Latent Trajectory Analysis
Sampling 1,000 people from the PCP 2016 Cohort
Trajectory Analysis:
For this analysis we will use a latent class mixed model to examine the effect that different weight cleaning algorithms would have on overall trajectories. Some things to consider: Since this is a linear method it makes the most sense to analyze those individuals where their trajectory is assumed to be decreasing or increasing, and thus this analysis only involves the MOVE 2016 cohort (could also do 2008). Second, based on experience with this data I am only assuming two latent groups or classes, this may not represent reality as people can presumably increase, decrease or maintain the same weight over time. Further analyses is warranted to find a better fitting model.
library(lcmm)
# variable 'subject' must be numeric
for (i in 1:length(me)) {
if (class(me[[i]]$PatientICN) == "factor") {
me[[i]]$PatientICN <- as.integer(as.character(me[[i]]$PatientICN))
}
}
# initial values/model
B <- lcmm(
Weight ~ time,
random = ~time,
subject = "PatientICN",
ng = 1,
idiag = TRUE,
data = me[[1]],
verbose = TRUE
)
lca.mods <- lapply(
me,
FUN = function(x) {
lcmm(
Weight ~ time,
random = ~time,
subject = "PatientICN",
mixture = ~time,
ng = 3,
idiag = TRUE,
data = x,
B = B
)
}
)
names(lca.mods) <- names(me)
for (i in 1:length(lca.mods)) lca.mods[[i]]$data <- me[[i]]Predicted classes
So, there is no guarantee that each class within an algorithms LCMM, will be the same class across algorithms. We can get this information from the signs of the coefficients on “time” in each model.
concept_classes <- c("Loss", "Maintain", "Gain")
traj_edies <- sapply(
lca.mods,
function(x) {
time_coefs <- coef(x)
which_coef <- grepl("time", names(time_coefs))
time_coefs <- time_coefs[which_coef]
traj_class <- sign(round(time_coefs, 3))
traj_class
}
) %>%
reshape2::melt() %>%
mutate(
concept_class = case_when(
value == -1 ~ concept_classes[1],
value == 0 ~ concept_classes[2],
value == 1 ~ concept_classes[3]
),
concept_class = factor(
concept_class,
levels = concept_classes,
labels = concept_classes
),
orig_class = case_when(
Var1 == "time class1" ~ "Class 1",
Var1 == "time class2" ~ "Class 2",
Var1 == "time class3" ~ "Class 3"
),
orig_class = factor(orig_class)
) %>%
select(-value, -Var1) %>%
rename(Algorithm = Var2)newdata <- data.frame(time = seq(0, 365, 5))
Ypred_lcmm <- lapply(
lca.mods,
function(x) {
y <- predictY(x, newdata, var.time = "time")
cbind(y$pred, y$time)
}
) %>%
dplyr::bind_rows(.id = "Algorithm") %>%
reshape2::melt(id.vars = c("Algorithm", "time")) %>%
mutate(
Algorithm = factor(Algorithm,
levels = names(me),
labels = names(me)),
orig_class = factor(variable,
labels = c("Class 1", "Class 2", "Class 3"))
) %>%
select(-variable) %>%
rename(Ypred = value) %>%
left_join(
traj_edies,
by = c("Algorithm", "orig_class")
)
Ypred_lcmm %>%
ggplot(
aes(
x = time, y = Ypred,
group = interaction(Algorithm, concept_class),
color = interaction(Algorithm, concept_class)
)
) %>%
add(geom_line(size = 1)) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(axis.title = element_text(), legend.position = "none")) %>%
add(xlim(0, 500)) %>%
add(directlabels::geom_dl(aes(label = interaction(Algorithm, concept_class)),
method = "last.polygons")) %>%
add(labs(
x = "Time (days)",
y = "Weight (Lbs.)",
title = "Class Specific Mean-Predicted Trajectory",
subtitle = "2016 Cohort - Latent Class Mixed Model",
color = ""
))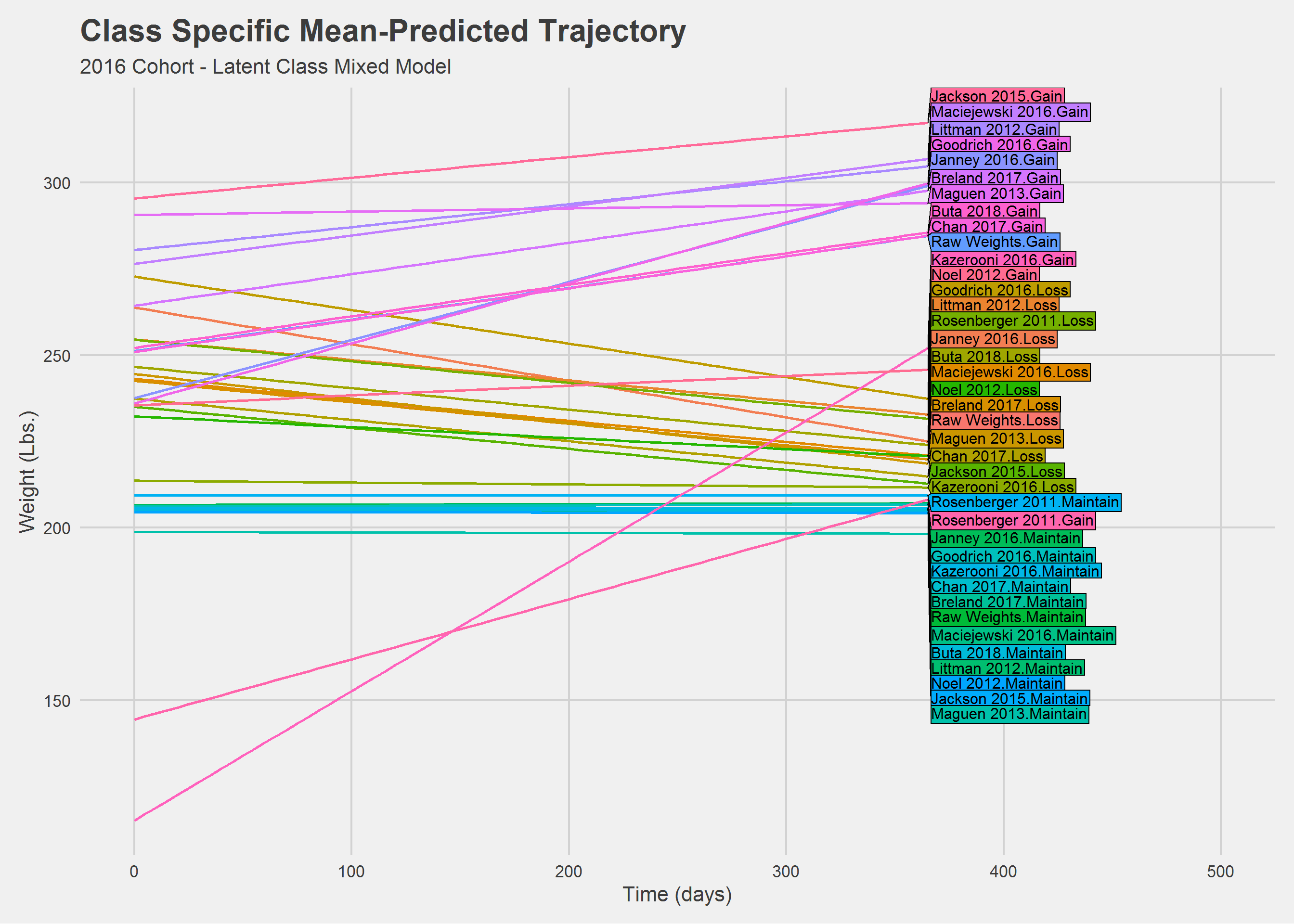
Hah! What a mess. Let’s look at things a little differently.
