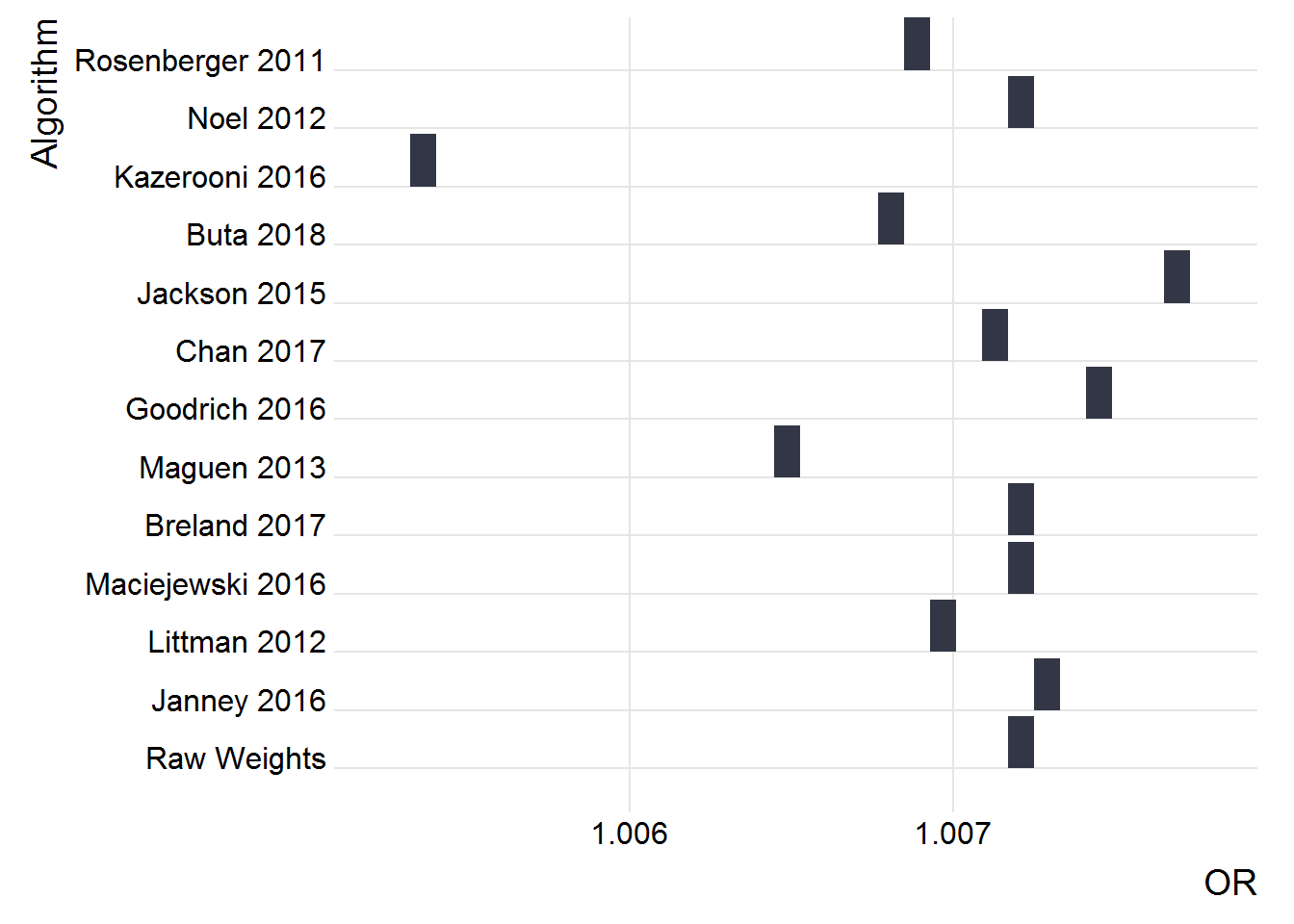
Weight as a Predictor
Weight is often chosen as a predictor in models developed in the healthcare sector. The idea here will be to “predict” “new-onset” diabetes as a function of weight. I am placing new-onset in quotations since it’s not so rigorously defined here, it is just whether or not there were two or more diabetes diagnosis codes after the patient’s index date, but not before said date. The emphasis here is not on building a practical model, just on components of estimation.
