
| 658 | 1222 | 0.6% |
| 667 | 1211 | 0.6% |
| 757 | 1161 | 0.6% |
| 405 | 892 | 0.5% |
| 438 | 966 | 0.5% |
| 459 | 902 | 0.5% |
| 502 | 971 | 0.5% |
| 503 | 977 | 0.5% |
| 531 | 949 | 0.5% |
| 581 | 962 | 0.5% |
| 607 | 1062 | 0.5% |
| 650 | 1077 | 0.5% |
| 654 | 1015 | 0.5% |
| 655 | 1031 | 0.5% |
| 662 | 1009 | 0.5% |
| 668 | 934 | 0.5% |
| 460 | 877 | 0.4% |
| 504 | 830 | 0.4% |
| 540 | 711 | 0.4% |
| 556 | 766 | 0.4% |
| 562 | 804 | 0.4% |
| 568 | 693 | 0.4% |
| 570 | 876 | 0.4% |
| 585 | 711 | 0.4% |
| 608 | 815 | 0.4% |
| 620 | 718 | 0.4% |
| 649 | 756 | 0.4% |
| 653 | 808 | 0.4% |
| 676 | 862 | 0.4% |
| 756 | 876 | 0.4% |
| 442 | 568 | 0.3% |
| 463 | 532 | 0.3% |
| 518 | 542 | 0.3% |
| 519 | 592 | 0.3% |
| 526 | 630 | 0.3% |
| 529 | 667 | 0.3% |
| 538 | 685 | 0.3% |
| 542 | 562 | 0.3% |
| 631 | 640 | 0.3% |
| 687 | 607 | 0.3% |
| 740 | 538 | 0.3% |
| 517 | 486 | 0.2% |
| 575 | 385 | 0.2% |
| 666 | 398 | 0.2% |
| 679 | 446 | 0.2% |
| 692 | 463 | 0.2% |
| 358 | 149 | 0.1% |
weightSamples %>%
distinct(PatientICN, Sta3n) %>%
group_by(Sta3n) %>%
count() %>%
ungroup() %>%
arrange(n) %>%
mutate(Sta3n = factor(Sta3n, Sta3n)) %>%
ggplot(aes(x = Sta3n, y = n)) %>%
add(geom_segment(aes(x = Sta3n, xend = Sta3n, y = 0, yend = n),
color = "grey")) %>%
add(geom_point(color = "black",
fill = "purple",
pch = 21,
size = 1.5,
alpha = 0.5)) %>%
add(theme_classic(10)) %>%
add(coord_flip()) %>%
add(labs(
y = "Number of Pts. in Sample",
x = "Sta3n"
))
Sta3n by person and sample year
weightSamples %>%
distinct(PatientICN, SampleYear, Sta3n) %>%
multTable(rc = "row", Sta3n, SampleYear) %>%
scrollTable()| Sta3n | 2008 | 2016 |
|---|---|---|
| 358 | 40.3% (60) | 59.7% (89) |
| 402 | 51.3% (722) | 48.7% (686) |
| 405 | 54.9% (491) | 45.1% (403) |
| 436 | 48.7% (582) | 51.3% (612) |
| 437 | 48.6% (530) | 51.4% (561) |
| 438 | 53.7% (522) | 46.3% (450) |
| 442 | 41.4% (237) | 58.6% (335) |
| 459 | 46.2% (419) | 53.8% (487) |
| 460 | 50.5% (445) | 49.5% (436) |
| 463 | 48.0% (257) | 52.0% (278) |
| 501 | 47.2% (722) | 52.8% (808) |
| 502 | 51.2% (500) | 48.8% (476) |
| 503 | 51.2% (504) | 48.8% (480) |
| 504 | 56.3% (469) | 43.7% (364) |
| 506 | 42.0% (551) | 58.0% (762) |
| 508 | 42.4% (1118) | 57.6% (1518) |
| 509 | 51.5% (565) | 48.5% (532) |
| 512 | 53.8% (853) | 46.2% (733) |
| 515 | 49.3% (671) | 50.7% (689) |
| 516 | 51.3% (1823) | 48.7% (1730) |
| 517 | 49.8% (242) | 50.2% (244) |
| 518 | 54.0% (293) | 46.0% (250) |
| 519 | 54.0% (321) | 46.0% (273) |
| 520 | 52.2% (990) | 47.8% (906) |
| 521 | 49.9% (880) | 50.1% (882) |
| 523 | 58.1% (751) | 41.9% (541) |
| 526 | 52.8% (335) | 47.2% (300) |
| 528 | 52.7% (2377) | 47.3% (2137) |
| 529 | 48.9% (328) | 51.1% (343) |
| 531 | 44.9% (428) | 55.1% (526) |
| 534 | 43.7% (809) | 56.3% (1043) |
| 537 | 54.9% (742) | 45.1% (610) |
| 538 | 54.1% (373) | 45.9% (316) |
| 539 | 47.1% (553) | 52.9% (620) |
| 540 | 55.6% (398) | 44.4% (318) |
| 541 | 50.0% (1759) | 50.0% (1756) |
| 542 | 57.7% (325) | 42.3% (238) |
| 544 | 47.6% (1210) | 52.4% (1331) |
| 546 | 53.3% (810) | 46.7% (710) |
| 548 | 55.6% (1210) | 44.4% (968) |
| 549 | 48.1% (1818) | 51.9% (1964) |
| 550 | 52.3% (617) | 47.7% (563) |
| 552 | 50.6% (573) | 49.4% (559) |
| 553 | 51.4% (643) | 48.6% (608) |
| 554 | 45.2% (1023) | 54.8% (1238) |
| 556 | 55.4% (426) | 44.6% (343) |
| 557 | 51.2% (596) | 48.8% (568) |
| 558 | 45.9% (760) | 54.1% (895) |
| 561 | 56.0% (1031) | 44.0% (810) |
| 562 | 50.9% (411) | 49.1% (397) |
| 564 | 50.8% (927) | 49.2% (899) |
| 565 | 46.0% (907) | 54.0% (1066) |
| 568 | 54.9% (383) | 45.1% (314) |
| 570 | 47.1% (415) | 52.9% (466) |
| 573 | 52.4% (2399) | 47.6% (2177) |
| 575 | 50.0% (194) | 50.0% (194) |
| 578 | 52.4% (923) | 47.6% (839) |
| 580 | 49.9% (1598) | 50.1% (1603) |
| 581 | 56.3% (543) | 43.7% (422) |
| 583 | 49.4% (841) | 50.6% (861) |
| 585 | 51.0% (364) | 49.0% (350) |
| 586 | 57.5% (894) | 42.5% (662) |
| 589 | 51.4% (2348) | 48.6% (2217) |
| 590 | 45.3% (554) | 54.7% (669) |
| 593 | 47.0% (708) | 53.0% (799) |
| 595 | 55.2% (849) | 44.8% (688) |
| 596 | 52.3% (633) | 47.7% (577) |
| 598 | 51.9% (885) | 48.1% (821) |
| 600 | 48.3% (708) | 51.7% (759) |
| 603 | 51.1% (775) | 48.9% (742) |
| 605 | 49.9% (1093) | 50.1% (1097) |
| 607 | 53.0% (565) | 47.0% (501) |
| 608 | 48.0% (393) | 52.0% (425) |
| 610 | 52.2% (784) | 47.8% (717) |
| 612 | 48.6% (1178) | 51.4% (1247) |
| 613 | 52.4% (598) | 47.6% (544) |
| 614 | 49.8% (739) | 50.2% (744) |
| 618 | 45.8% (1080) | 54.2% (1279) |
| 619 | 46.7% (653) | 53.3% (744) |
| 620 | 56.9% (409) | 43.1% (310) |
| 621 | 50.9% (956) | 49.1% (923) |
| 623 | 50.9% (621) | 49.1% (599) |
| 626 | 46.8% (1203) | 53.2% (1368) |
| 629 | 48.7% (621) | 51.3% (653) |
| 630 | 55.2% (684) | 44.8% (555) |
| 631 | 38.3% (247) | 61.7% (398) |
| 632 | 54.9% (602) | 45.1% (494) |
| 635 | 52.9% (920) | 47.1% (820) |
| 636 | 52.3% (2279) | 47.7% (2077) |
| 637 | 44.8% (566) | 55.2% (697) |
| 640 | 49.6% (913) | 50.4% (928) |
| 642 | 53.8% (781) | 46.2% (671) |
| 644 | 45.2% (1037) | 54.8% (1257) |
| 646 | 50.4% (912) | 49.6% (899) |
| 648 | 44.9% (924) | 55.1% (1136) |
| 649 | 51.2% (389) | 48.8% (371) |
| 650 | 54.8% (594) | 45.2% (490) |
| 652 | 42.8% (646) | 57.2% (863) |
| 653 | 48.8% (396) | 51.2% (416) |
| 654 | 50.3% (514) | 49.7% (507) |
| 655 | 47.3% (490) | 52.7% (545) |
| 656 | 51.1% (588) | 48.9% (563) |
| 657 | 51.6% (1891) | 48.4% (1777) |
| 658 | 50.5% (619) | 49.5% (606) |
| 659 | 50.1% (1096) | 49.9% (1092) |
| 660 | 50.6% (666) | 49.4% (650) |
| 662 | 50.6% (512) | 49.4% (500) |
| 663 | 44.8% (951) | 55.2% (1171) |
| 664 | 49.4% (1020) | 50.6% (1046) |
| 666 | 49.6% (199) | 50.4% (202) |
| 667 | 55.6% (677) | 44.4% (540) |
| 668 | 46.3% (434) | 53.7% (504) |
| 671 | 48.6% (1154) | 51.4% (1222) |
| 672 | 55.3% (1295) | 44.7% (1045) |
| 673 | 61.0% (1873) | 39.0% (1197) |
| 674 | 47.7% (1236) | 52.3% (1353) |
| 675 | 0.0% (0) | 100.0% (1619) |
| 676 | 54.6% (474) | 45.4% (394) |
| 678 | 53.0% (846) | 47.0% (749) |
| 679 | 50.7% (228) | 49.3% (222) |
| 687 | 48.5% (295) | 51.5% (313) |
| 688 | 44.7% (575) | 55.3% (712) |
| 689 | 54.6% (1012) | 45.4% (841) |
| 691 | 53.3% (1406) | 46.7% (1233) |
| 692 | 57.1% (266) | 42.9% (200) |