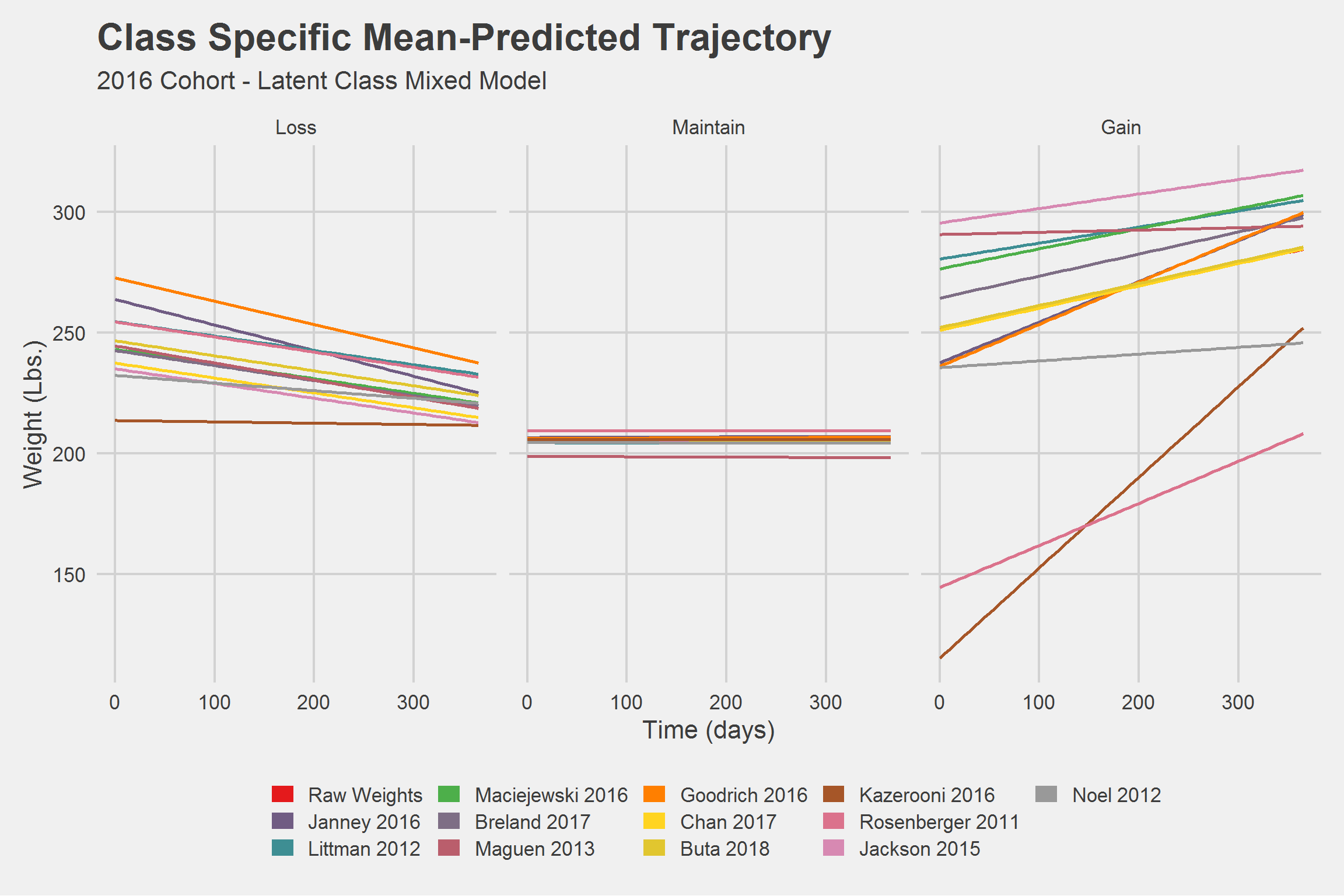
This next one certainly won’t help anyone,
An Illustration:
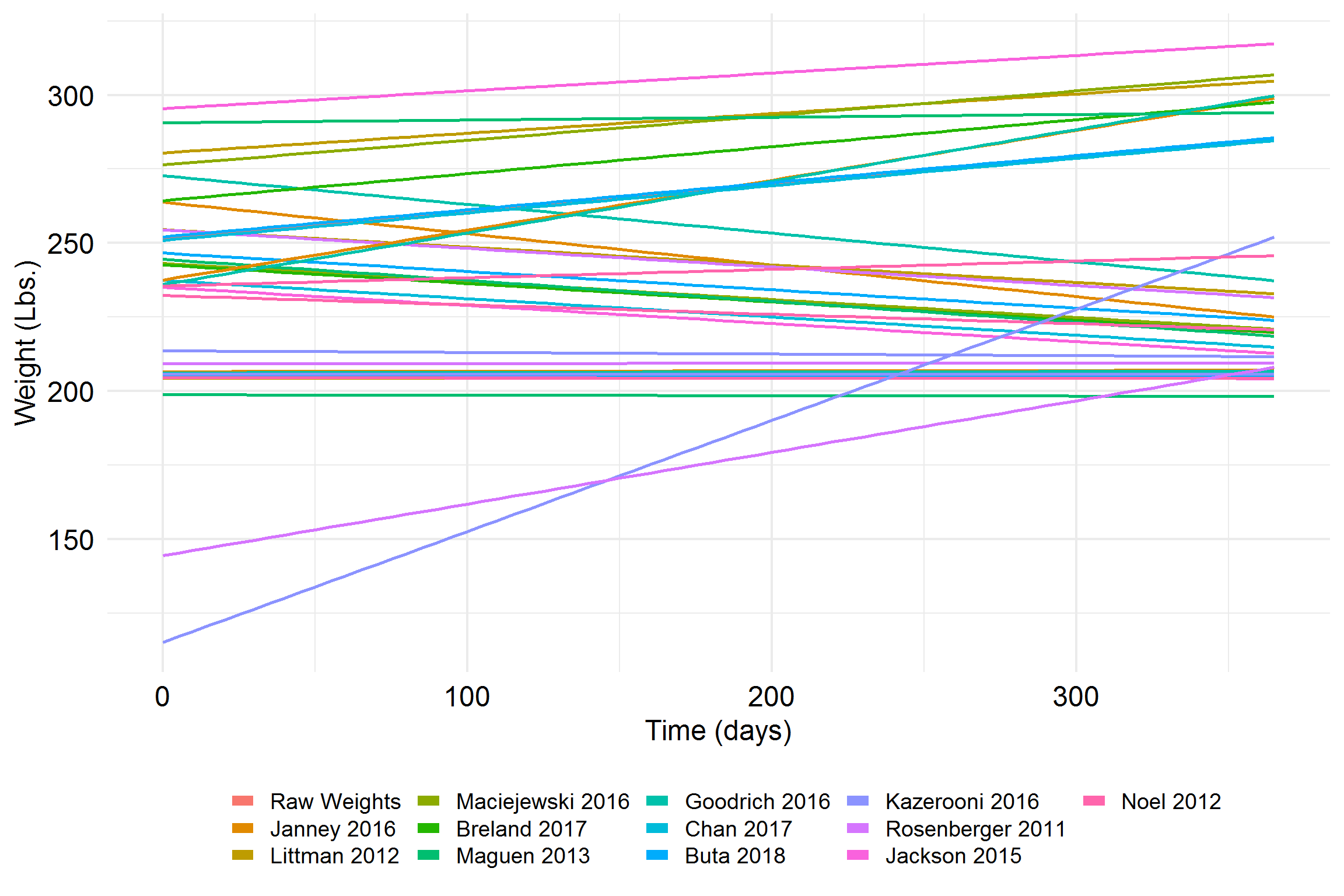
library(gganimate)
Ypred_lcmm %>%
ggplot(
aes(
x = time,
y = Ypred,
color = Algorithm,
group = interaction(Algorithm, concept_class)
)
) %>%
add(geom_line(size = 1)) %>%
add(theme_minimal(20)) %>%
add(theme(legend.position = "none")) %>%
add(labs(
title = 'Algorithm: {closest_state}',
x = "Time (days)",