for (i in 1:2) {
samp <- weight.ls[[i]] %>% distinct(PatientICN) %>% sample_n(5000)
weight.ls[[i]] <- weight.ls[[i]] %>% filter(PatientICN %in% samp[[1]])
}
weight.ls <- lapply(
weight.ls,
function(x) {
x <- x %>%
mutate(
NewDiabetes = ifelse(DiabetesTiming == "Diabetes After", 1, 0),
NewDiabetes = factor(NewDiabetes, 0:1, c("No Diabetes", "Diabetes"))
)
}
)
getDiabData.f <- function(df, Algorithm = "Raw Weights") {
timePoints.f(df) %>%
filter(measureTime == "t_0") %>%
distinct(PatientICN, Weight, NewDiabetes) %>%
mutate(Algorithm = Algorithm) %>%
na.omit()
}
#----------------------------------- Raw Data --------------------------------#
raw.ls <- lapply(weight.ls, getDiabData.f)
#--------------------------------- Janney 2016 -------------------------------#
janney2016.ls <- lapply(
weight.ls,
FUN = function(x) {
Janney2016.f(
DF = x,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime",
startPoint = "VisitDateTime"
) %>%
filter(!is.na(Weight_OR)) %>%
select(
PatientICN,
WeightDateTime,
VisitDateTime,
Weight_OR,
NewDiabetes
) %>%
rename(Weight = Weight_OR)
}
)
janney2016.ls <- lapply(
janney2016.ls,
getDiabData.f,
Algorithm = algos.fac[2]
)
#-------------------------------- Littman 2012 -------------------------------#
littman2012.ls <- lapply(
weight.ls,
FUN = function(x) {
Littman2012.f(
DF = x,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime"