| 693 | 57.2% (796) | 42.8% (595) |
| 695 | 51.7% (988) | 48.3% (923) |
| 740 | 0.0% (0) | 100.0% (538) |
| 756 | 48.9% (430) | 51.1% (449) |
| 757 | 47.2% (551) | 52.8% (616) |
Sta6a
By Sta6a
weightSamples %>%
tabyl(Sta6a) %>%
adorn_pct_formatting() %>%
arrange(desc(percent)) %>%
scrollTable()| Sta6a | n | percent |
|---|---|---|
| 580 | 25896 | 1.1% |
| 549 | 24649 | 1.0% |
| 672 | 23379 | 1.0% |
| 516 | 22071 | 0.9% |
| 652 | 19348 | 0.8% |
| 673 | 18963 | 0.8% |
| 501 | 16058 | 0.7% |
| 583 | 17062 | 0.7% |
| 586 | 15912 | 0.7% |
| 618 | 16242 | 0.7% |
| 695 | 16179 | 0.7% |
| 521 | 13752 | 0.6% |
| 537 | 13578 | 0.6% |
| 548 | 14932 | 0.6% |
| 558 | 14214 | 0.6% |
| 570 | 13442 | 0.6% |
| 578 | 14320 | 0.6% |
| 600 | 14308 | 0.6% |
| 644 | 15349 | 0.6% |
| 678 | 13943 | 0.6% |
| 508 | 11145 | 0.5% |
| 546 | 11970 | 0.5% |
| 552 | 11577 | 0.5% |
| 553 | 12605 | 0.5% |
| 554 | 11889 | 0.5% |
| 589 | 11218 | 0.5% |
| 605 | 13284 | 0.5% |
| 621 | 12607 | 0.5% |
| 635 | 12800 | 0.5% |
| 657 | 10967 | 0.5% |
| 658 | 11084 | 0.5% |
| 663 | 11930 | 0.5% |
| 674 | 12684 | 0.5% |
| 688 | 12354 | 0.5% |
| 691 | 12356 | 0.5% |
| 402 | 8789 | 0.4% |
| 509A0 | 10829 | 0.4% |
| 512 | 9960 | 0.4% |
| 516BZ | 9704 | 0.4% |
| 526 | 9663 | 0.4% |
| 534 | 9219 | 0.4% |
| 541 | 10799 | 0.4% |
| 544 | 8655 | 0.4% |
| 549BY | 9415 | 0.4% |
| 564 | 9198 | 0.4% |
| 573 | 10644 | 0.4% |
| 589A4 | 8746 | 0.4% |
| 590 | 10488 | 0.4% |
| 593 | 10805 | 0.4% |
| 598A0 | 9092 | 0.4% |
| 612A4 | 9331 | 0.4% |
| 626 | 9953 | 0.4% |
| 630 | 10048 | 0.4% |
| 632 | 9856 | 0.4% |
| 636 | 9961 | 0.4% |
| 637 | 10764 | 0.4% |
| 642 | 9793 | 0.4% |
| 654 | 9694 | 0.4% |
| 656 | 8680 | 0.4% |
| 659 | 10897 | 0.4% |
| 663A4 | 9975 | 0.4% |
| 667 | 9494 | 0.4% |
| 668 | 8884 | 0.4% |
| 671 | 10495 | 0.4% |
| 671BY | 9704 | 0.4% |
| 689 | 10750 | 0.4% |
| 437 | 7180 | 0.3% |
| 438 | 7963 | 0.3% |
| 504 | 7059 | 0.3% |
| 506 | 7647 | 0.3% |
| 508QF | 6239 | 0.3% |
| 517 | 7241 | 0.3% |
| 520 | 6653 | 0.3% |
| 528 | 7992 | 0.3% |
| 528A7 | 7952 | 0.3% |
| 531 | 8079 | 0.3% |
| 539 | 7432 | 0.3% |
| 541GG | 6468 | 0.3% |
| 546BZ | 6332 | 0.3% |
| 556 | 8062 | 0.3% |
| 557 | 7949 | 0.3% |
| 561 | 6838 | 0.3% |
| 561A4 | 6499 | 0.3% |
| 564BY | 6384 | 0.3% |
| 565 | 7677 | 0.3% |
| 565GL | 6890 | 0.3% |
| 573BY | 8238 | 0.3% |
| 581 | 8439 | 0.3% |
| 589A5 | 7471 | 0.3% |
| 595 | 7780 | 0.3% |
| 596 | 8419 | 0.3% |
| 607 | 6858 | 0.3% |
| 610A4 | 6373 | 0.3% |
| 614 | 7031 | 0.3% |
| 623 | 6159 | 0.3% |
| 629 | 7525 | 0.3% |
| 630A4 | 8218 | 0.3% |
| 636A6 | 8076 | 0.3% |
| 640 | 6387 | 0.3% |
| 648 | 6265 | 0.3% |
| 650 | 8272 | 0.3% |
| 655 | 8008 | 0.3% |
| 659BY | 6566 | 0.3% |
| 660 | 7619 | 0.3% |
| 662 | 8456 | 0.3% |
| 664 | 7012 | 0.3% |
| 664BY | 6195 | 0.3% |
| 673BZ | 6241 | 0.3% |
| 674BY | 8177 | 0.3% |
| 689A4 | 7142 | 0.3% |
| 691A4 | 8265 | 0.3% |
| 693 | 8150 | 0.3% |
| 695BY | 6099 | 0.3% |
| 756 | 7114 | 0.3% |
| 757 | 8053 | 0.3% |
| 405 | 5235 | 0.2% |
| 459 | 5146 | 0.2% |
| 460 | 4527 | 0.2% |
| 502 | 5642 | 0.2% |
| 503 | 3798 | 0.2% |
| 506GA | 4577 | 0.2% |
| 508GA | 4917 | 0.2% |
| 515 | 4220 | 0.2% |
| 515BY | 3910 | 0.2% |
| 516GA | 3644 | 0.2% |
| 516GE | 3795 | 0.2% |
| 520BZ | 5923 | 0.2% |
| 521GA | 3831 | 0.2% |
| 523 | 4073 | 0.2% |
| 523A5 | 5046 | 0.2% |
| 528A8 | 5229 | 0.2% |
| 528GE | 3647 | 0.2% |
| 529 | 3671 | 0.2% |
| 537BY | 5203 | 0.2% |
| 537GD | 4655 | 0.2% |
| 538 | 5722 | 0.2% |
| 540 | 5995 | 0.2% |
| 541BY | 5391 | 0.2% |
| 541BZ | 4808 | 0.2% |
| 541GD | 4005 | 0.2% |
| 542 | 3705 | 0.2% |
| 544BZ | 3639 | 0.2% |
| 549A4 | 4982 | 0.2% |
| 550 | 4740 | 0.2% |
| 554GE | 4855 | 0.2% |
| 558GA | 5019 | 0.2% |
| 558GB | 4753 | 0.2% |
| 561BZ | 4085 | 0.2% |
| 562 | 4719 | 0.2% |
| 564GB | 4090 | 0.2% |
| 573A4 | 5818 | 0.2% |
| 573GD | 3979 | 0.2% |
| 573GF | 5153 | 0.2% |
| 575 | 4432 | 0.2% |
| 580BY | 4063 | 0.2% |
| 585 | 4442 | 0.2% |
| 589A7 | 6040 | 0.2% |
| 593GD | 4582 | 0.2% |
| 595GA | 3803 | 0.2% |
| 596A4 | 4707 | 0.2% |
| 603GB | 3942 | 0.2% |
| 603GC | 4137 | 0.2% |
| 603GE | 4296 | 0.2% |
| 608 | 5530 | 0.2% |
| 612B4 | 5353 | 0.2% |
| 612GF | 3812 | 0.2% |
| 612GH | 3896 | 0.2% |
| 613 | 5306 | 0.2% |
| 614GF | 5023 | 0.2% |
| 619 | 3850 | 0.2% |
| 619A4 | 5739 | 0.2% |
| 623BY | 5634 | 0.2% |
| 626A4 | 5673 | 0.2% |
| 626GF | 4202 | 0.2% |
| 629BY | 5138 | 0.2% |
| 636A8 | 5788 | 0.2% |
| 644BY | 5231 | 0.2% |
| 646 | 5637 | 0.2% |
| 646A4 | 3748 | 0.2% |
| 648A4 | 5771 | 0.2% |
| 649 | 4245 | 0.2% |
| 653 | 4317 | 0.2% |
| 657A0 | 4909 | 0.2% |
| 657A5 | 4820 | 0.2% |
| 657GJ | 4659 | 0.2% |
| 659GA | 4046 | 0.2% |
| 672B0 | 5399 | 0.2% |
| 672BZ | 4782 | 0.2% |
| 673GA | 3635 | 0.2% |
| 674A4 | 5475 | 0.2% |
| 675GA | 5724 | 0.2% |
| 675GG | 4182 | 0.2% |
| 676 | 3679 | 0.2% |
| 679 | 4539 | 0.2% |
| 691GE | 3992 | 0.2% |
| Missing | 2447 | 0.1% |
| 402GD | 2109 | 0.1% |
| 402HB | 3201 | 0.1% |
| 405HA | 1474 | 0.1% |
| 436 | 3260 | 0.1% |
| 436GB | 1307 | 0.1% |
| 436GC | 1643 | 0.1% |
| 436GF | 1598 | 0.1% |
| 436GH | 2030 | 0.1% |
| 437GB | 1403 | 0.1% |
| 438GC | 1266 | 0.1% |
| 442 | 3535 | 0.1% |
| 442GC | 1267 | 0.1% |
| 463 | 3138 | 0.1% |
| 502GA | 1429 | 0.1% |
| 502GB | 2532 | 0.1% |
| 503GA | 1265 | 0.1% |
| 503GB | 1217 | 0.1% |
| 504BY | 3358 | 0.1% |
| 508GE | 2315 | 0.1% |
| 508GF | 2707 | 0.1% |
| 508GG | 1711 | 0.1% |
| 508GH | 3221 | 0.1% |
| 508GI | 1213 | 0.1% |
| 509GA | 1303 | 0.1% |
| 512A5 | 2285 | 0.1% |
| 512GA | 1367 | 0.1% |
| 512GC | 1862 | 0.1% |
| 512GD | 2156 | 0.1% |
| 516GC | 2900 | 0.1% |
| 516GD | 2988 | 0.1% |
| 516GF | 2781 | 0.1% |
| 516GH | 1468 | 0.1% |
| 518 | 2623 | 0.1% |
| 519 | 2235 | 0.1% |
| 519HC | 1363 | 0.1% |
| 520GA | 3505 | 0.1% |
| 520GB | 1921 | 0.1% |
| 520GC | 1284 | 0.1% |
| 521GC | 1300 | 0.1% |
| 521GD | 2586 | 0.1% |
| 521GE | 1516 | 0.1% |
| 523A4 | 3217 | 0.1% |
| 523BY | 1632 | 0.1% |
| 523BZ | 3133 | 0.1% |
| 528A4 | 1662 | 0.1% |
| 528A5 | 3016 | 0.1% |
| 528A6 | 2864 | 0.1% |
| 528G9 | 1214 | 0.1% |
| 528GM | 3034 | 0.1% |
| 528GN | 1795 | 0.1% |
| 528GO | 1907 | 0.1% |
| 528GQ | 1406 | 0.1% |
| 528GT | 1592 | 0.1% |
| 534BY | 2984 | 0.1% |
| 534GB | 2746 | 0.1% |
| 534GC | 1407 | 0.1% |