y = "Weight (lbs.)"
)) %>%
add(transition_states(
states = Algorithm,
transition_length = 1,
state_length = 1
))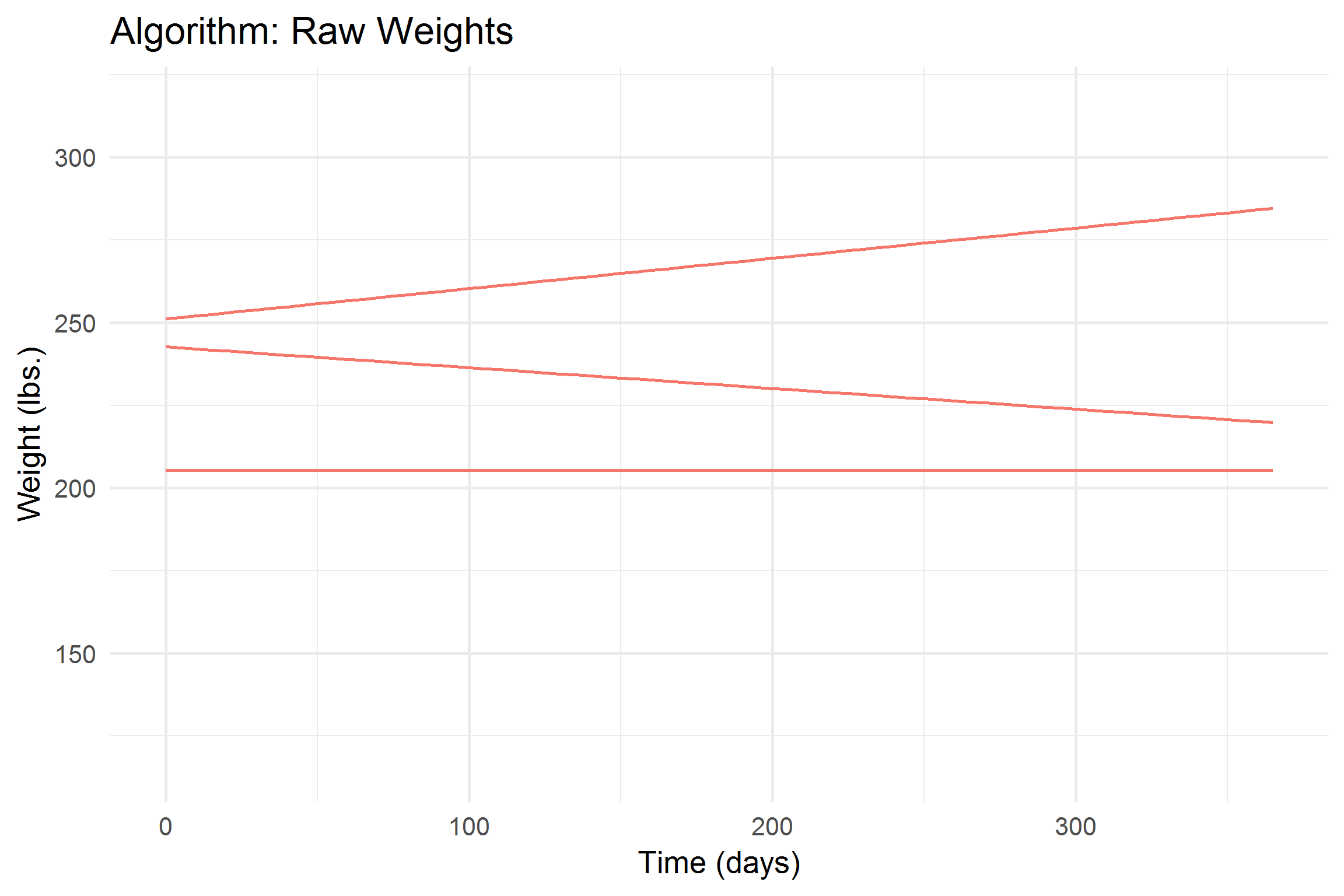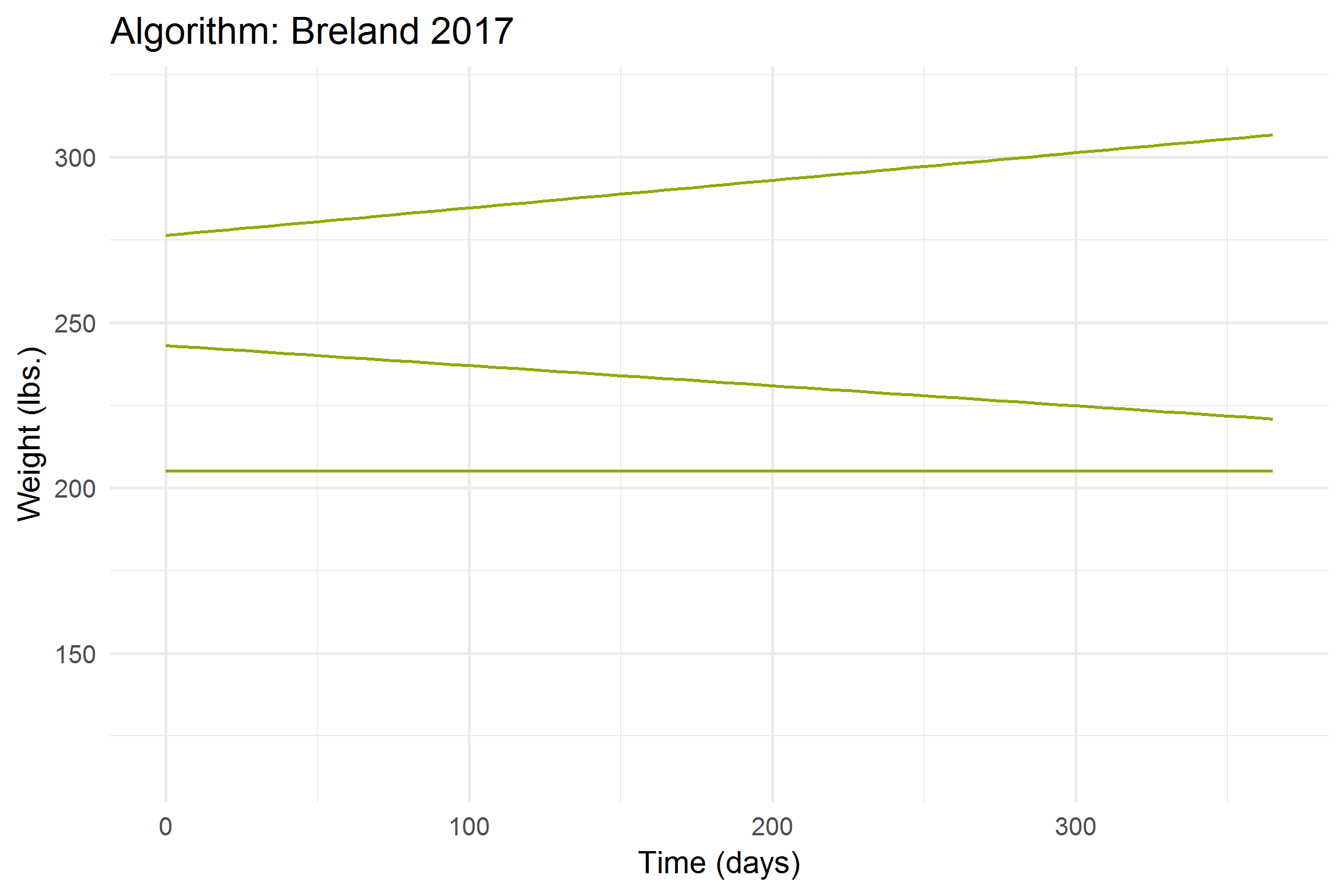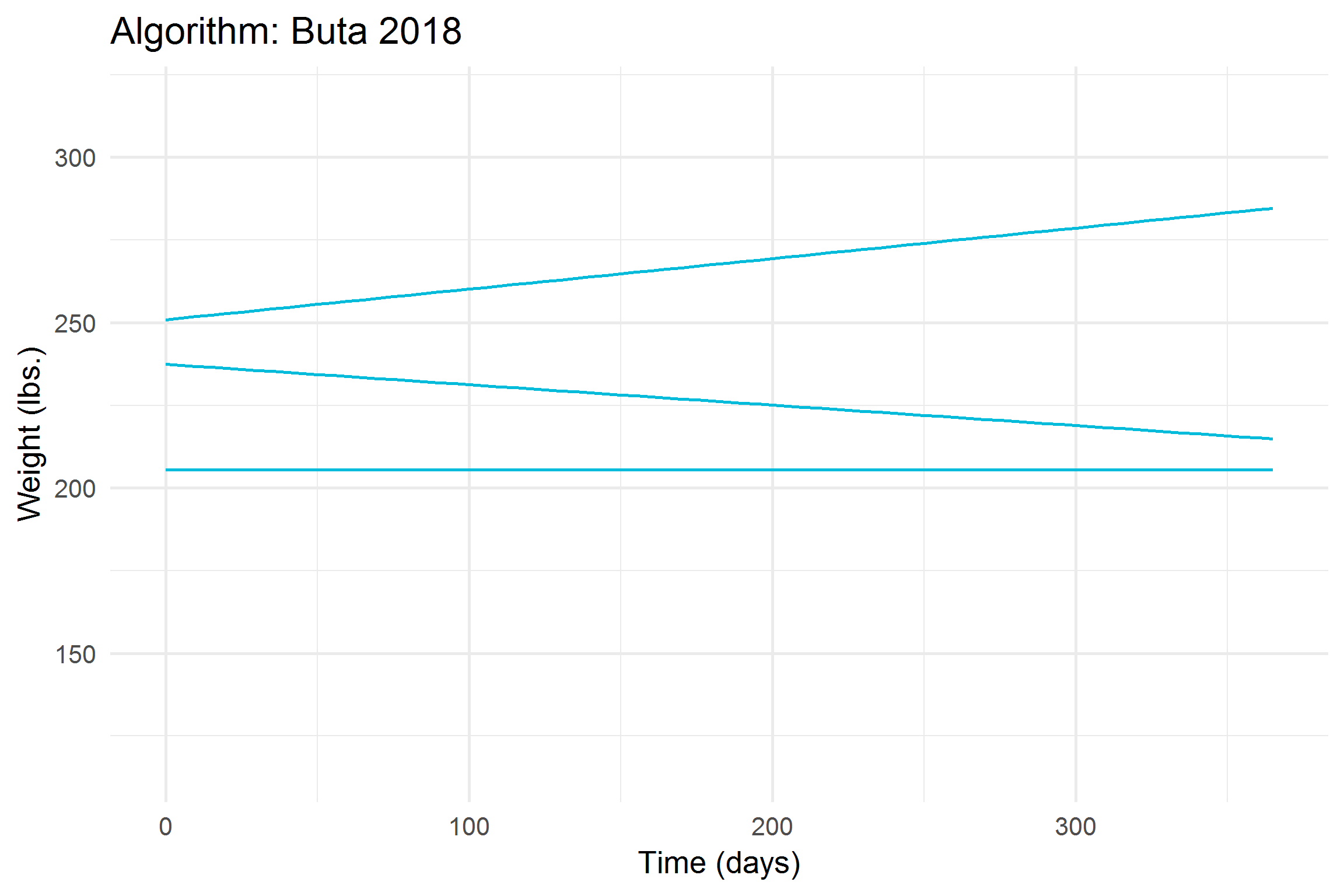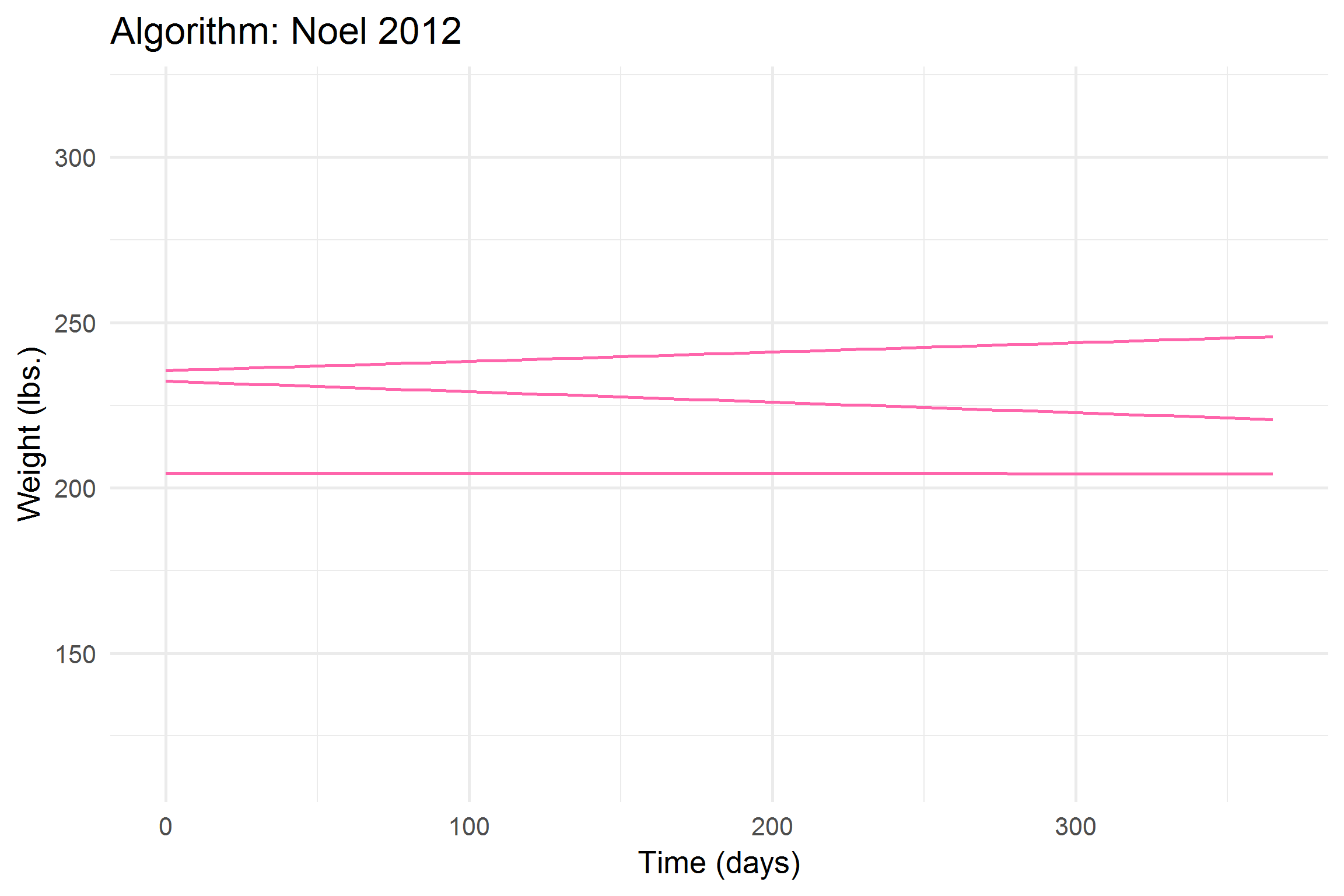
Let’s break it down a bit,
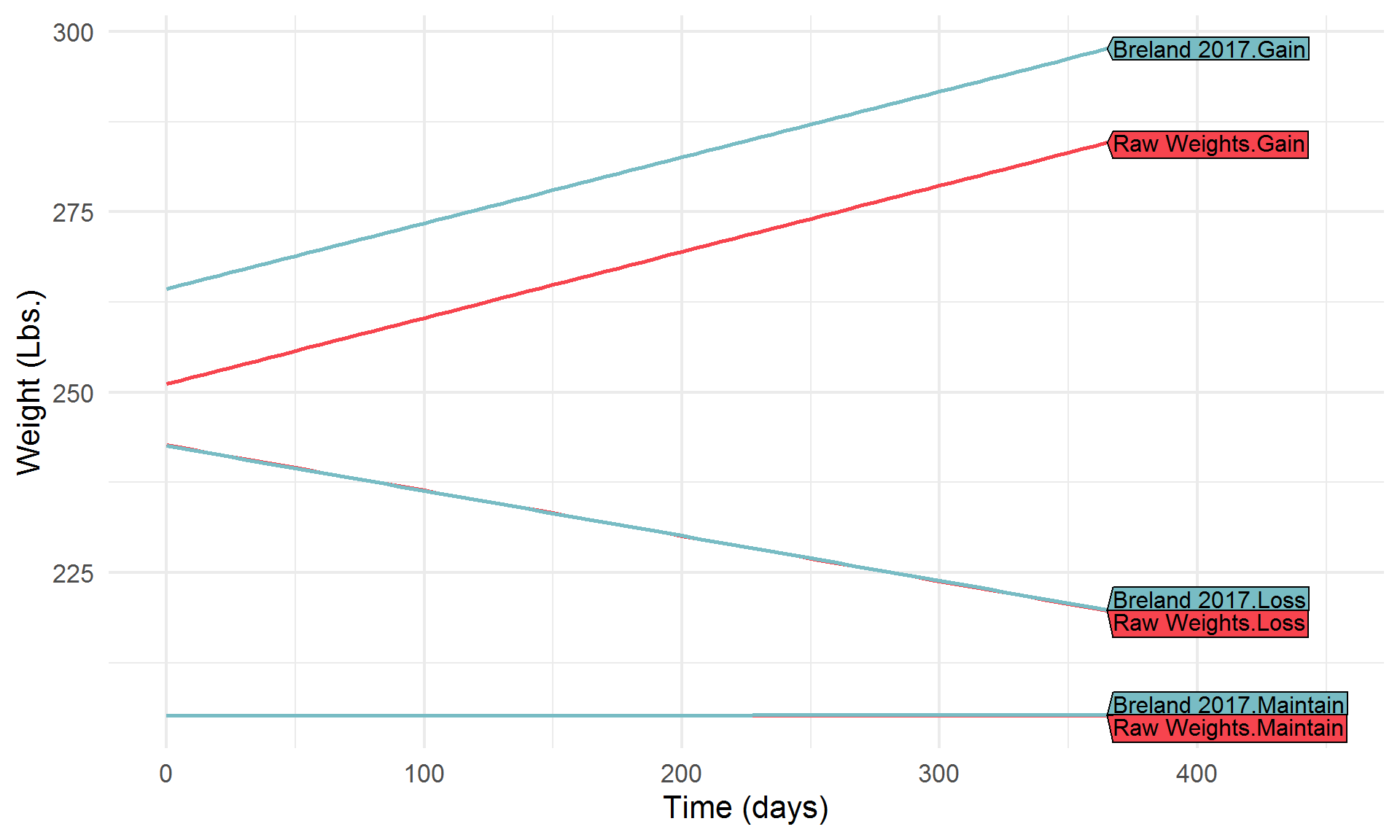
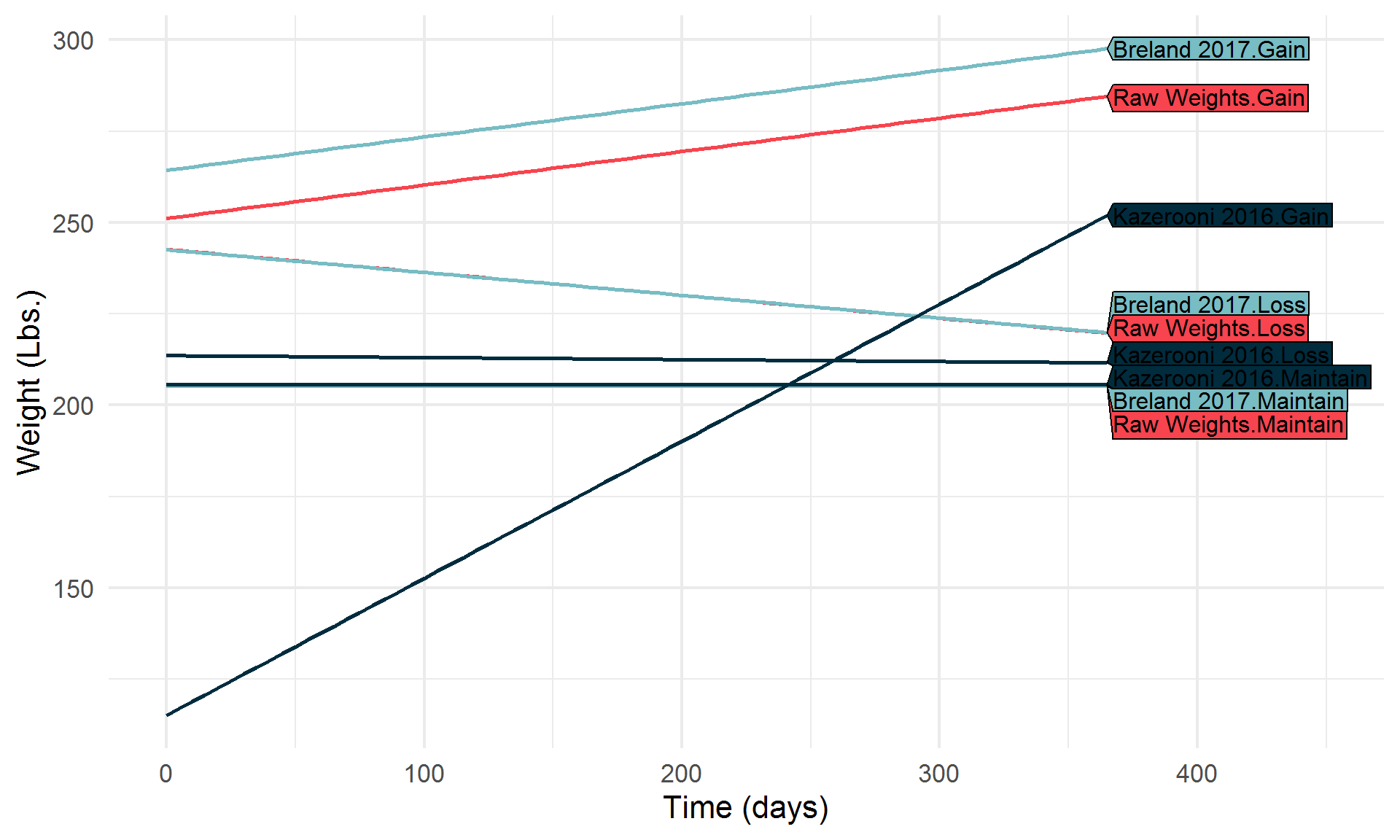
Highlight a few algorithms of interest,
Ypred_lcmm %>%
ggplot(
aes(
x = time,
y = Ypred,
group = interaction(Algorithm, concept_class),
color = Algorithm
)
) %>%
add(geom_line(size = 1)) %>%
add(theme_minimal(16)) %>%
add(theme(axis.title = element_text(), legend.position = "none")) %>%
add(xlim(0, 450)) %>%
add(gghighlight::gghighlight(
Algorithm %in% algos,
label_key = Algorithm
)) %>%
add(scale_color_manual(values = COLS[c(1, 3, 4)])) %>%
add(labs(
x = "Time (days)",
y = "Weight (Lbs.)"