) %>%
select(
PatientICN,
VisitDateTime,
OutputMeasurement,
WeightDateTime,
NewDiabetes
) %>%
filter(!is.na(OutputMeasurement)) %>%
rename(Weight = OutputMeasurement)
}
)
littman2012.ls <- lapply(
littman2012.ls,
getDiabData.f,
Algorithm = algos.fac[3]
)
#------------------------------ Maciejewski 2016 -----------------------------#
maciejewski2016.2008 <- maciejewski %>%
filter(SampleYear == "2008" & IO == "Output") %>%
mutate(PatientICN = as.character(PatientICN)) %>%
left_join(
weight.ls[["PCP2008"]] %>%
distinct(PatientICN, NewDiabetes),
by = "PatientICN"
) %>%
mutate(
WeightDateTime = lubridate::as_datetime(WeightDate, tz = "UTC")
) %>%
select(-IO, -WeightDate, -SampleYear)
maciejewski2016.2016 <- maciejewski %>%
filter(SampleYear == "2016" & IO == "Output") %>%
mutate(PatientICN = as.character(PatientICN)) %>%
left_join(
weight.ls[["PCP2016"]] %>%
distinct(PatientICN, NewDiabetes),
by = "PatientICN"
) %>%
mutate(
WeightDateTime = lubridate::as_datetime(WeightDate, tz = "UTC")
) %>%
select(-IO, -WeightDate, -SampleYear)
rm(maciejewski) # takes up a LOT of space
maciejewski2016.ls <- list(
PCP2008 = maciejewski2016.2008,
PCP2016 = maciejewski2016.2016
)
rm(maciejewski2016.2008, maciejewski2016.2016)
maciejewski2016.ls <- lapply(
maciejewski2016.ls,
getDiabData.f,
Algorithm = algos.fac[4]
)