| 534GD | 3098 | 0.1% |
| 537HA | 1241 | 0.1% |
| 538GB | 1497 | 0.1% |
| 539GA | 1382 | 0.1% |
| 539GB | 2248 | 0.1% |
| 539GC | 1447 | 0.1% |
| 539GD | 1394 | 0.1% |
| 539GE | 1375 | 0.1% |
| 540GB | 1219 | 0.1% |
| 541A0 | 2598 | 0.1% |
| 541GB | 3445 | 0.1% |
| 541GC | 2161 | 0.1% |
| 541GE | 2007 | 0.1% |
| 541GF | 3257 | 0.1% |
| 541GH | 1412 | 0.1% |
| 541GI | 1958 | 0.1% |
| 541GJ | 1495 | 0.1% |
| 541GK | 1838 | 0.1% |
| 541GL | 2249 | 0.1% |
| 544GB | 2062 | 0.1% |
| 544GC | 1828 | 0.1% |
| 544GD | 1467 | 0.1% |
| 546GD | 1335 | 0.1% |
| 548GA | 1343 | 0.1% |
| 548GB | 2176 | 0.1% |
| 548GC | 1869 | 0.1% |
| 549GD | 1720 | 0.1% |
| 550BY | 2740 | 0.1% |
| 550GA | 1215 | 0.1% |
| 550GD | 1512 | 0.1% |
| 552GA | 1308 | 0.1% |
| 552GB | 1580 | 0.1% |
| 552GC | 1329 | 0.1% |
| 554GB | 3255 | 0.1% |
| 554GC | 3480 | 0.1% |
| 554GD | 2122 | 0.1% |
| 556GC | 1426 | 0.1% |
| 557GA | 2353 | 0.1% |
| 557GB | 1609 | 0.1% |
| 558GC | 1940 | 0.1% |
| 561GD | 2395 | 0.1% |
| 565GA | 1381 | 0.1% |
| 565GC | 2090 | 0.1% |
| 568 | 3124 | 0.1% |
| 568A4 | 1689 | 0.1% |
| 568GA | 1484 | 0.1% |
| 570GB | 1712 | 0.1% |
| 573BZ | 3300 | 0.1% |
| 573GA | 1273 | 0.1% |
| 573GE | 1955 | 0.1% |
| 573GG | 2444 | 0.1% |
| 573GH | 1254 | 0.1% |
| 573GI | 3273 | 0.1% |
| 578GA | 1751 | 0.1% |
| 578GG | 2286 | 0.1% |
| 580BZ | 3275 | 0.1% |
| 580GC | 2957 | 0.1% |
| 580GD | 3412 | 0.1% |
| 580GE | 1606 | 0.1% |
| 580GG | 1318 | 0.1% |
| 580GH | 1935 | 0.1% |
| 581GA | 2040 | 0.1% |
| 581GB | 2691 | 0.1% |
| 583GA | 1700 | 0.1% |
| 586GB | 1351 | 0.1% |
| 586GD | 2351 | 0.1% |
| 589A6 | 3422 | 0.1% |
| 590GB | 1935 | 0.1% |
| 593GB | 1539 | 0.1% |
| 593GC | 1677 | 0.1% |
| 593GE | 2929 | 0.1% |
| 593GF | 2652 | 0.1% |
| 593GG | 2878 | 0.1% |
| 595GC | 1660 | 0.1% |
| 595GD | 1648 | 0.1% |
| 595GE | 2277 | 0.1% |
| 596GA | 1891 | 0.1% |
| 598 | 3249 | 0.1% |
| 598GA | 1571 | 0.1% |
| 598GC | 2120 | 0.1% |
| 600GB | 1332 | 0.1% |
| 603 | 1291 | 0.1% |
| 603GA | 2216 | 0.1% |
| 605GA | 1736 | 0.1% |
| 605GB | 1609 | 0.1% |
| 605GC | 2110 | 0.1% |
| 605GE | 1273 | 0.1% |
| 607HA | 2439 | 0.1% |
| 610 | 3123 | 0.1% |
| 610GA | 2083 | 0.1% |
| 610GB | 1239 | 0.1% |
| 612BY | 3479 | 0.1% |
| 612GG | 2187 | 0.1% |
| 613GA | 1562 | 0.1% |
| 613GB | 1689 | 0.1% |
| 613GC | 1721 | 0.1% |
| 613GF | 1245 | 0.1% |
| 614GA | 1401 | 0.1% |
| 614GB | 1251 | 0.1% |
| 614GE | 3601 | 0.1% |
| 618BY | 2324 | 0.1% |
| 618GD | 1287 | 0.1% |
| 619GB | 1831 | 0.1% |
| 619GF | 2845 | 0.1% |
| 619QB | 2006 | 0.1% |
| 620 | 1704 | 0.1% |
| 620A4 | 2487 | 0.1% |
| 621BY | 2979 | 0.1% |
| 626GE | 1905 | 0.1% |
| 629GA | 1765 | 0.1% |
| 629GB | 1886 | 0.1% |
| 629GC | 1856 | 0.1% |
| 629GD | 1321 | 0.1% |
| 630A5 | 1537 | 0.1% |
| 631 | 2264 | 0.1% |
| 631BY | 2120 | 0.1% |
| 632GA | 1895 | 0.1% |
| 632HD | 1265 | 0.1% |
| 635GA | 2910 | 0.1% |
| 635GB | 1563 | 0.1% |
| 635QB | 1788 | 0.1% |
| 636A4 | 3484 | 0.1% |
| 636A5 | 3327 | 0.1% |
| 636GC | 1579 | 0.1% |
| 636GF | 3199 | 0.1% |
| 636GH | 1505 | 0.1% |
| 636GI | 1377 | 0.1% |
| 636GK | 1724 | 0.1% |
| 640A4 | 2010 | 0.1% |
| 640BY | 3257 | 0.1% |
| 640HA | 2264 | 0.1% |
| 640HB | 2350 | 0.1% |
| 640HC | 3281 | 0.1% |
| 642GA | 1432 | 0.1% |
| 642GC | 2323 | 0.1% |
| 642GD | 1534 | 0.1% |
| 644GA | 3195 | 0.1% |
| 644GB | 1370 | 0.1% |
| 644GE | 2873 | 0.1% |
| 646GA | 1515 | 0.1% |
| 646GB | 2066 | 0.1% |
| 646GC | 1417 | 0.1% |
| 646GD | 1253 | 0.1% |
| 648GA | 1632 | 0.1% |
| 648GB | 2650 | 0.1% |
| 648GE | 2838 | 0.1% |
| 648GF | 1630 | 0.1% |
| 648GG | 1573 | 0.1% |
| 650GB | 1304 | 0.1% |
| 652GA | 2446 | 0.1% |
| 653BY | 3108 | 0.1% |
| 653GA | 1230 | 0.1% |
| 656GA | 1851 | 0.1% |
| 657A4 | 3364 | 0.1% |
| 657GA | 1413 | 0.1% |
| 657GB | 1743 | 0.1% |
| 657GD | 1372 | 0.1% |
| 657GH | 1436 | 0.1% |
| 657GI | 1256 | 0.1% |
| 657GL | 1797 | 0.1% |
| 658GB | 2512 | 0.1% |
| 659BZ | 1415 | 0.1% |
| 660GA | 1551 | 0.1% |
| 662GA | 2328 | 0.1% |
| 662GC | 1689 | 0.1% |
| 663GA | 2919 | 0.1% |
| 663GB | 1526 | 0.1% |
| 663GC | 1470 | 0.1% |
| 664GB | 2626 | 0.1% |
| 664GC | 1679 | 0.1% |
| 664GD | 1779 | 0.1% |
| 666 | 2171 | 0.1% |
| 667GA | 2984 | 0.1% |
| 667GB | 2281 | 0.1% |
| 667GC | 2293 | 0.1% |
| 668GB | 1428 | 0.1% |
| 671A4 | 3539 | 0.1% |
| 671GB | 1243 | 0.1% |
| 671GF | 1930 | 0.1% |
| 671GK | 2846 | 0.1% |
| 671GO | 2841 | 0.1% |
| 672GC | 2196 | 0.1% |
| 673BY | 2129 | 0.1% |
| 673GB | 3446 | 0.1% |
| 673GC | 2515 | 0.1% |
| 673GF | 1289 | 0.1% |
| 674GA | 1314 | 0.1% |
| 674GB | 1486 | 0.1% |
| 674GC | 2124 | 0.1% |
| 674GD | 1911 | 0.1% |
| 675 | 3057 | 0.1% |
| 675GB | 3291 | 0.1% |
| 676GA | 1289 | 0.1% |
| 676GC | 1793 | 0.1% |
| 678GA | 2344 | 0.1% |
| 678GB | 1296 | 0.1% |
| 678GC | 1430 | 0.1% |
| 678GF | 2851 | 0.1% |
| 678GG | 1213 | 0.1% |
| 687 | 2172 | 0.1% |
| 688GA | 1238 | 0.1% |
| 689HC | 1548 | 0.1% |
| 691GB | 1418 | 0.1% |
| 691GD | 2960 | 0.1% |
| 691GG | 1359 | 0.1% |
| 691GL | 1385 | 0.1% |
| 691GM | 2167 | 0.1% |
| 692 | 2133 | 0.1% |
| 693B4 | 3375 | 0.1% |
| 693GA | 1309 | 0.1% |
| 695GA | 2039 | 0.1% |
| 695GC | 1508 | 0.1% |
| 695GD | 2424 | 0.1% |
| 740GA | 1249 | 0.1% |
| 740GB | 2163 | 0.1% |
| 740GC | 1437 | 0.1% |
| 757GA | 1706 | 0.1% |
| 358 | 1089 | 0.0% |
| 402GA | 881 | 0.0% |
| 402GB | 440 | 0.0% |
| 402GC | 881 | 0.0% |
| 402GE | 891 | 0.0% |
| 402GF | 278 | 0.0% |
| 402HC | 589 | 0.0% |
| 402HL | 110 | 0.0% |
| 402QA | 70 | 0.0% |
| 402QB | 51 | 0.0% |
| 405GA | 646 | 0.0% |
| 405GC | 200 | 0.0% |
| 405HC | 834 | 0.0% |
| 405HE | 168 | 0.0% |
| 405HF | 632 | 0.0% |
| 436GA | 280 | 0.0% |
| 436GD | 628 | 0.0% |
| 436GI | 161 | 0.0% |
| 436GJ | 389 | 0.0% |
| 436GK | 234 | 0.0% |
| 436GL | 246 | 0.0% |
| 436GM | 171 | 0.0% |
| 436HC | 139 | 0.0% |
| 437GA | 413 | 0.0% |
| 437GC | 541 | 0.0% |
| 437GD | 804 | 0.0% |
| 437GE | 697 | 0.0% |
| 437GF | 275 | 0.0% |
| 437GI | 778 | 0.0% |
| 438GA | 724 | 0.0% |
| 438GD | 914 | 0.0% |
| 438GE | 128 | 0.0% |
| 438GF | 406 | 0.0% |
| 442BU | 13 | 0.0% |
| 442GB | 106 | 0.0% |
| 442GD | 1040 | 0.0% |
| 442HK | 45 | 0.0% |
| 442QA | 62 | 0.0% |
| 442QB | 33 | 0.0% |
| 459GA | 690 | 0.0% |
| 459GB | 607 | 0.0% |
| 459GC | 425 | 0.0% |
| 459GD | 348 | 0.0% |
| 459GE | 872 | 0.0% |
| 459GF | 436 | 0.0% |