))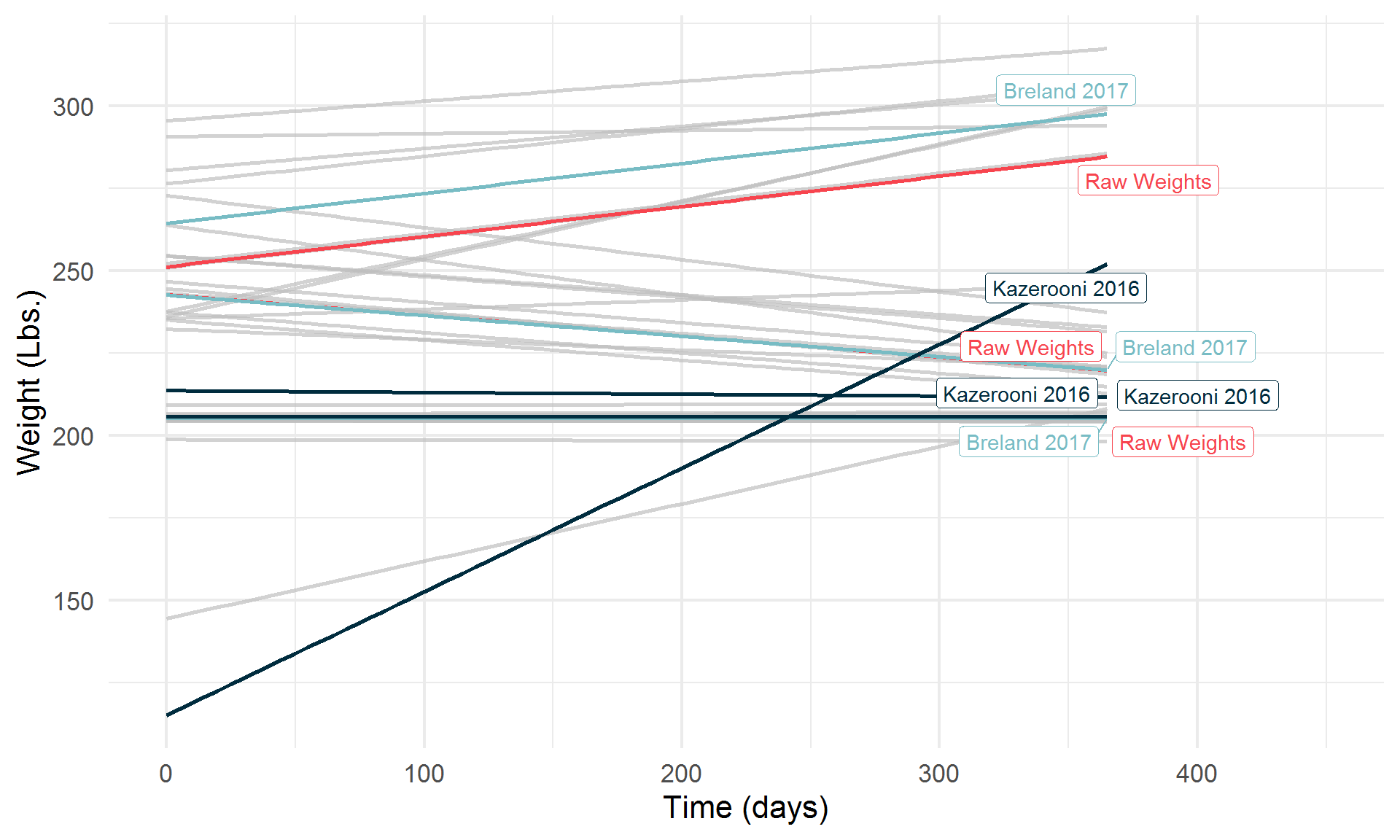
By Class
Looking only at Class 1: Loss
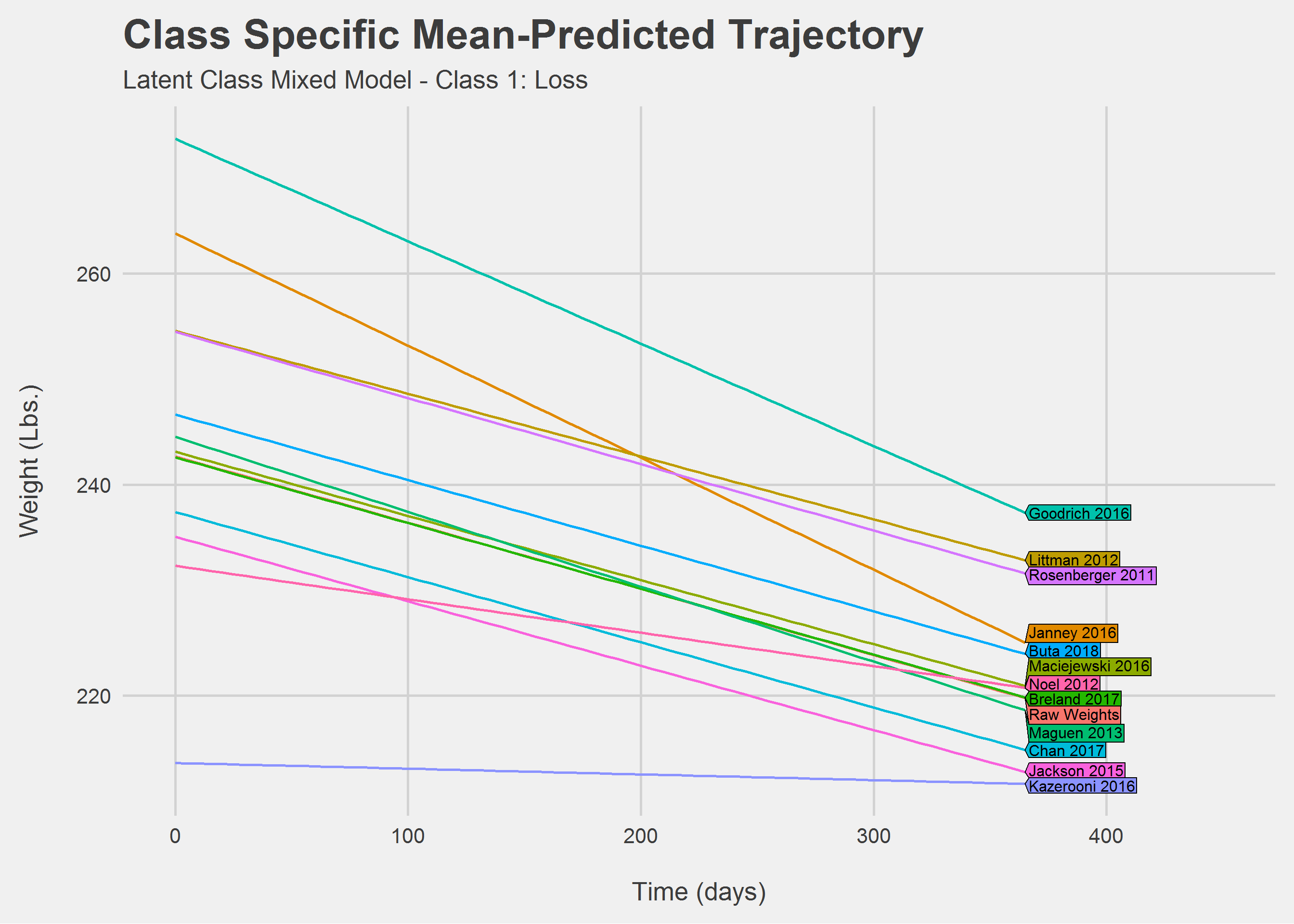
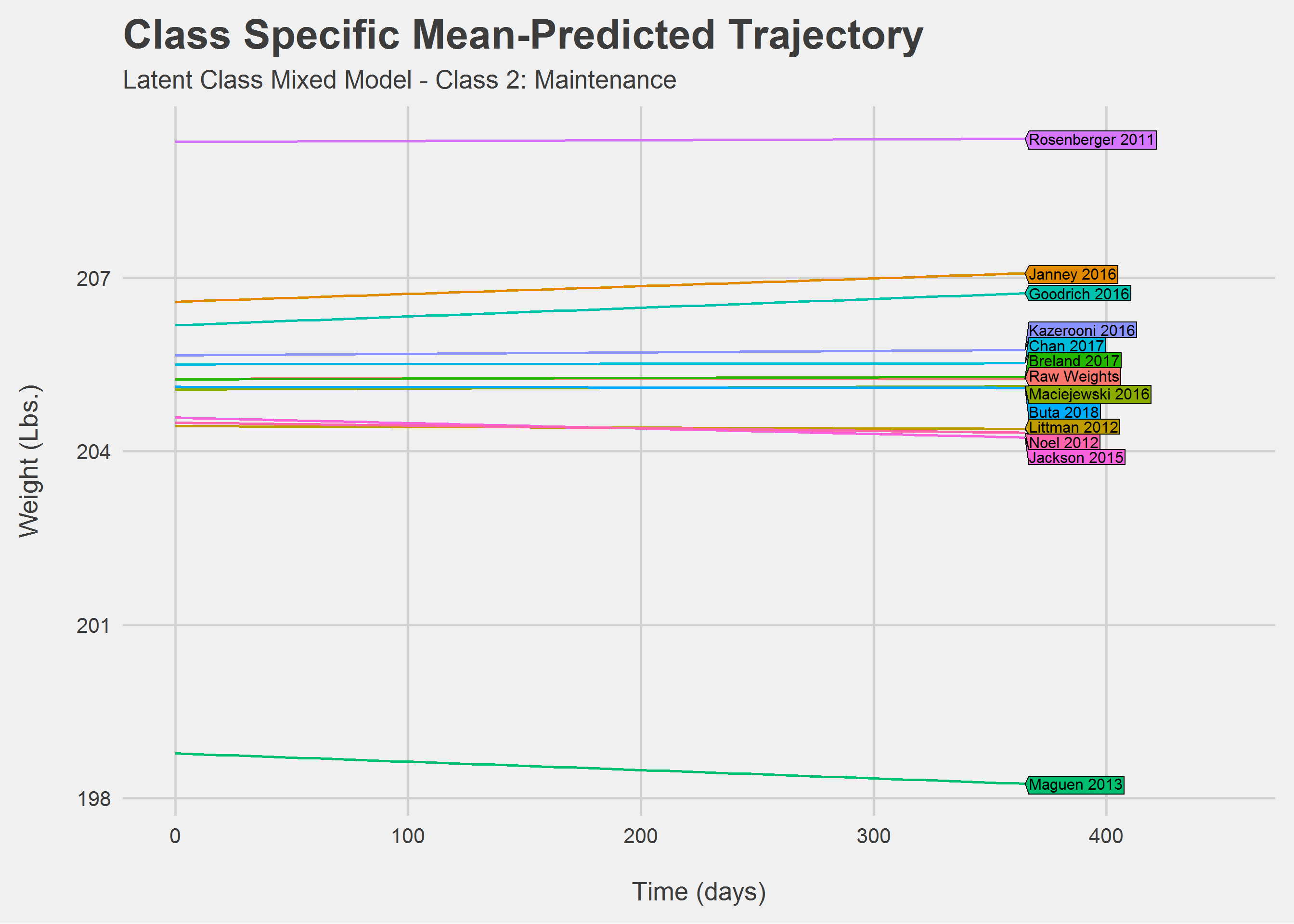
Looking only at Class 2: Maintainence
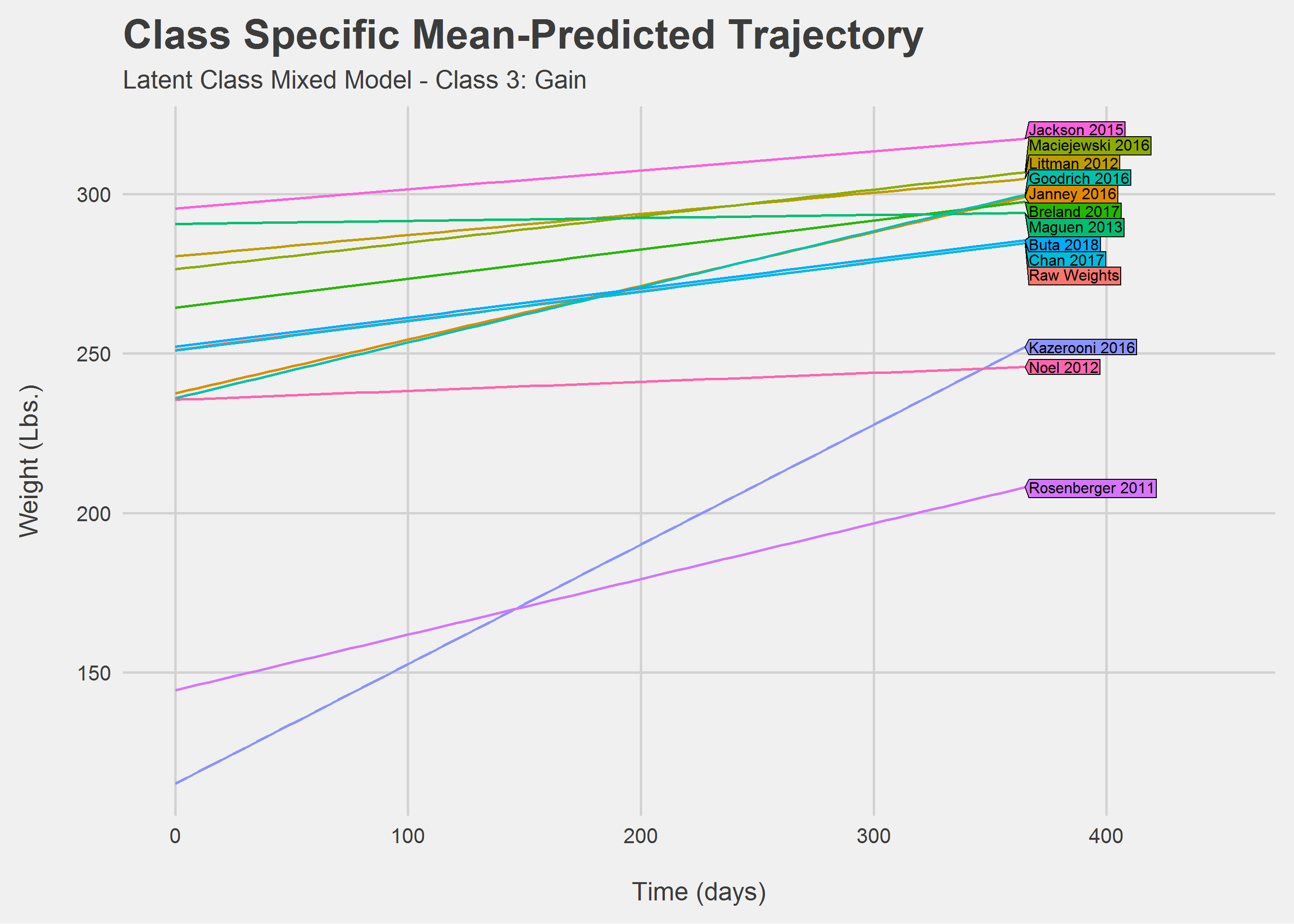
Looking only at Class 3: Gain
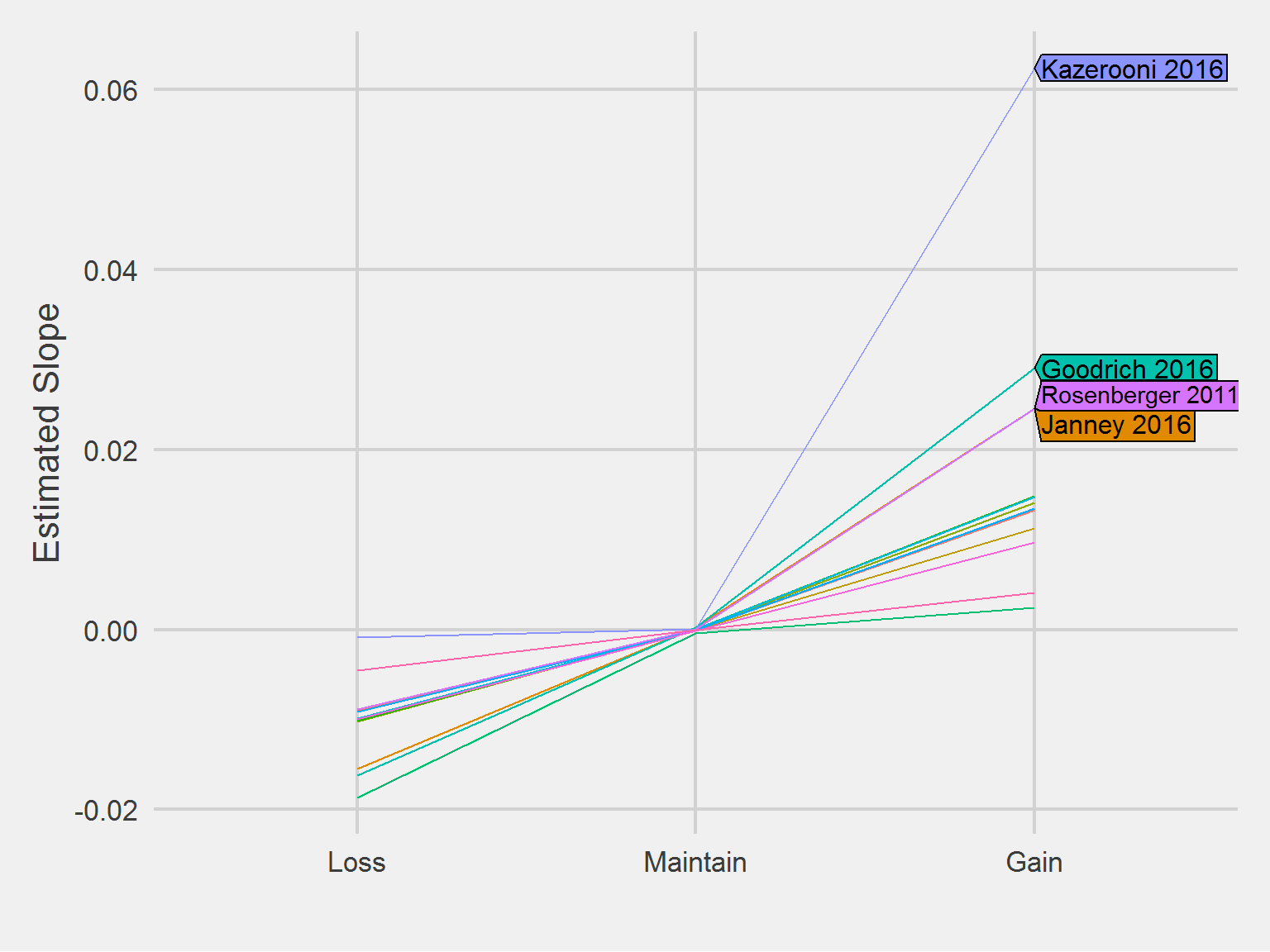
Comparing slopes, numerically
slopes <- sapply(
lca.mods,
function(x) {
time_coefs <- coef(x)
which_coef <- grepl("time", names(time_coefs))
time_coefs <- time_coefs[which_coef]
time_coefs <- round(time_coefs, 5)
time_coefs
}
) %>%
reshape2::melt() %>%
mutate(
orig_class = case_when(
Var1 == "time class1" ~ "Class 1",
Var1 == "time class2" ~ "Class 2",
Var1 == "time class3" ~ "Class 3"
),
orig_class = factor(orig_class)
) %>%
select(-Var1) %>%
rename(Algorithm = Var2, Slope = value) %>%
full_join(traj_edies, by = c("Algorithm", "orig_class"))
slopes %>%
select(-orig_class) %>%
select(Algorithm, concept_class, Slope) %>%
tableStyle(digits = 5)| Algorithm | concept_class | Slope |
|---|---|---|
| Raw Weights | Gain | 0.01322 |
| Raw Weights | Loss | -0.00906 |
| Raw Weights | Maintain | 0.00000 |
| Janney 2016 | Maintain | 0.00020 |
| Janney 2016 | Gain | 0.02454 |
| Janney 2016 | Loss | -0.01548 |
| Littman 2012 | Gain | 0.01123 |
| Littman 2012 | Loss | -0.01005 |
| Littman 2012 | Maintain | -0.00002 |
| Maciejewski 2016 | Gain | 0.01405 |
| Maciejewski 2016 | Loss | -0.01026 |
| Maciejewski 2016 | Maintain | 0.00003 |
| Breland 2017 | Loss | -0.01013 |
| Breland 2017 | Gain | 0.01482 |
| Breland 2017 | Maintain | 0.00002 |
| Maguen 2013 | Gain | 0.00248 |
| Maguen 2013 | Maintain | -0.00038 |
| Maguen 2013 | Loss | -0.01869 |
| Goodrich 2016 | Gain | 0.02909 |
| Goodrich 2016 | Maintain | 0.00025 |
| Goodrich 2016 | Loss | -0.01617 |
| Chan 2017 | Gain | 0.01470 |
| Chan 2017 | Loss | -0.00983 |
| Chan 2017 | Maintain | 0.00001 |
| Buta 2018 | Gain | 0.01346 |
| Buta 2018 | Loss | -0.00913 |
| Buta 2018 | Maintain | -0.00001 |
| Kazerooni 2016 | Maintain | 0.00004 |
| Kazerooni 2016 | Loss | -0.00090 |
| Kazerooni 2016 | Gain | 0.06236 |
| Rosenberger 2011 | Gain | 0.02456 |
| Rosenberger 2011 | Maintain | 0.00002 |
| Rosenberger 2011 | Loss | -0.00884 |
| Jackson 2015 | Maintain | -0.00015 |
| Jackson 2015 | Gain | 0.00972 |
| Jackson 2015 | Loss | -0.00991 |
| Noel 2012 | Maintain | -0.00007 |
| Noel 2012 | Gain | 0.00406 |
| Noel 2012 | Loss | -0.00457 |
algos <- c("Janney 2016", "Goodrich 2016", "Rosenberger 2011", "Kazerooni 2016")
slopes %>%
ggplot(aes(
x = concept_class,
y = Slope,
group = Algorithm,
color = Algorithm
)) %>%
add(geom_line()) %>%
add(directlabels::geom_dl(
data = slopes %>%
filter(Algorithm %in% algos),
aes(label = Algorithm),
method = list("last.polygons")
)) %>%
add(theme_fivethirtyeight(16)) %>%
add(theme(legend.position = "none", axis.title = element_text())) %>%
add(labs(
x = "",
y = "Estimated Slope"
))
Membership Posterior Probabilities
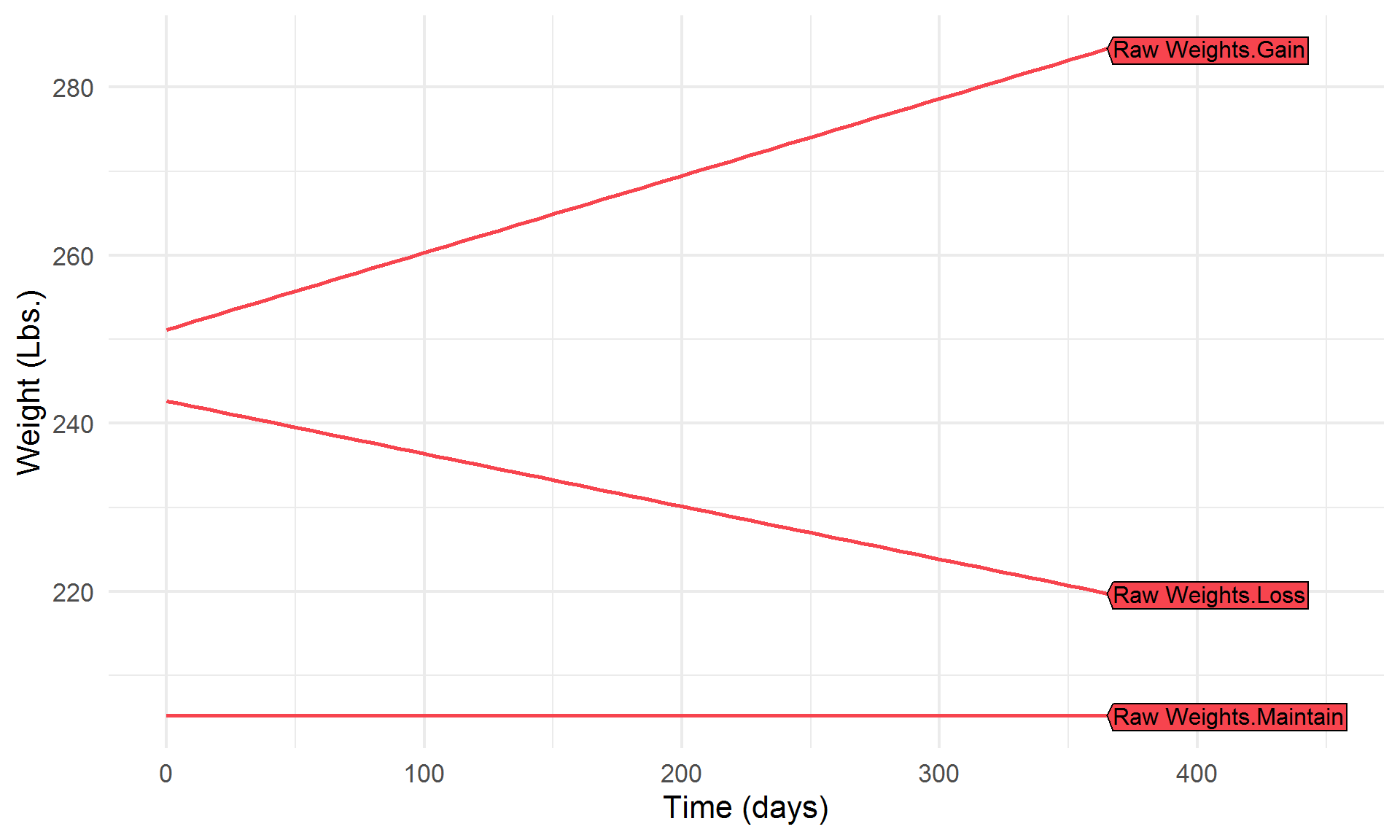
For each algorithm weight was modeled with a random slope and intercept. To illustrate the concept, the output from the Raw Weight (non-algorithm) is displayed:
General latent class mixed model
fitted by maximum likelihood method
lcmm(fixed = Weight ~ time, mixture = ~time, random = ~time,
subject = "PatientICN", ng = 3, idiag = TRUE, data = x)
Statistical Model:
Dataset: x
Number of subjects: 993
Number of observations: 7329
Number of latent classes: 3
Number of parameters: 11
Link function: linear
Iteration process:
Convergence criteria satisfied
Number of iterations: 51
Convergence criteria: parameters= 9.6e-05
: likelihood= 2e-06
: second derivatives= 9.8e-07