#-------------------------------- Breland 2017 --------------------------------#
breland2017.ls <- lapply(
weight.ls,
FUN = function(x) {
Breland2017.f(
DF = x,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime"
) %>%
filter(!is.na(measures_aug_)) %>%
select(
PatientICN,
VisitDateTime,
WeightDateTime,
measures_aug_,
NewDiabetes
) %>%
rename(Weight = measures_aug_)
}
)
breland2017.ls <- lapply(
breland2017.ls,
getDiabData.f,
Algorithm = algos.fac[5]
)
#-------------------------------- Maguen 2013 --------------------------------#
maguen2013.ls <- lapply(
weight.ls,
FUN = function(x) {
Maguen2013.f(
DF = x,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime",
variables = c("AgeAtVisit", "Gender")
) %>%
filter(!is.na(Output)) %>%
select(
PatientICN,
VisitDateTime,
WeightDateTime,
Output,
NewDiabetes
) %>%
rename(Weight = Output)
}
)
maguen2013.ls <- lapply(
maguen2013.ls,
getDiabData.f,
Algorithm = algos.fac[6]
)
#------------------------------- Goodrich 2016 -------------------------------#
goodrich2016.ls <- lapply(
weight.ls,
FUN = function(x) {
Goodrich2016.f(
DF = x,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime",
startPoint = "VisitDateTime"
) %>%
filter(!is.na(output)) %>%
select(
PatientICN,
VisitDateTime,
WeightDateTime,
output,
NewDiabetes
) %>%
rename(Weight = output)
}
)
goodrich2016.ls <- lapply(
goodrich2016.ls,
getDiabData.f,