| 459GG | 720 | 0.0% |
| 460GA | 928 | 0.0% |
| 460GC | 674 | 0.0% |
| 460GD | 286 | 0.0% |
| 460HE | 752 | 0.0% |
| 460HG | 648 | 0.0% |
| 460HK | 34 | 0.0% |
| 463GA | 560 | 0.0% |
| 463GB | 640 | 0.0% |
| 463GC | 221 | 0.0% |
| 463GE | 0 | 0.0% |
| 501G2 | 343 | 0.0% |
| 501GA | 813 | 0.0% |
| 501GB | 843 | 0.0% |
| 501GC | 501 | 0.0% |
| 501GD | 628 | 0.0% |
| 501GE | 670 | 0.0% |
| 501GH | 637 | 0.0% |
| 501GI | 572 | 0.0% |
| 501GJ | 693 | 0.0% |
| 501GK | 869 | 0.0% |
| 501GM | 753 | 0.0% |
| 501HB | 414 | 0.0% |
| 502GE | 327 | 0.0% |
| 502GF | 506 | 0.0% |
| 502GG | 155 | 0.0% |
| 503GC | 1120 | 0.0% |
| 503GD | 195 | 0.0% |
| 503GE | 263 | 0.0% |
| 504BZ | 704 | 0.0% |
| 504GA | 160 | 0.0% |
| 504HB | 84 | 0.0% |
| 506GB | 832 | 0.0% |
| 506GC | 1086 | 0.0% |
| 508GJ | 636 | 0.0% |
| 508GK | 812 | 0.0% |
| 509 | 155 | 0.0% |
| 509GB | 882 | 0.0% |
| 512GE | 504 | 0.0% |
| 512GF | 1069 | 0.0% |
| 512GG | 699 | 0.0% |
| 512QA | 0 | 0.0% |
| 515GA | 1040 | 0.0% |
| 515GB | 1139 | 0.0% |
| 515GC | 544 | 0.0% |
| 516GB | 1021 | 0.0% |
| 517GB | 243 | 0.0% |
| 517QA | 183 | 0.0% |
| 518GA | 466 | 0.0% |
| 518GB | 523 | 0.0% |
| 518GE | 197 | 0.0% |
| 518GG | 216 | 0.0% |
| 519GA | 1153 | 0.0% |
| 519GB | 374 | 0.0% |
| 519GD | 33 | 0.0% |
| 519HD | 15 | 0.0% |
| 519HF | 862 | 0.0% |
| 521GB | 513 | 0.0% |
| 521GF | 969 | 0.0% |
| 521GG | 920 | 0.0% |
| 521GH | 610 | 0.0% |
| 521GI | 1142 | 0.0% |
| 521GJ | 434 | 0.0% |
| 523GA | 548 | 0.0% |
| 523GB | 990 | 0.0% |
| 523GC | 490 | 0.0% |
| 523GD | 169 | 0.0% |
| 526GA | 778 | 0.0% |
| 526GB | 393 | 0.0% |
| 526GC | 257 | 0.0% |
| 526GD | 248 | 0.0% |
| 528G1 | 136 | 0.0% |
| 528G2 | 221 | 0.0% |
| 528G3 | 605 | 0.0% |
| 528G4 | 895 | 0.0% |
| 528G5 | 665 | 0.0% |
| 528G6 | 508 | 0.0% |
| 528G7 | 681 | 0.0% |
| 528G8 | 390 | 0.0% |
| 528GB | 789 | 0.0% |
| 528GC | 729 | 0.0% |
| 528GD | 721 | 0.0% |
| 528GK | 481 | 0.0% |
| 528GL | 898 | 0.0% |
| 528GP | 755 | 0.0% |
| 528GR | 860 | 0.0% |
| 528GV | 786 | 0.0% |
| 528GW | 714 | 0.0% |
| 528GX | 700 | 0.0% |
| 528GY | 980 | 0.0% |
| 528GZ | 940 | 0.0% |
| 528J1 | 117 | 0.0% |
| 529GA | 552 | 0.0% |
| 529GB | 364 | 0.0% |
| 529GC | 272 | 0.0% |
| 529GD | 304 | 0.0% |
| 529GF | 398 | 0.0% |
| 531GE | 1007 | 0.0% |
| 531GG | 763 | 0.0% |
| 534GE | 889 | 0.0% |
| 534GF | 0 | 0.0% |
| 534QA | 275 | 0.0% |
| 537GA | 902 | 0.0% |
| 538GA | 722 | 0.0% |
| 538GC | 626 | 0.0% |
| 538GD | 802 | 0.0% |
| 538GE | 498 | 0.0% |
| 539GF | 574 | 0.0% |
| 539QA | 18 | 0.0% |
| 540GA | 573 | 0.0% |
| 540GC | 518 | 0.0% |
| 540GD | 413 | 0.0% |
| 540HK | 27 | 0.0% |
| 542GA | 1039 | 0.0% |
| 542GE | 879 | 0.0% |
| 544GE | 632 | 0.0% |
| 544GF | 1062 | 0.0% |
| 544GG | 798 | 0.0% |
| 546GA | 47 | 0.0% |
| 546GB | 844 | 0.0% |
| 546GC | 1074 | 0.0% |
| 546GE | 580 | 0.0% |
| 546GF | 1188 | 0.0% |
| 546GG | 101 | 0.0% |
| 546GH | 509 | 0.0% |
| 548GD | 865 | 0.0% |
| 548GE | 1070 | 0.0% |
| 548GF | 780 | 0.0% |
| 548QA | 0 | 0.0% |
| 549GA | 785 | 0.0% |
| 549GC | 518 | 0.0% |
| 549GE | 704 | 0.0% |
| 549GF | 577 | 0.0% |
| 549GH | 493 | 0.0% |
| 549GJ | 652 | 0.0% |
| 549GL | 58 | 0.0% |
| 549HA | 155 | 0.0% |
| 549QC | 732 | 0.0% |
| 550GC | 686 | 0.0% |
| 550GF | 256 | 0.0% |
| 552GD | 1209 | 0.0% |
| 553GA | 1190 | 0.0% |
| 553GB | 1160 | 0.0% |
| 554A5 | 1089 | 0.0% |
| 554GF | 480 | 0.0% |
| 554GG | 560 | 0.0% |
| 554GH | 249 | 0.0% |
| 554GI | 125 | 0.0% |
| 556GA | 746 | 0.0% |
| 556GD | 1150 | 0.0% |
| 557GC | 213 | 0.0% |
| 557GE | 300 | 0.0% |
| 557GF | 43 | 0.0% |
| 557HA | 360 | 0.0% |
| 561GA | 707 | 0.0% |
| 561GB | 548 | 0.0% |
| 561GE | 613 | 0.0% |
| 561GF | 686 | 0.0% |
| 561GG | 76 | 0.0% |
| 561GH | 802 | 0.0% |
| 561GI | 889 | 0.0% |
| 561GJ | 650 | 0.0% |
| 562GA | 921 | 0.0% |
| 562GB | 765 | 0.0% |
| 562GC | 425 | 0.0% |
| 562GD | 515 | 0.0% |
| 562GE | 623 | 0.0% |
| 564GA | 929 | 0.0% |
| 564GC | 818 | 0.0% |
| 564GD | 65 | 0.0% |
| 564GE | 551 | 0.0% |
| 565GD | 577 | 0.0% |
| 565GE | 669 | 0.0% |
| 565GF | 303 | 0.0% |
| 565GG | 136 | 0.0% |
| 568GB | 315 | 0.0% |
| 568HA | 33 | 0.0% |
| 568HB | 100 | 0.0% |
| 568HC | 100 | 0.0% |
| 568HF | 20 | 0.0% |
| 568HH | 391 | 0.0% |
| 568HJ | 158 | 0.0% |
| 568HM | 49 | 0.0% |
| 568HP | 300 | 0.0% |
| 570GA | 1105 | 0.0% |
| 570GC | 352 | 0.0% |
| 573GJ | 557 | 0.0% |
| 573GK | 953 | 0.0% |
| 573GL | 511 | 0.0% |
| 573GM | 412 | 0.0% |
| 573GN | 238 | 0.0% |
| 573QJ | 1011 | 0.0% |
| 575GA | 435 | 0.0% |
| 575GB | 67 | 0.0% |
| 578GC | 984 | 0.0% |
| 578GD | 1114 | 0.0% |
| 578GE | 1144 | 0.0% |
| 578GF | 1098 | 0.0% |
| 580GF | 728 | 0.0% |
| 581GD | 62 | 0.0% |
| 581GE | 125 | 0.0% |
| 583GB | 1107 | 0.0% |
| 583GC | 419 | 0.0% |
| 583GF | 89 | 0.0% |
| 585GA | 441 | 0.0% |
| 585GB | 1037 | 0.0% |
| 585GC | 605 | 0.0% |
| 585GD | 419 | 0.0% |
| 585HA | 741 | 0.0% |
| 585HB | 384 | 0.0% |
| 586GA | 959 | 0.0% |
| 586GC | 1041 | 0.0% |
| 586GE | 1006 | 0.0% |
| 586GF | 645 | 0.0% |
| 586GG | 364 | 0.0% |
| 589BU | 0 | 0.0% |
| 589G1 | 874 | 0.0% |
| 589G2 | 379 | 0.0% |
| 589G3 | 24 | 0.0% |
| 589G4 | 591 | 0.0% |
| 589G5 | 537 | 0.0% |
| 589G7 | 364 | 0.0% |
| 589G8 | 713 | 0.0% |
| 589GB | 770 | 0.0% |
| 589GC | 549 | 0.0% |
| 589GD | 746 | 0.0% |
| 589GE | 977 | 0.0% |
| 589GF | 746 | 0.0% |
| 589GH | 848 | 0.0% |
| 589GI | 981 | 0.0% |
| 589GJ | 620 | 0.0% |
| 589GM | 192 | 0.0% |
| 589GN | 163 | 0.0% |
| 589GP | 58 | 0.0% |
| 589GQ | 4 | 0.0% |
| 589GR | 700 | 0.0% |
| 589GU | 234 | 0.0% |
| 589GV | 149 | 0.0% |
| 589GW | 844 | 0.0% |
| 589GX | 922 | 0.0% |
| 589GY | 541 | 0.0% |
| 589GZ | 402 | 0.0% |
| 589HK | 34 | 0.0% |
| 589JA | 291 | 0.0% |
| 589JB | 445 | 0.0% |
| 589JD | 183 | 0.0% |
| 589JE | 31 | 0.0% |
| 589JF | 898 | 0.0% |
| 590GC | 325 | 0.0% |
| 590GD | 648 | 0.0% |
| 593GH | 260 | 0.0% |
| 595GF | 611 | 0.0% |
| 596GB | 588 | 0.0% |
| 596GC | 309 | 0.0% |
| 596GD | 742 | 0.0% |
| 598GB | 694 | 0.0% |
| 598GD | 790 | 0.0% |
| 598GE | 849 | 0.0% |
| 598GF | 602 | 0.0% |
| 598GG | 738 | 0.0% |
| 598GH | 342 | 0.0% |
| 600GA | 1016 | 0.0% |
| 600GC | 311 | 0.0% |
| 600GD | 1013 | 0.0% |
| 600GE | 832 | 0.0% |
| 603GD | 1070 | 0.0% |
| 603GF | 918 | 0.0% |
| 603GG | 617 | 0.0% |
| 603GH | 271 | 0.0% |
| 605BZ | 363 | 0.0% |