Goodness-of-fit statistics:
maximum log-likelihood: -27768.14
AIC: 55558.27
BIC: 55612.18
Maximum Likelihood Estimates:
Fixed effects in the class-membership model:
(the class of reference is the last class)
coef Se Wald p-value
intercept class1 -4.52848 0.34988 -12.943 0.00000
intercept class2 -2.93765 0.22929 -12.812 0.00000
Fixed effects in the longitudinal model:
coef Se Wald p-value
intercept class1 (not estimated) 0
intercept class2 -1.22305 2.76107 -0.443 0.65779
intercept class3 -6.62143 2.47619 -2.674 0.00749
time class1 0.01322 0.00100 13.272 0.00000
time class2 -0.00906 0.00067 -13.428 0.00000
time class3 0.00000 0.00011 0.022 0.98252
Variance-covariance matrix of the random-effects:
intercept time
intercept 42.42468
time 0.00000 0
Residual standard error (not estimated) = 1
Parameters of the link function:
coef Se Wald p-value
Linear 1 (intercept) 251.15091 17.02209 14.754 0.00000
Linear 2 (std err) 6.93123 0.06530 106.151 0.00000Each class has a slightly different slope on time. The posterior classifications can be examined from each algorithm:
Posterior classification:
class1 class2 class3
N 9.00 33.00 951.00
% 0.91 3.32 95.77
Posterior classification table:
--> mean of posterior probabilities in each class
prob1 prob2 prob3
class1 0.9631 0.0000 0.0369
class2 0.0000 0.9081 0.0919
class3 0.0015 0.0205 0.9780
Posterior probabilities above a threshold (%):
class1 class2 class3
prob>0.7 100.00 90.91 98.21
prob>0.8 88.89 84.85 96.64
prob>0.9 88.89 75.76 94.43
Posterior classification:
class1 class2 class3
N 958.00 3.00 14.00
% 98.26 0.31 1.44
Posterior classification table:
--> mean of posterior probabilities in each class
prob1 prob2 prob3
class1 0.9891 0.0013 0.0096
class2 0.0015 0.9985 0.0000
class3 0.1426 0.0000 0.8574
Posterior probabilities above a threshold (%):
class1 class2 class3
prob>0.7 99.69 100 78.57
prob>0.8 98.96 100 71.43
prob>0.9 97.29 100 57.14
Posterior classification:
class1 class2 class3
N 7.00 32.0 930.00
% 0.72 3.3 95.98
Posterior classification table:
--> mean of posterior probabilities in each class
prob1 prob2 prob3
class1 0.9923 0.0000 0.0077
class2 0.0000 0.8442 0.1558
class3 0.0045 0.0177 0.9778
Posterior probabilities above a threshold (%):
class1 class2 class3
prob>0.7 100 75.00 98.17
prob>0.8 100 65.62 96.34
prob>0.9 100 59.38 93.66
Posterior classification:
class1 class2 class3
N 7.0 38.00 948.00
% 0.7 3.83 95.47
Posterior classification table:
--> mean of posterior probabilities in each class
prob1 prob2 prob3
class1 0.9847 0.0000 0.0153
class2 0.0000 0.8570 0.1430
class3 0.0017 0.0187 0.9795
Posterior probabilities above a threshold (%):
class1 class2 class3
prob>0.7 100 76.32 98.31
prob>0.8 100 68.42 97.05
prob>0.9 100 60.53 94.51
Posterior classification:
class1 class2 class3
N 35.00 8.00 950.00
% 3.52 0.81 95.67
Posterior classification table:
--> mean of posterior probabilities in each class
prob1 prob2 prob3
class1 0.8810 0.0000 0.1190
class2 0.0000 0.9684 0.0316
class3 0.0191 0.0014 0.9795
Posterior probabilities above a threshold (%):
class1 class2 class3
prob>0.7 82.86 100.0 98.21
prob>0.8 77.14 100.0 96.95
prob>0.9 65.71 87.5 94.42
Posterior classification:
class1 class2 class3
N 63.00 902.00 14.00
% 6.44 92.13 1.43
Posterior classification table:
--> mean of posterior probabilities in each class
prob1 prob2 prob3
class1 0.7793 0.2164 0.0043
class2 0.0364 0.9591 0.0045
class3 0.0165 0.1173 0.8662
Posterior probabilities above a threshold (%):
class1 class2 class3
prob>0.7 66.67 96.34 78.57
prob>0.8 46.03 93.35 57.14
prob>0.9 34.92 87.80 57.14
Posterior classification:
class1 class2 class3
N 3.00 958.00 14.00