Algorithm = algos.fac[7]
)
#----------------------------- Chan & Raffa 2017 -----------------------------#
chan2017.ls <- lapply(
weight.ls,
FUN = function(x) {
Chan2017.f(
DF = x,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime"
) %>%
filter(!is.na(measures_aug_)) %>%
select(
PatientICN,
VisitDateTime,
WeightDateTime,
measures_aug_,
NewDiabetes
) %>%
rename(Weight = measures_aug_)
}
)
chan2017.ls <- lapply(
chan2017.ls,
getDiabData.f,
Algorithm = algos.fac[8]
)
#------------------------------- Jackson 2015 --------------------------------#
jackson2015.ls <- lapply(
weight.ls,
FUN = function(x) {
Jackson2015.f(
DF = x,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime",
startPoint = "VisitDateTime"
) %>%
filter(measureTime == "t_0") %>%
select(PatientICN, output) %>%
rename(Weight = output) %>%
left_join(
x %>%
distinct(PatientICN, NewDiabetes),
by = "PatientICN"
) %>%
mutate(Algorithm = algos.fac[9])
}
)
#--------------------------------- Buta 2018 ---------------------------------#
buta2018.ls <- lapply(
weight.ls,
FUN = function(x) {
Buta2018.f(
DF = x,
id = "PatientICN",
measures = "BMI",
tmeasures = "WeightDateTime"
) %>%
filter(!is.na(BMI))
}
)
buta2018.ls <- lapply(
buta2018.ls,
getDiabData.f,
Algorithm = algos.fac[10]
)
#--------------------------- Kazerooni & Lim 2016 ----------------------------#
kazerooni2016.ls <- lapply(