| 605GD | 764 | 0.0% |
| 607GC | 1042 | 0.0% |
| 607GD | 622 | 0.0% |
| 607GE | 912 | 0.0% |
| 607GF | 553 | 0.0% |
| 608GA | 468 | 0.0% |
| 608GC | 794 | 0.0% |
| 608GD | 320 | 0.0% |
| 608HA | 704 | 0.0% |
| 610GC | 665 | 0.0% |
| 610GD | 540 | 0.0% |
| 612GD | 990 | 0.0% |
| 612GE | 788 | 0.0% |
| 612GI | 506 | 0.0% |
| 613BU | 0 | 0.0% |
| 613GD | 114 | 0.0% |
| 613GE | 547 | 0.0% |
| 613GG | 419 | 0.0% |
| 614AA | 198 | 0.0% |
| 614GC | 361 | 0.0% |
| 614GD | 820 | 0.0% |
| 614GG | 543 | 0.0% |
| 614GI | 316 | 0.0% |
| 614GN | 264 | 0.0% |
| 618GA | 1026 | 0.0% |
| 618GB | 787 | 0.0% |
| 618GE | 1016 | 0.0% |
| 618GG | 823 | 0.0% |
| 618GH | 908 | 0.0% |
| 618GI | 1034 | 0.0% |
| 618GJ | 386 | 0.0% |
| 618GK | 258 | 0.0% |
| 618QA | 1 | 0.0% |
| 619GA | 638 | 0.0% |
| 619GD | 425 | 0.0% |
| 619GE | 282 | 0.0% |
| 620BU | 138 | 0.0% |
| 620GA | 1038 | 0.0% |
| 620GB | 434 | 0.0% |
| 620GD | 575 | 0.0% |
| 620GE | 476 | 0.0% |
| 620GF | 280 | 0.0% |
| 620GG | 421 | 0.0% |
| 620GH | 144 | 0.0% |
| 621BU | 271 | 0.0% |
| 621GA | 487 | 0.0% |
| 621GC | 480 | 0.0% |
| 621GD | 384 | 0.0% |
| 621GG | 380 | 0.0% |
| 621GI | 443 | 0.0% |
| 621GJ | 497 | 0.0% |
| 621GK | 160 | 0.0% |
| 623GA | 541 | 0.0% |
| 623GB | 369 | 0.0% |
| 626GA | 333 | 0.0% |
| 626GC | 551 | 0.0% |
| 626GD | 299 | 0.0% |
| 626GG | 560 | 0.0% |
| 626GH | 1114 | 0.0% |
| 626GI | 35 | 0.0% |
| 626GJ | 902 | 0.0% |
| 626GK | 469 | 0.0% |
| 626GL | 257 | 0.0% |
| 626GM | 572 | 0.0% |
| 626GN | 135 | 0.0% |
| 626GO | 18 | 0.0% |
| 626QB | 27 | 0.0% |
| 629GE | 141 | 0.0% |
| 629GF | 185 | 0.0% |
| 630BZ | 453 | 0.0% |
| 630GA | 19 | 0.0% |
| 630GB | 583 | 0.0% |
| 630GC | 70 | 0.0% |
| 631GC | 467 | 0.0% |
| 631GD | 604 | 0.0% |
| 631GE | 897 | 0.0% |
| 631GF | 282 | 0.0% |
| 631QB | 0 | 0.0% |
| 632HA | 82 | 0.0% |
| 632HB | 691 | 0.0% |
| 632HC | 189 | 0.0% |
| 635GC | 207 | 0.0% |
| 635GD | 878 | 0.0% |
| 635GE | 328 | 0.0% |
| 635GF | 46 | 0.0% |
| 635GG | 228 | 0.0% |
| 635HB | 266 | 0.0% |
| 636A7 | 374 | 0.0% |
| 636BU | 44 | 0.0% |
| 636GA | 319 | 0.0% |
| 636GB | 958 | 0.0% |
| 636GD | 267 | 0.0% |
| 636GG | 830 | 0.0% |
| 636GJ | 676 | 0.0% |
| 636GL | 362 | 0.0% |
| 636GM | 105 | 0.0% |
| 636GN | 1132 | 0.0% |
| 636GP | 155 | 0.0% |
| 636GQ | 404 | 0.0% |
| 636GR | 477 | 0.0% |
| 636GS | 443 | 0.0% |
| 636GT | 414 | 0.0% |
| 636GU | 247 | 0.0% |
| 637GA | 656 | 0.0% |
| 637GB | 804 | 0.0% |
| 637GC | 909 | 0.0% |
| 640A0 | 53 | 0.0% |
| 640GA | 353 | 0.0% |
| 640GB | 1149 | 0.0% |
| 640GC | 598 | 0.0% |
| 642BU | 0 | 0.0% |
| 642GE | 5 | 0.0% |
| 644GC | 155 | 0.0% |
| 644GD | 351 | 0.0% |
| 644GF | 174 | 0.0% |
| 644GG | 314 | 0.0% |
| 646A5 | 935 | 0.0% |
| 646GE | 1013 | 0.0% |
| 648GD | 456 | 0.0% |
| 649GA | 499 | 0.0% |
| 649GB | 300 | 0.0% |
| 649GC | 741 | 0.0% |
| 649GD | 326 | 0.0% |
| 649GE | 679 | 0.0% |
| 649QC | 21 | 0.0% |
| 650GA | 964 | 0.0% |
| 650GD | 872 | 0.0% |
| 650GE | 20 | 0.0% |
| 652GB | 201 | 0.0% |
| 652GE | 1208 | 0.0% |
| 652GF | 327 | 0.0% |
| 653GB | 594 | 0.0% |
| 654GA | 1093 | 0.0% |
| 654GB | 1058 | 0.0% |
| 654GC | 825 | 0.0% |
| 654GD | 232 | 0.0% |
| 654QC | 0 | 0.0% |
| 655GA | 1148 | 0.0% |
| 655GB | 965 | 0.0% |
| 655GC | 882 | 0.0% |
| 655GD | 633 | 0.0% |
| 655GE | 745 | 0.0% |
| 655GF | 135 | 0.0% |
| 655GG | 249 | 0.0% |
| 655GH | 190 | 0.0% |
| 655GI | 155 | 0.0% |
| 656GB | 556 | 0.0% |
| 656GC | 362 | 0.0% |
| 657GE | 22 | 0.0% |
| 657GF | 1041 | 0.0% |
| 657GG | 762 | 0.0% |
| 657GK | 847 | 0.0% |
| 657GM | 1011 | 0.0% |
| 657GN | 81 | 0.0% |
| 657GO | 484 | 0.0% |
| 657GP | 955 | 0.0% |
| 657GQ | 391 | 0.0% |
| 657GR | 257 | 0.0% |
| 657GS | 188 | 0.0% |
| 657GT | 613 | 0.0% |
| 657GU | 247 | 0.0% |
| 657GV | 236 | 0.0% |
| 658GA | 488 | 0.0% |
| 658GC | 832 | 0.0% |
| 658GD | 419 | 0.0% |
| 658GE | 666 | 0.0% |
| 659GB | 164 | 0.0% |
| 660GB | 1188 | 0.0% |
| 660GC | 70 | 0.0% |
| 660GD | 327 | 0.0% |
| 660GE | 750 | 0.0% |
| 660GG | 836 | 0.0% |
| 660GI | 36 | 0.0% |
| 660GJ | 812 | 0.0% |
| 660GK | 96 | 0.0% |
| 662GD | 1146 | 0.0% |
| 662GE | 925 | 0.0% |
| 662GF | 560 | 0.0% |
| 662GG | 380 | 0.0% |
| 663GD | 648 | 0.0% |
| 663HK | 62 | 0.0% |
| 664GA | 485 | 0.0% |
| 666GB | 905 | 0.0% |
| 666GC | 505 | 0.0% |
| 666GD | 381 | 0.0% |
| 666GE | 389 | 0.0% |
| 666GF | 274 | 0.0% |
| 667QA | 6 | 0.0% |
| 668GA | 494 | 0.0% |
| 668HK | 145 | 0.0% |
| 671BZ | 1149 | 0.0% |
| 671GA | 80 | 0.0% |
| 671GD | 28 | 0.0% |
| 671GE | 0 | 0.0% |
| 671GH | 267 | 0.0% |
| 671GI | 13 | 0.0% |
| 671GL | 597 | 0.0% |
| 671GN | 121 | 0.0% |
| 671GP | 30 | 0.0% |
| 671GQ | 311 | 0.0% |
| 672GA | 287 | 0.0% |
| 672GB | 271 | 0.0% |
| 672GD | 1014 | 0.0% |
| 672GE | 1105 | 0.0% |
| 672QC | 61 | 0.0% |
| 673GD | 188 | 0.0% |
| 673GE | 305 | 0.0% |
| 674HB | 214 | 0.0% |
| 675GC | 912 | 0.0% |
| 675GD | 661 | 0.0% |
| 675GE | 580 | 0.0% |
| 675GF | 594 | 0.0% |
| 676GD | 1210 | 0.0% |
| 676GE | 283 | 0.0% |
| 678GD | 298 | 0.0% |
| 678GE | 1155 | 0.0% |
| 679HK | 141 | 0.0% |
| 687GA | 1016 | 0.0% |
| 687GB | 638 | 0.0% |
| 687GC | 242 | 0.0% |
| 687HA | 930 | 0.0% |
| 688GB | 608 | 0.0% |
| 688GC | 296 | 0.0% |
| 688GD | 891 | 0.0% |
| 688GE | 965 | 0.0% |
| 689GA | 672 | 0.0% |
| 689GB | 605 | 0.0% |
| 689GC | 580 | 0.0% |
| 689GD | 615 | 0.0% |
| 689GE | 526 | 0.0% |
| 689PA | 60 | 0.0% |
| 691GC | 945 | 0.0% |
| 691GF | 386 | 0.0% |
| 691GK | 1111 | 0.0% |
| 692GA | 406 | 0.0% |
| 692GB | 149 | 0.0% |
| 693GB | 995 | 0.0% |
| 693GC | 220 | 0.0% |
| 693GE | 114 | 0.0% |
| 693GF | 608 | 0.0% |
| 693GG | 731 | 0.0% |
| 740 | 772 | 0.0% |
| 740GD | 380 | 0.0% |
| 756GA | 1083 | 0.0% |
| 756GB | 755 | 0.0% |
| 757GB | 1092 | 0.0% |
| 757GC | 765 | 0.0% |
| 757GD | 1044 | 0.0% |
Bariatric Surgery Patients
By Person
weightSamples %>%
distinct(PatientICN, SampleYear, Bariatric) %>%
select(-PatientICN) %>%
multTable(rc = "col", Bariatric, SampleYear) %>%
tableStyle()| Bariatric | 2008 | 2016 |
|---|---|---|
| No | 100.0% (98771) | 100.0% (98922) |
| Yes | 0.0% (15) | 0.0% (36) |
By Weight Sample
| Bariatric | 2008 | 2016 |
|---|---|---|
| No | 100.0% (1213797) | 99.9% (1207228) |
| Yes | 0.0% (596) | 0.1% (1506) |
Weight trajectories from just the bariatric surgery patients.