% 0.31 98.26 1.44
Posterior classification table:
--> mean of posterior probabilities in each class
prob1 prob2 prob3
class1 0.9983 0.0017 0.0000
class2 0.0012 0.9874 0.0114
class3 0.0000 0.1175 0.8825
Posterior probabilities above a threshold (%):
class1 class2 class3
prob>0.7 100 99.37 85.71
prob>0.8 100 98.23 71.43
prob>0.9 100 96.97 71.43
Posterior classification:
class1 class2 class3
N 9.00 38.00 922.00
% 0.93 3.92 95.15
Posterior classification table:
--> mean of posterior probabilities in each class
prob1 prob2 prob3
class1 0.9635 0.0000 0.0365
class2 0.0000 0.8557 0.1443
class3 0.0013 0.0175 0.9812
Posterior probabilities above a threshold (%):
class1 class2 class3
prob>0.7 100.00 76.32 98.48
prob>0.8 88.89 68.42 97.29
prob>0.9 88.89 57.89 95.34
Posterior classification:
class1 class2 class3
N 9.00 33.00 868.00
% 0.99 3.63 95.38
Posterior classification table:
--> mean of posterior probabilities in each class
prob1 prob2 prob3
class1 0.9668 0.0000 0.0332
class2 0.0000 0.8972 0.1028
class3 0.0014 0.0204 0.9782
Posterior probabilities above a threshold (%):
class1 class2 class3
prob>0.7 100.00 84.85 98.16
prob>0.8 88.89 81.82 96.31
prob>0.9 88.89 72.73 94.47
Posterior classification:
class1 class2 class3
N 0 262.00 1.00
% 0 99.62 0.38
Posterior classification table:
--> mean of posterior probabilities in each class
prob1 prob2 prob3
class1 NaN NaN NaN
class2 0.0037 0.9963 0
class3 0.0000 0.0000 1
Posterior probabilities above a threshold (%):
class1 class2 class3
prob>0.7 NaN 100 100
prob>0.8 NaN 100 100
prob>0.9 NaN 100 100
Posterior classification:
class1 class2 class3
N 2.0 634.0 21.0
% 0.3 96.5 3.2
Posterior classification table:
--> mean of posterior probabilities in each class
prob1 prob2 prob3
class1 1 0.0000 0.0000
class2 0 0.9870 0.0130
class3 0 0.2019 0.7981
Posterior probabilities above a threshold (%):
class1 class2 class3
prob>0.7 100 98.42 66.67
prob>0.8 100 97.79 57.14
prob>0.9 100 96.21 42.86
Posterior classification:
class1 class2 class3
N 940.00 11.00 26.00
% 96.21 1.13 2.66
Posterior classification table:
--> mean of posterior probabilities in each class
prob1 prob2 prob3
class1 0.9726 0.0081 0.0193
class2 0.0487 0.9510 0.0002
class3 0.1397 0.0000 0.8603
Posterior probabilities above a threshold (%):
class1 class2 class3
prob>0.7 98.30 90.91 88.46
prob>0.8 96.38 90.91 69.23
prob>0.9 93.51 90.91 53.85
Posterior classification:
class1 class2 class3
N 919.0 35.0 46.0
% 91.9 3.5 4.6
Posterior classification table:
--> mean of posterior probabilities in each class
prob1 prob2 prob3
class1 0.9537 0.0247 0.0216
class2 0.1849 0.8150 0.0002
class3 0.1795 0.0000 0.8205
Posterior probabilities above a threshold (%):
class1 class2 class3
prob>0.7 96.41 65.71 71.74
prob>0.8 92.82 51.43 65.22
prob>0.9 86.18 40.00 43.48
Posterior Classifications, by Algorithm
post_class <- vector("list", length(pprobs))
for(i in seq_along(pprobs)) {
post_class[[i]] <- reshape2::melt(pprobs[[i]][[1]])
post_class[[i]]$Algorithm <- names(pprobs)[[i]]
}
post_class %>%
bind_rows() %>%
mutate(
orig_class = case_when(
Var2 == "class1" ~ "Class 1",
Var2 == "class2" ~ "Class 2",
Var2 == "class3" ~ "Class 3"
)
) %>%
left_join(traj_edies, by = c("Algorithm", "orig_class")) %>%
select(Algorithm, concept_class, value, Var1) %>%
reshape2::dcast(Algorithm + concept_class ~ Var1, value.var = "value") %>%
scrollTable()| Algorithm | concept_class | N | % |
|---|---|---|---|
| Breland 2017 | Loss | 35 | 3.52 |
| Breland 2017 | Maintain | 950 | 95.67 |
| Breland 2017 | Gain | 8 | 0.81 |
| Buta 2018 | Loss | 33 | 3.63 |
| Buta 2018 | Maintain | 868 | 95.38 |
| Buta 2018 | Gain | 9 | 0.99 |