weight.ls,
FUN = function(x) {
Kazerooni2016.f(
DF = x,
id = "PatientICN",
measures = "Weight",
tmeasures = "WeightDateTime",
startPoint = "VisitDateTime"
)
}
)
kazerooni2016.ls <- lapply(
kazerooni2016.ls,
getDiabData.f,
Algorithm = algos.fac[11]
)
#--------------------------------- Noel 2012 ---------------------------------#
noel2012.ls <- lapply(
weight.ls,
FUN = function(x) {
Noel2012.f(
DF = x,
id = "PatientICN",
measure = "Weight",
tmeasures = "WeightDateTime"
) %>%
select(
PatientICN,
VisitDateTime,
WeightDateTime,
Qmedian,
NewDiabetes
) %>%
rename(Weight = Qmedian)
}
)
noel2012.ls <- lapply(
noel2012.ls,
getDiabData.f,
Algorithm = algos.fac[12]
)
#------------------------------ Rosenberger 2011 -----------------------------#
rosenberger2011.ls <- lapply(
weight.ls,
FUN = function(x) {
Rosenberger2011.f(
DF = x,
id = "PatientICN",
tmeasures = "WeightDateTime",
startPoint = "VisitDateTime",
pad = 1
) %>%
select(
PatientICN,
VisitDateTime,
WeightDateTime,
Weight,
NewDiabetes
)
}
)
rosenberger2011.ls <- lapply(
rosenberger2011.ls,
getDiabData.f,
algos.fac[13]
)
#------------------------------ Stack Together -------------------------------#
eval.ls <- vector("list", length(weight.ls))
eval.ls[[1]] <- bind_rows(
raw.ls[[1]],
janney2016.ls[[1]],
littman2012.ls[[1]],
maciejewski2016.ls[[1]],
breland2017.ls[[1]],
maguen2013.ls[[1]],
goodrich2016.ls[[1]],