bar_sub <- weightSamples %>%
filter(Bariatric == "Yes" & SampleYear == "2016") %>%
mutate(
WeightDate = as.Date(WeightDateTime, "%Y-%m-%d", tz = "UTC"),
BariatricDate = as.Date(BariatricDateTime, "%Y-%m-%d", tz = "UTC")
) %>%
select(PatientICN, WeightDate, Weight, BariatricDate)
post_bar <- bar_sub %>%
filter(WeightDate >= BariatricDate) %>%
group_by(PatientICN) %>%
arrange(PatientICN, WeightDate) %>%
mutate(
time = as.numeric(WeightDate - BariatricDate)
) %>%
distinct() %>%
ungroup() %>%
select(PatientICN, Weight, time)
keys <- c("PatientICN", "time")
post_bar <- data.table::as.data.table(post_bar)
post_bar <- post_bar[, list(mweight = mean(Weight)), keys]
p <- post_bar %>%
ggplot(aes(x = time, y = mweight, group = PatientICN)) %>%
add(geom_line(alpha = 0.2)) %>%
add(geom_smooth(aes(group = 1),
method = "gam",
formula = y ~ s(x, bs = "cs"),
se = FALSE,
color = "darkred",
size = 2)) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(labs(
x = 'Days Since Bariatric Surgery',
y = ''
)) %>%
add(ylim(100, 400))
all_bar <- bar_sub %>%
group_by(PatientICN) %>%
arrange(PatientICN, WeightDate) %>%
mutate(time = as.numeric(WeightDate - BariatricDate)) %>%
distinct() %>%
ungroup() %>%
select(PatientICN, Weight, time)
all_bar <- data.table::as.data.table(all_bar)
all_bar <- all_bar[, list(mweight = mean(Weight)), keys]
q <- all_bar %>%
ggplot(aes(x = time, y = mweight, group = PatientICN)) %>%
add(geom_line(alpha = 0.2)) %>%
add(geom_smooth(aes(group = 1),
method = "gam",
formula = y ~ s(x, bs = "cs"),
se = FALSE,
color = "darkred",
size = 2)) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(labs(
x = 'Days',
y = 'Weight (lbs.)',
caption = "Days = 0 denotes Bariatric Surgery Date"
)) %>%
add(ylim(100, 400))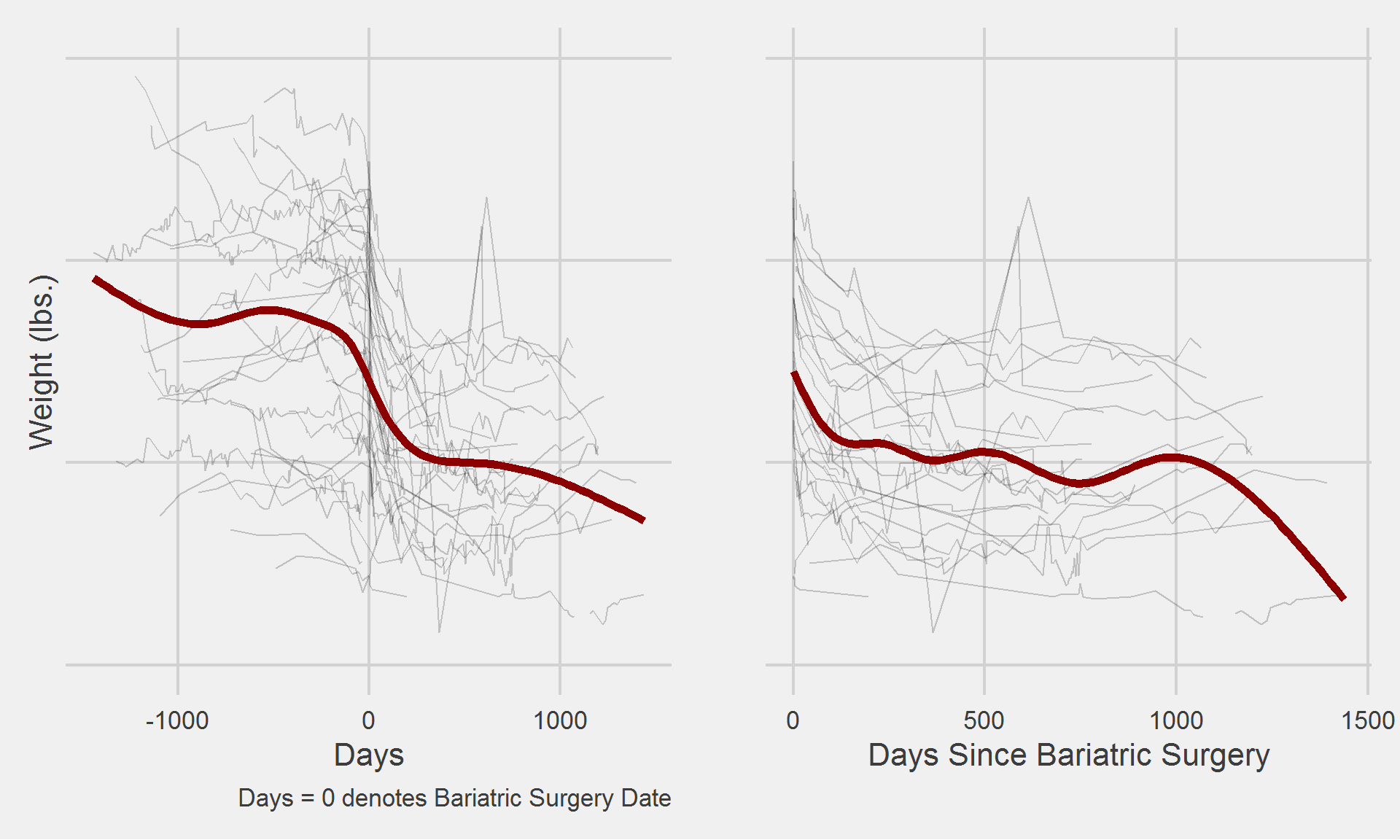
I’m just going to play here and make a GAM model, adjusting for person level effects
mod1 <- gam(mweight ~ s(time) + s(PatientICN, bs = "re"),
data = post_bar %>%
mutate(PatientICN = factor(PatientICN)))
summary(mod1)
Family: gaussian
Link function: identity
Formula:
mweight ~ s(time) + s(PatientICN, bs = "re")
Parametric coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 213.718 9.748 21.93 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Approximate significance of smooth terms:
edf Ref.df F p-value
s(time) 7.915 8.694 51.59 <2e-16 ***
s(PatientICN) 31.612 32.000 113.63 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
R-sq.(adj) = 0.861 Deviance explained = 86.9%
GCV = 288.7 Scale est. = 272.16 n = 707Plot the first smooth component against weight, it looks different because it’s not on the original weight scale.
plot(sm(viz, 1)) %>%
add(l_fitLine(color = "red")) %>%
add(l_rug(mapping = aes(x = x, y = y), alpha = 0.8)) %>%
add(l_ciLine(mul = 5, color = "blue", linetype = 2)) %>%
add(l_points(shape = 19, size = 1, alpha = 0.1)) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text()))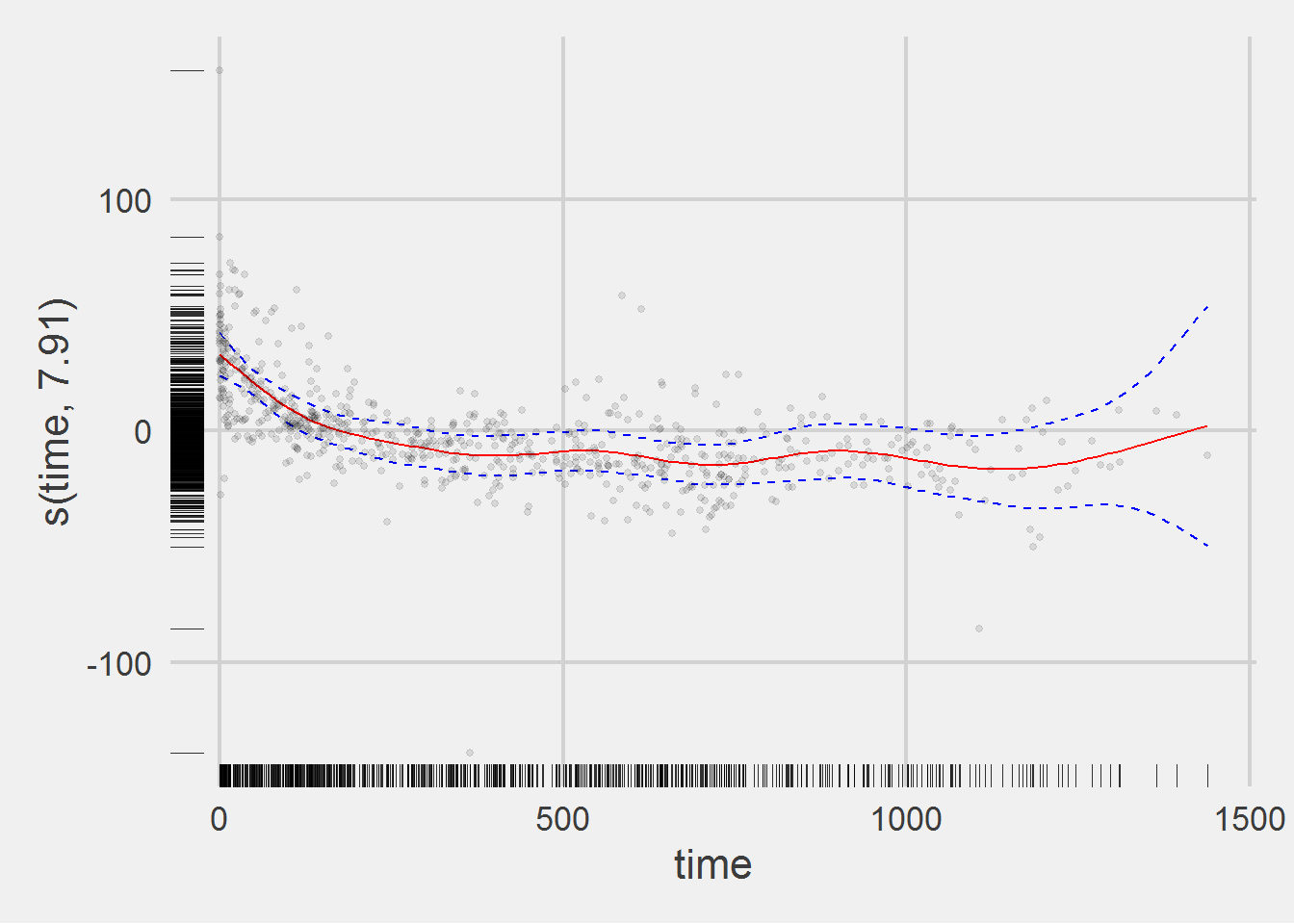
Or just for fun, we’ll plot the underlying density
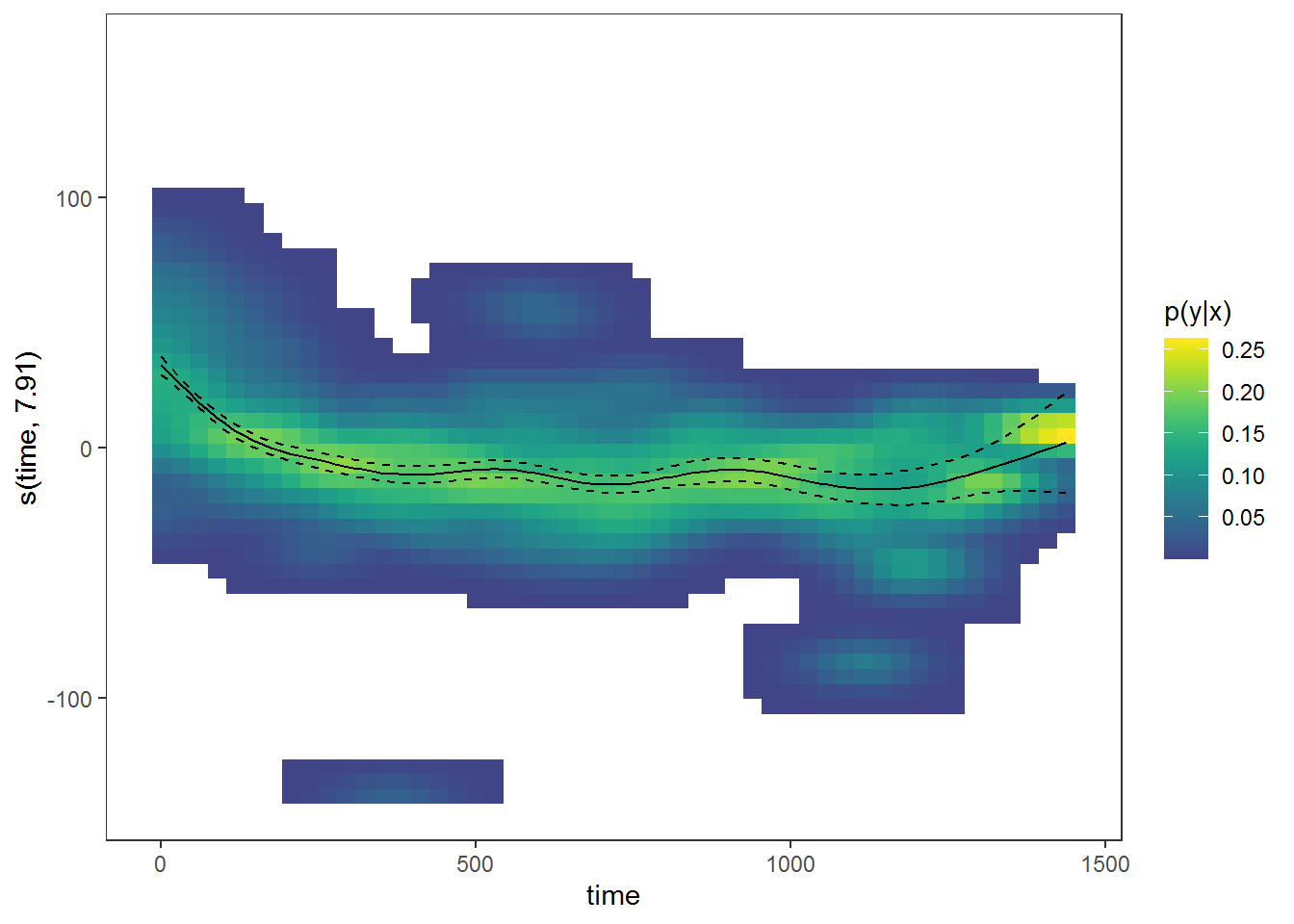
Inpatient Weights
By Person
weightSamples %>%
distinct(PatientICN, SampleYear, InptWeight) %>%
select(-PatientICN) %>%
multTable(rc = "col", InptWeight, SampleYear) %>%
tableStyle()| InptWeight | 2008 | 2016 |
|---|---|---|
| Outpatient | 99.7% (98783) | 99.4% (98957) |
| Inpatient | 0.3% (282) | 0.6% (567) |
By Weight Sample
| InptWeight | 2008 | 2016 |
|---|---|---|
| Outpatient | 99.7% (1211169) | 99.6% (1204096) |
| Inpatient | 0.3% (3224) | 0.4% (4638) |
Outpatient vs. Inpatient