| Chan 2017 | Loss | 38 | 3.92 |
| Chan 2017 | Maintain | 922 | 95.15 |
| Chan 2017 | Gain | 9 | 0.93 |
| Goodrich 2016 | Loss | 14 | 1.44 |
| Goodrich 2016 | Maintain | 958 | 98.26 |
| Goodrich 2016 | Gain | 3 | 0.31 |
| Jackson 2015 | Loss | 26 | 2.66 |
| Jackson 2015 | Maintain | 940 | 96.21 |
| Jackson 2015 | Gain | 11 | 1.13 |
| Janney 2016 | Loss | 14 | 1.44 |
| Janney 2016 | Maintain | 958 | 98.26 |
| Janney 2016 | Gain | 3 | 0.31 |
| Kazerooni 2016 | Loss | 262 | 99.62 |
| Kazerooni 2016 | Maintain | 0 | 0.00 |
| Kazerooni 2016 | Gain | 1 | 0.38 |
| Littman 2012 | Loss | 32 | 3.30 |
| Littman 2012 | Maintain | 930 | 95.98 |
| Littman 2012 | Gain | 7 | 0.72 |
| Maciejewski 2016 | Loss | 38 | 3.83 |
| Maciejewski 2016 | Maintain | 948 | 95.47 |
| Maciejewski 2016 | Gain | 7 | 0.70 |
| Maguen 2013 | Loss | 14 | 1.43 |
| Maguen 2013 | Maintain | 902 | 92.13 |
| Maguen 2013 | Gain | 63 | 6.44 |
| Noel 2012 | Loss | 46 | 4.60 |
| Noel 2012 | Maintain | 919 | 91.90 |
| Noel 2012 | Gain | 35 | 3.50 |
| Raw Weights | Loss | 33 | 3.32 |
| Raw Weights | Maintain | 951 | 95.77 |
| Raw Weights | Gain | 9 | 0.91 |
| Rosenberger 2011 | Loss | 21 | 3.20 |
| Rosenberger 2011 | Maintain | 634 | 96.50 |
| Rosenberger 2011 | Gain | 2 | 0.30 |
meanPP <- vector("list", length(pprobs))
for(i in seq_along(pprobs)) {
meanPP[[i]] <- reshape2::melt(pprobs[[i]][[2]])
meanPP[[i]]$Algorithm <- names(pprobs)[[i]]
}
meanPP %>%
bind_rows() %>%
mutate(
orig_class = case_when(
Var1 == "class1" ~ "Class 1",
Var1 == "class2" ~ "Class 2",
Var1 == "class3" ~ "Class 3"
)
) %>%
left_join(traj_edies, by = c("Algorithm", "orig_class")) %>%
select(Algorithm, concept_class, value, Var2) %>%
rename(cProb = Var2) %>%
reshape2::dcast(Algorithm + cProb ~ concept_class, value.var = "value") %>%
scrollTable()| Algorithm | cProb | Loss | Maintain | Gain |
|---|---|---|---|---|
| Breland 2017 | prob1 | 0.88 | 0.02 | 0.00 |
| Breland 2017 | prob2 | 0.00 | 0.00 | 0.97 |
| Breland 2017 | prob3 | 0.12 | 0.98 | 0.03 |
| Buta 2018 | prob1 | 0.00 | 0.00 | 0.97 |
| Buta 2018 | prob2 | 0.90 | 0.02 | 0.00 |
| Buta 2018 | prob3 | 0.10 | 0.98 | 0.03 |
| Chan 2017 | prob1 | 0.00 | 0.00 | 0.96 |
| Chan 2017 | prob2 | 0.86 | 0.02 | 0.00 |
| Chan 2017 | prob3 | 0.14 | 0.98 | 0.04 |
| Goodrich 2016 | prob1 | 0.00 | 0.00 | 1.00 |
| Goodrich 2016 | prob2 | 0.12 | 0.99 | 0.00 |
| Goodrich 2016 | prob3 | 0.88 | 0.01 | 0.00 |
| Jackson 2015 | prob1 | 0.14 | 0.97 | 0.05 |
| Jackson 2015 | prob2 | 0.00 | 0.01 | 0.95 |
| Jackson 2015 | prob3 | 0.86 | 0.02 | 0.00 |
| Janney 2016 | prob1 | 0.14 | 0.99 | 0.00 |
| Janney 2016 | prob2 | 0.00 | 0.00 | 1.00 |
| Janney 2016 | prob3 | 0.86 | 0.01 | 0.00 |
| Kazerooni 2016 | prob1 | 0.00 | NaN | 0.00 |
| Kazerooni 2016 | prob2 | 1.00 | NaN | 0.00 |
| Kazerooni 2016 | prob3 | 0.00 | NaN | 1.00 |
| Littman 2012 | prob1 | 0.00 | 0.00 | 0.99 |
| Littman 2012 | prob2 | 0.84 | 0.02 | 0.00 |
| Littman 2012 | prob3 | 0.16 | 0.98 | 0.01 |
| Maciejewski 2016 | prob1 | 0.00 | 0.00 | 0.98 |
| Maciejewski 2016 | prob2 | 0.86 | 0.02 | 0.00 |
| Maciejewski 2016 | prob3 | 0.14 | 0.98 | 0.02 |
| Maguen 2013 | prob1 | 0.02 | 0.04 | 0.78 |
| Maguen 2013 | prob2 | 0.12 | 0.96 | 0.22 |
| Maguen 2013 | prob3 | 0.87 | 0.00 | 0.00 |
| Noel 2012 | prob1 | 0.18 | 0.95 | 0.18 |
| Noel 2012 | prob2 | 0.00 | 0.02 | 0.82 |
| Noel 2012 | prob3 | 0.82 | 0.02 | 0.00 |
| Raw Weights | prob1 | 0.00 | 0.00 | 0.96 |
| Raw Weights | prob2 | 0.91 | 0.02 | 0.00 |
| Raw Weights | prob3 | 0.09 | 0.98 | 0.04 |
| Rosenberger 2011 | prob1 | 0.00 | 0.00 | 1.00 |
| Rosenberger 2011 | prob2 | 0.20 | 0.99 | 0.00 |
| Rosenberger 2011 | prob3 | 0.80 | 0.01 | 0.00 |