chan2017.ls[[1]],
jackson2015.ls[[1]],
buta2018.ls[[1]],
kazerooni2016.ls[[1]],
noel2012.ls[[1]],
rosenberger2011.ls[[1]]
) %>%
mutate(Algorithm = factor(Algorithm, algos.fac, algos.fac))
eval.ls[[2]] <- bind_rows(
raw.ls[[2]],
janney2016.ls[[2]],
littman2012.ls[[2]],
maciejewski2016.ls[[2]],
breland2017.ls[[2]],
maguen2013.ls[[2]],
goodrich2016.ls[[2]],
chan2017.ls[[2]],
jackson2015.ls[[2]],
buta2018.ls[[2]],
kazerooni2016.ls[[2]],
noel2012.ls[[2]],
rosenberger2011.ls[[2]]
) %>%
mutate(Algorithm = factor(Algorithm, algos.fac, algos.fac))
names(eval.ls) <- c("PCP 2008", "PCP 2016")
rm(
breland2017.ls,
buta2018.ls,
chan2017.ls,
goodrich2016.ls,
jackson2015.ls,
janney2016.ls,
kazerooni2016.ls,
littman2012.ls,
maciejewski2016.ls,
maguen2013.ls,
noel2012.ls,
raw.ls,
rosenberger2011.ls
)
pred_diab_model <- function(df) {
glm(NewDiabetes ~ Weight, data = df, family = binomial(link = logit))
}
eval.ls <- lapply(
eval.ls,
FUN = function(df) {
df %>%
group_by(Algorithm) %>%
nest() %>%
mutate(
model = map(data, pred_diab_model),
estimates = map(model, broom::tidy)
) %>%
unnest(estimates) %>%
filter(term == "Weight") %>%
select(-data, -model, -term) %>%
ungroup() %>%
mutate(
Type = case_when(
Algorithm == "Raw Weights" ~ 1,
Algorithm %in% algos.fac[c(2, 7, 9, 11:13)] ~ 2,
Algorithm %in% algos.fac[c(3:6, 8, 10)] ~ 3
),