inpt <- weightSamples %>%
filter(InptWeight == "Inpatient") %>%
distinct(PatientICN) %>%
left_join(weightSamples, by = "PatientICN") %>%
filter(!is.na(Weight)) %>%
select(PatientICN, WeightDateTime, Weight, InptWeight)
inpt %>%
group_by(InptWeight) %>%
summarize(
n = n(),
mean = mean(Weight),
SD = sd(Weight),
median = median(Weight),
Q1 = fivenum(Weight)[2],
Q3 = fivenum(Weight)[4]
) %>%
tableStyle()| InptWeight | n | mean | SD | median | Q1 | Q3 |
|---|---|---|---|---|---|---|
| Outpatient | 29925 | 216.91 | 59.64 | 211.4 | 174.0 | 247.0 |
| Inpatient | 7862 | 213.11 | 64.64 | 195.6 | 170.7 | 250.8 |
inpt %>%
ggplot(aes(x = InptWeight, y = Weight)) %>%
add(geom_boxplot()) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text()))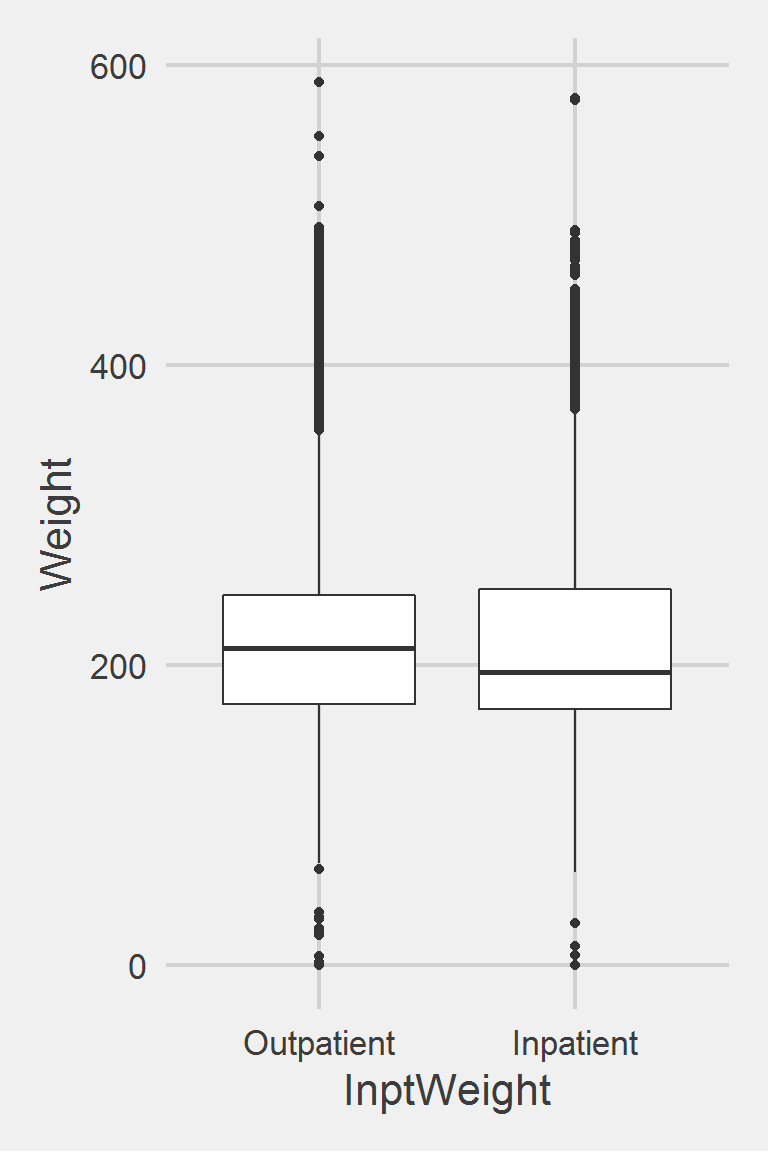
Diabetes
weightSamples %>%
distinct(PatientICN, Diabetic) %>%
tabyl(Diabetic) %>%
adorn_pct_formatting() %>%
tableStyle()| Diabetic | n | percent |
|---|---|---|
| Non-Diabetic | 139678 | 71.0% |
| Diabetic | 56947 | 29.0% |
How many weights collected for diabetics vs. non-diabetics
| Diabetic | n | percent |
|---|---|---|
| Non-Diabetic | 1490724 | 61.5% |
| Diabetic | 932403 | 38.5% |
So, diabetics make up 1/3 of the sample, but account for 50% of all weights collected. That does make some sense, diabetics are more heavily monitored. We should not see any significant differences between the 2008 and 2016 cohorts,
weightSamples %>%
distinct(PatientICN, SampleYear, Diabetic) %>%
multTable(rc = "col", Diabetic, SampleYear) %>%
tableStyle()| Diabetic | 2008 | 2016 |
|---|---|---|
| Non-Diabetic | 71.4% (70485) | 70.6% (69891) |
| Diabetic | 28.6% (28301) | 29.4% (29067) |
\(\approx 30\)% of each sample.
Timing of diabetic diagnoses codes, patient-level
weightSamples %>%
distinct(PatientICN, DiabetesTiming) %>%
tabyl(DiabetesTiming) %>%
adorn_pct_formatting() %>%
tableStyle()| DiabetesTiming | n | percent |
|---|---|---|
| Non-Diabetic | 139678 | 71.0% |
| Diabetes After | 11848 | 6.0% |
| Diabetes Before | 1292 | 0.7% |
| Diabetes Before and After | 43807 | 22.3% |
We collected diabetes so as to have some sort of outcome to evaluate choice of weight as a predictor of diabetes. The formulation we have at the moment is to choose “New” onset diabetic diagnoses, following each veterans index date (general PCP visit date). So, in a logistic regression the outcome would be “Diabetes After”, with a single predictor of weight which will vary by algorithm, time-point closest to index date and the window around that date.
Weight Distribution
weightSamples %>%
group_by(SampleYear) %>%
summarize(
n = n(),
mean = mean(Weight),
SD = sd(Weight)
) %>%
tableStyle()| SampleYear | n | mean | SD |
|---|---|---|---|
| 2008 | 1214393 | 202.66 | 47.65 |
| 2016 | 1208734 | 207.57 | 48.55 |
weightSamples %>%
ggplot(aes(y = Weight, x = SampleYear)) %>%
add(geom_boxplot()) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(labs(
x = 'Sample Year',
y = 'Weight (lbs.)'
))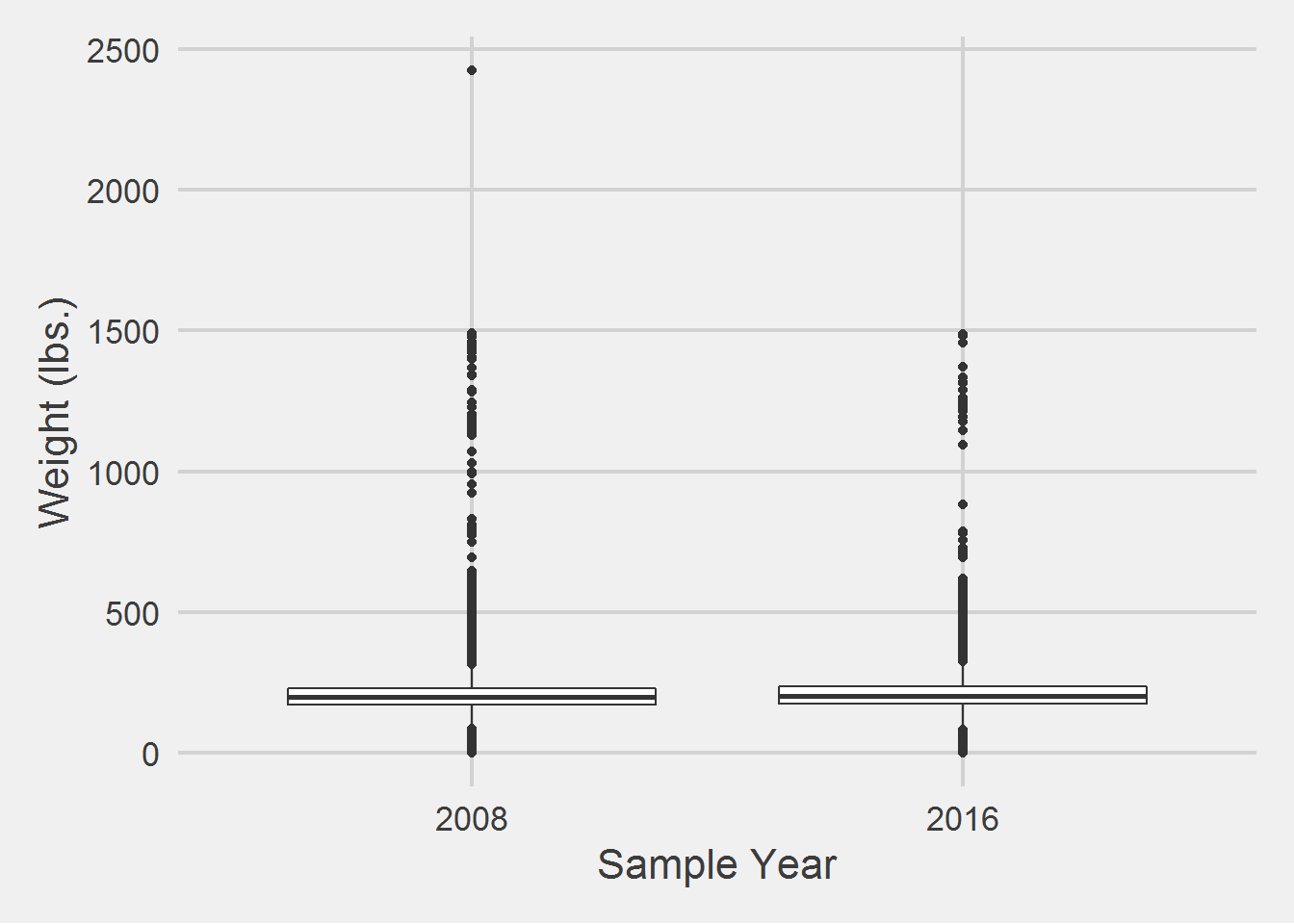
Weight Distribution, by Diabetic Status
weightSamples %>%
group_by(Diabetic) %>%
summarize(
n = n(),
mean = mean(Weight),
SD = sd(Weight)
) %>%
tableStyle()| Diabetic | n | mean | SD |
|---|---|---|---|
| Non-Diabetic | 1490724 | 196.02 | 44.30 |
| Diabetic | 932403 | 219.64 | 50.47 |
By Diabetic status
weightSamples %>%
ggplot(aes(y = Weight, x = Diabetic)) %>%
add(geom_boxplot()) %>%
add(facet_wrap(~SampleYear, scales = "free")) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(labs(
x = 'Sample Year',
y = 'Weight (lbs.)'
))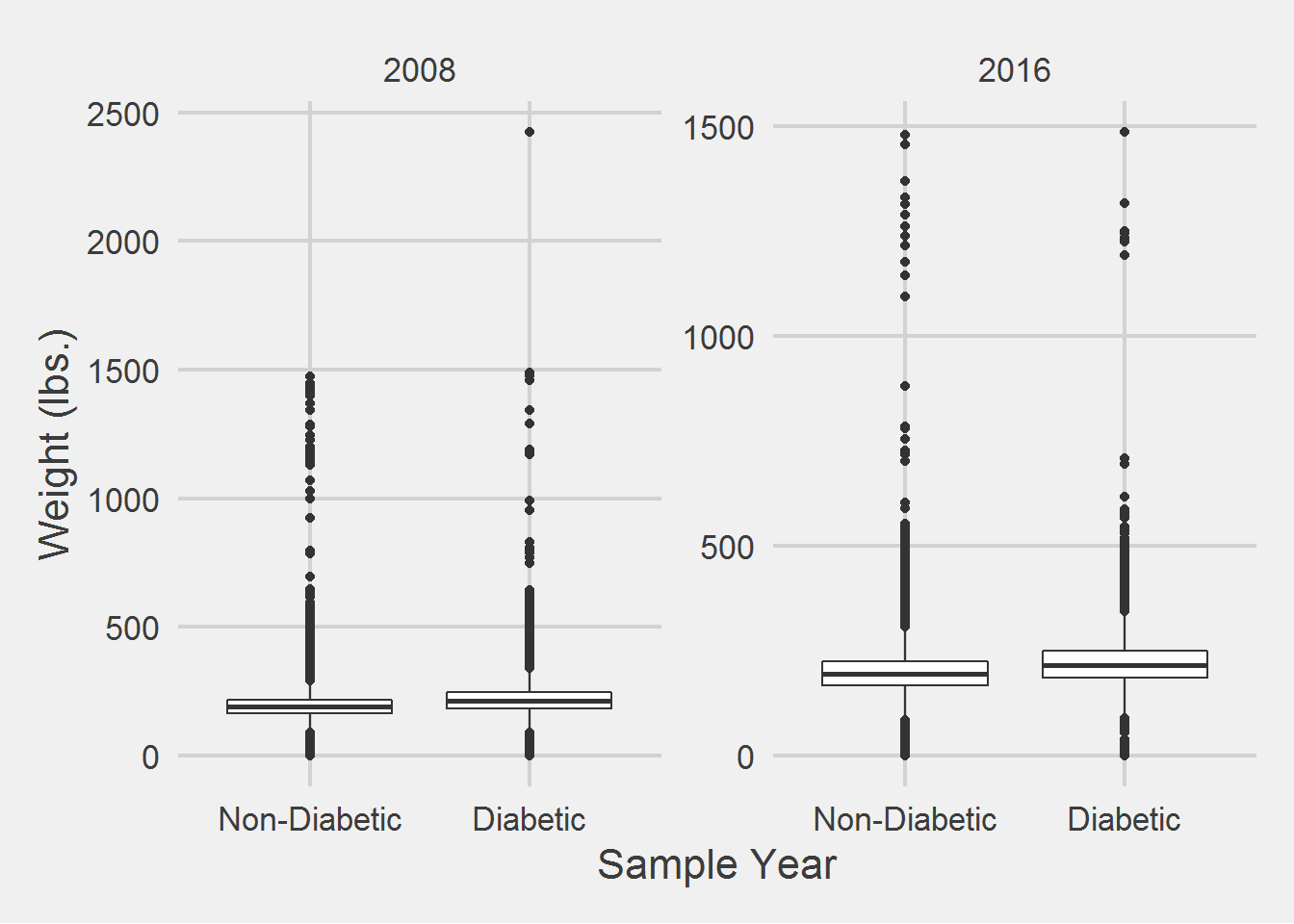
How many weights, per person?
weightSamples %>%
filter(!is.na(Weight)) %>%
group_by(SampleYear, PatientICN) %>%
count() %>%
ungroup() %>%
group_by(SampleYear) %>%
summarize(
mean = mean(n),
SD = sd(n),
min = min(n),
Q1 = fivenum(n)[2],
median = median(n),
Q3 = fivenum(n)[4],
max = max(n)
) %>%
tableStyle()| SampleYear | mean | SD | min | Q1 | median | Q3 | max |
|---|---|---|---|---|---|---|---|
| 2008 | 12.29 | 15.97 | 1 | 5 | 9 | 15 | 1479 |
| 2016 | 12.21 | 24.85 | 1 | 5 | 8 | 15 | 4981 |
I find it very hard to believe that anyone has >1,000 weight measurements. Let’s sleuth,
# weightSamples %>%
# filter(!is.na(Weight)) %>%
# group_by(SampleYear, PatientICN) %>%
# count() %>%
# filter(n > 1000) %>%
# View()
# weightSamples %>%
# filter(PatientICN == [REDACTED]) %>%
# View()Alright, so, all those with > 1,000 values actually have very few real duplicate values, however, the 1 person with 4,981 measurements actually only has 20 real non-duplicate measurements.
Height Distribution
heightSamples %>%
group_by(SampleYear) %>%
summarize(
n = n(),
mean = mean(Height),
SD = sd(Height)
) %>%
tableStyle()| SampleYear | n | mean | SD |
|---|---|---|---|
| 2008 | 514356 | 69.26 | 3.34 |
| 2016 | 511366 | 69.34 | 3.31 |
heightSamples %>%
ggplot(aes(y = Height, x = SampleYear)) %>%
add(geom_boxplot()) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(labs(
x = 'Sample Year',
y = 'Height (in.)'
))
BMI Distribution
BMI Vignette
Compute Mode of Height
# Calculate mode(height) for each individual
ModeOfHeight <- heightSamples %>%
filter(Height > 0) %>%
select(PatientICN, Height)
# convert to data.table for the quickness
ModeOfHeight <- data.table::setDT(ModeOfHeight)
ModeOfHeight <- data.table::setkey(
ModeOfHeight[,
list(freq = .N),
by = list(PatientICN, Height)
],
by = PatientICN, freq
)[J(unique(PatientICN)), mult = "last"]
ModeOfHeight %>%
rename(HeightMode = Height) %>%
select(-freq) %>%
left_join(
heightSamples %>%
select(
PatientICN,
Height,
SampleYear
),
by = "PatientICN"
) %>%
tidyr::gather(
HeightType,
Measurement,
HeightMode:Height,
factor_key = TRUE
) %>%
distinct() %>%
mutate(
HeightType = factor(
HeightType,
levels = c("HeightMode", "Height"),
labels = c("Mode(Height)", "Height")
)
) %>%
ggplot(aes(
y = Measurement,
x = SampleYear,
fill = HeightType
)) %>%
add(geom_boxplot()) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(scale_fill_brewer(palette = "Set1")) %>%
add(labs(
x = "Sample Year",
y = "Height (in.)",
fill = ""
))
BMI_samples <- ModeOfHeight %>%
select(-freq) %>%
left_join(
weightSamples %>%
select(
PatientICN,
WeightDateTime,
Weight,
SampleYear
),
by = "PatientICN"
) %>%
mutate(
BMI = 703 * Weight / (Height ^ 2)
)BMI_samples %>%
group_by(SampleYear) %>%
summarize(
n = n(),
mean = mean(BMI),
SD = sd(BMI)
) %>%
tableStyle()| SampleYear | n | mean | SD |
|---|---|---|---|
| 2008 | 1168801 | 29.64 | 7.03 |
| 2016 | 1167096 | 30.28 | 7.75 |
Vignettes
Weight Vignette
samp <- weightSamples %>%
distinct(SampleYear, PatientICN) %>%
group_by(SampleYear) %>%
sample_n(8) %>%
pull(PatientICN)
Set1 <- RColorBrewer::brewer.pal(9, "Set1")
weightSamples %>%
filter(PatientICN %in% samp & !is.na(Weight)) %>%
ggplot(aes(x = WeightDateTime, y = Weight)) %>%
add(geom_line(aes(color = SampleYear), size = 1)) %>%
add(geom_point(
aes(fill = SampleYear),
color = "black",
pch = 21,
size = 2
)) %>%
add(scale_fill_manual(values = Set1[1:2])) %>%
add(scale_color_manual(values = Set1[1:2])) %>%
add(facet_wrap(~PatientICN, scales = "free")) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(labs(
x = 'Weight Measurement Time',
y = 'Weight (lbs.)'
))
Height Vignette
samp <- heightSamples %>%
distinct(SampleYear, PatientICN) %>%
group_by(SampleYear) %>%
sample_n(8) %>%
pull(PatientICN)
heightSamples %>%
filter(PatientICN %in% samp & !is.na(Height)) %>%
ggplot(aes(x = HeightDateTime, y = Height)) %>%
add(geom_line(aes(color = SampleYear), size = 1)) %>%
add(geom_point(
aes(fill = SampleYear),
color = "black",
pch = 21,
size = 2
)) %>%
add(scale_fill_manual(values = Set1[1:2])) %>%
add(scale_color_manual(values = Set1[1:2])) %>%
add(facet_wrap(~PatientICN, scales = "free")) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(labs(
x = 'Height Measurement Time',
y = 'Height (in.)'
))
BMI Vignette
samp <- BMI_samples %>%
distinct(SampleYear, PatientICN) %>%
group_by(SampleYear) %>%
sample_n(8) %>%
pull(PatientICN)
BMI_samples %>%
filter(PatientICN %in% samp & !is.na(BMI)) %>%
ggplot(aes(x = WeightDateTime, y = BMI)) %>%
add(geom_line(aes(color = SampleYear), size = 1)) %>%
add(geom_point(
aes(fill = SampleYear),
color = "black",
pch = 21,
size = 2
)) %>%
add(scale_fill_manual(values = Set1[1:2])) %>%
add(scale_color_manual(values = Set1[1:2])) %>%
add(facet_wrap(~PatientICN, scales = "free")) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text())) %>%
add(labs(
x = 'BMI Measurement Time',
y = 'BMI',
caption = "BMI = 703 X [Weight (lbs.)] / [Height (in.) ^ 2]"
))
These should mirror the weight trajectories, if they were the same sample patients …