Type = factor(Type, 1:3, c("Raw", "Time-Periods", "Time-Series")),
OR = exp(estimate),
LB = exp(estimate - 1.96 * std.error),
UB = exp(estimate + 1.96 * std.error)
)
}
)
eval.df <- bind_rows(eval.ls, .id = "Cohort")col.f <- colorRampPalette(RColorBrewer::brewer.pal(8, "Dark2"))
colors <- deframe(ggthemes::ggthemes_data[["fivethirtyeight"]])
p <- eval.df %>%
ggplot(aes(x = Algorithm, y = OR)) %>%
add(geom_pointrange(
aes(ymin = LB, ymax = UB, fill = Algorithm),
pch = 21,
color = "black",
size = 1,
fatten = 5
)) %>%
add(facet_grid(Type ~ Cohort, space = "free", scales = "free_y")) %>%
add(theme(
text = element_text(size = 24),
axis.text = element_text(color = "black"),
legend.position = "none",
panel.grid = element_blank(),
strip.background = element_blank()
)) %>%
add(scale_fill_manual(values = col.f(13))) %>%
add(labs(
x = "",
y = "OR (95% CI)"
)) %>%
add(coord_flip())
p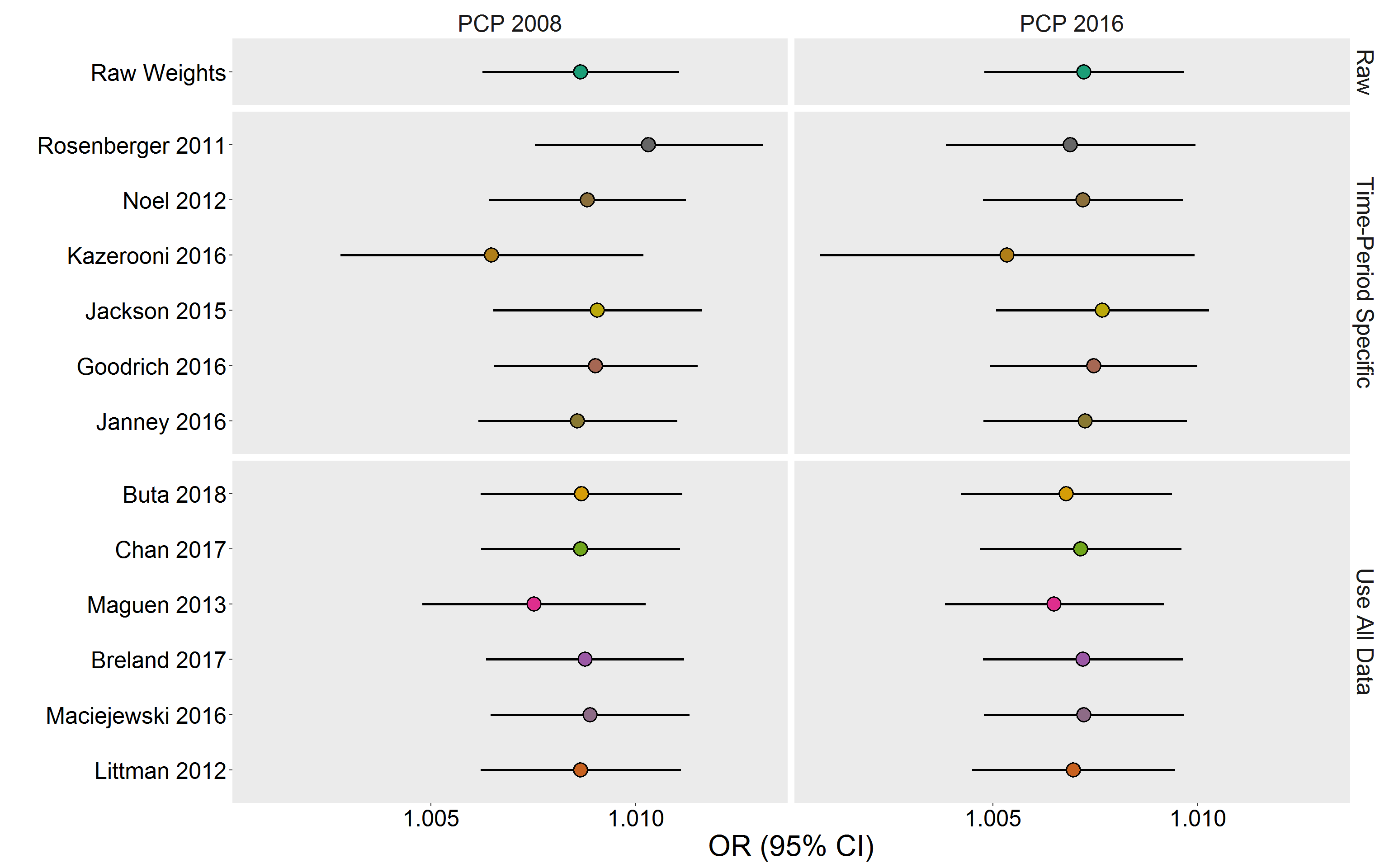
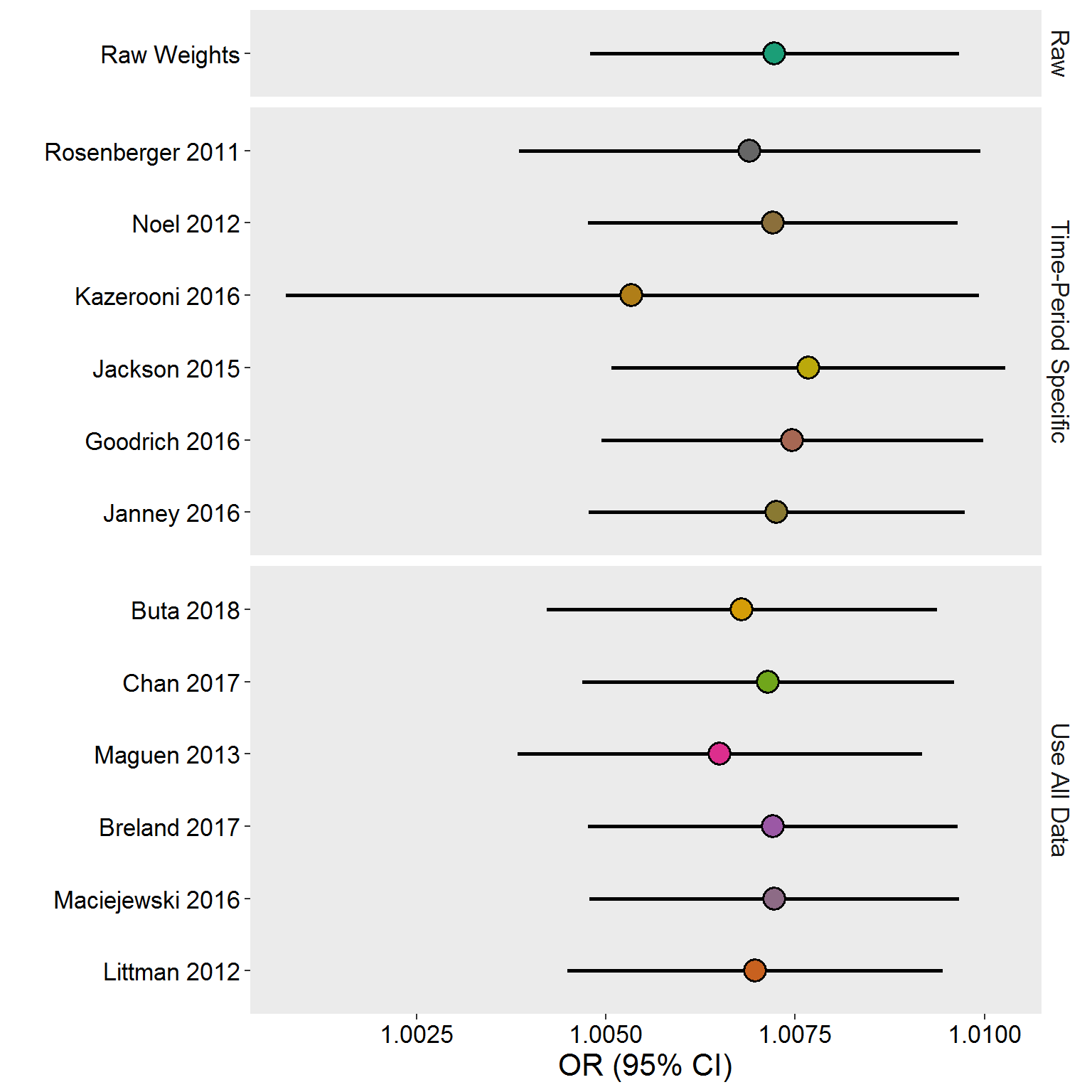
Just the 2016 Cohort
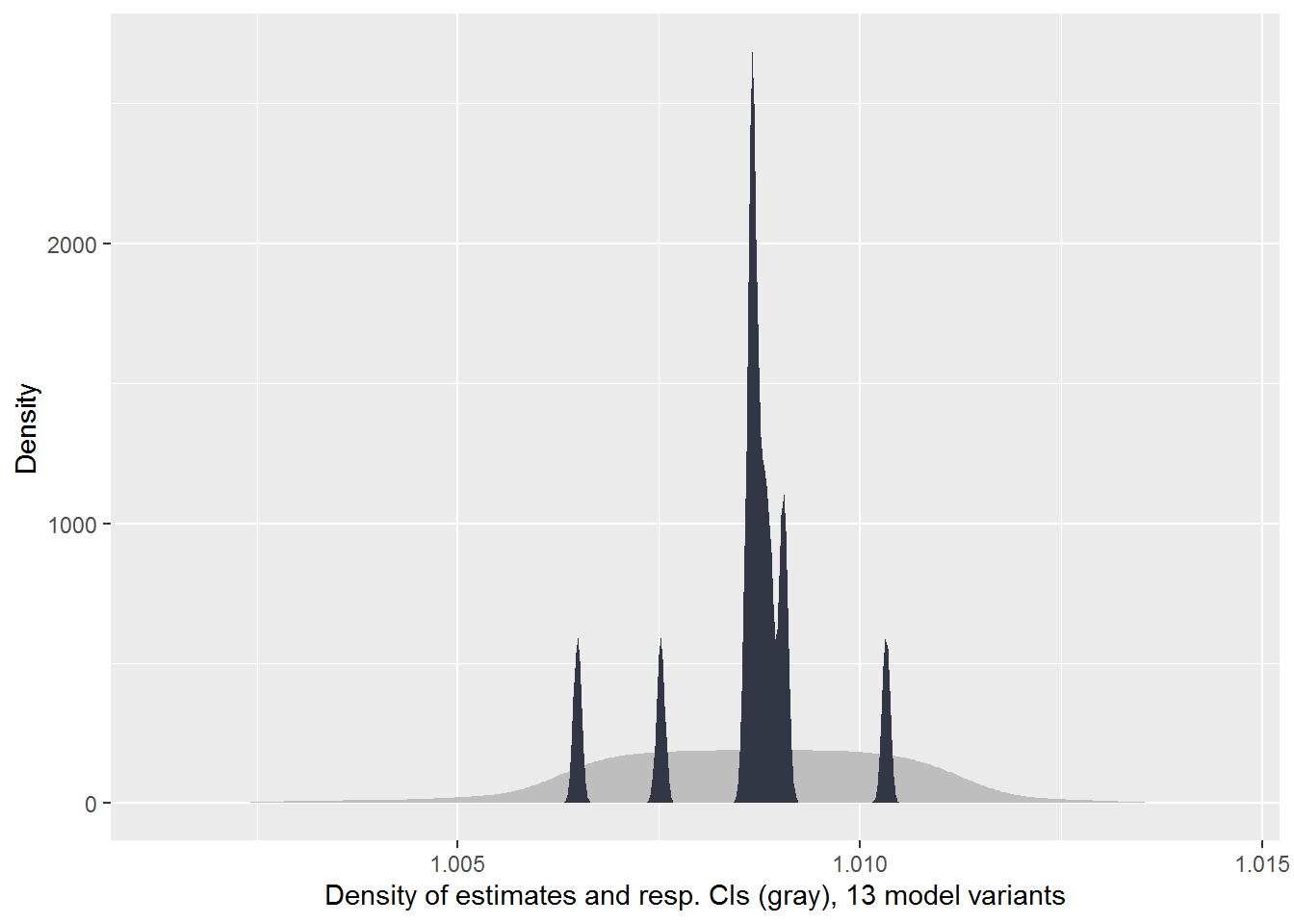
Researcher Degrees of Freedom Analysis
PCP 2008 Cohort
plot_rdf_estimate_density(
eval.df %>% filter(Cohort == "PCP 2008"),
est = "OR",
lb = "LB",
ub = "UB",
color = "#333745"
)
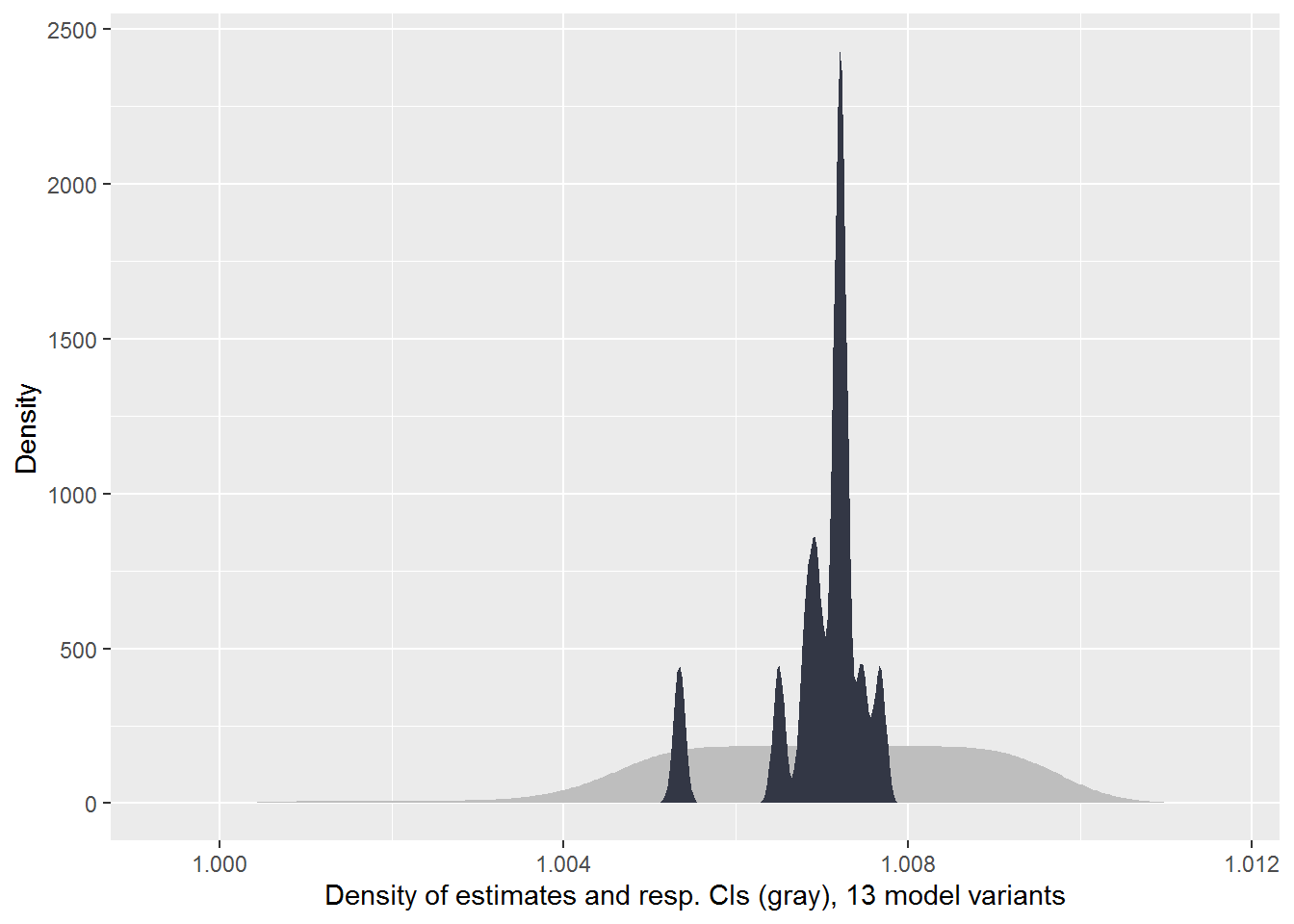
PCP 2016 Cohort
plot_rdf_estimate_density(
eval.df %>% filter(Cohort == "PCP 2016"),
est = "OR",
lb = "LB",
ub = "UB",
color = "#333745"
)
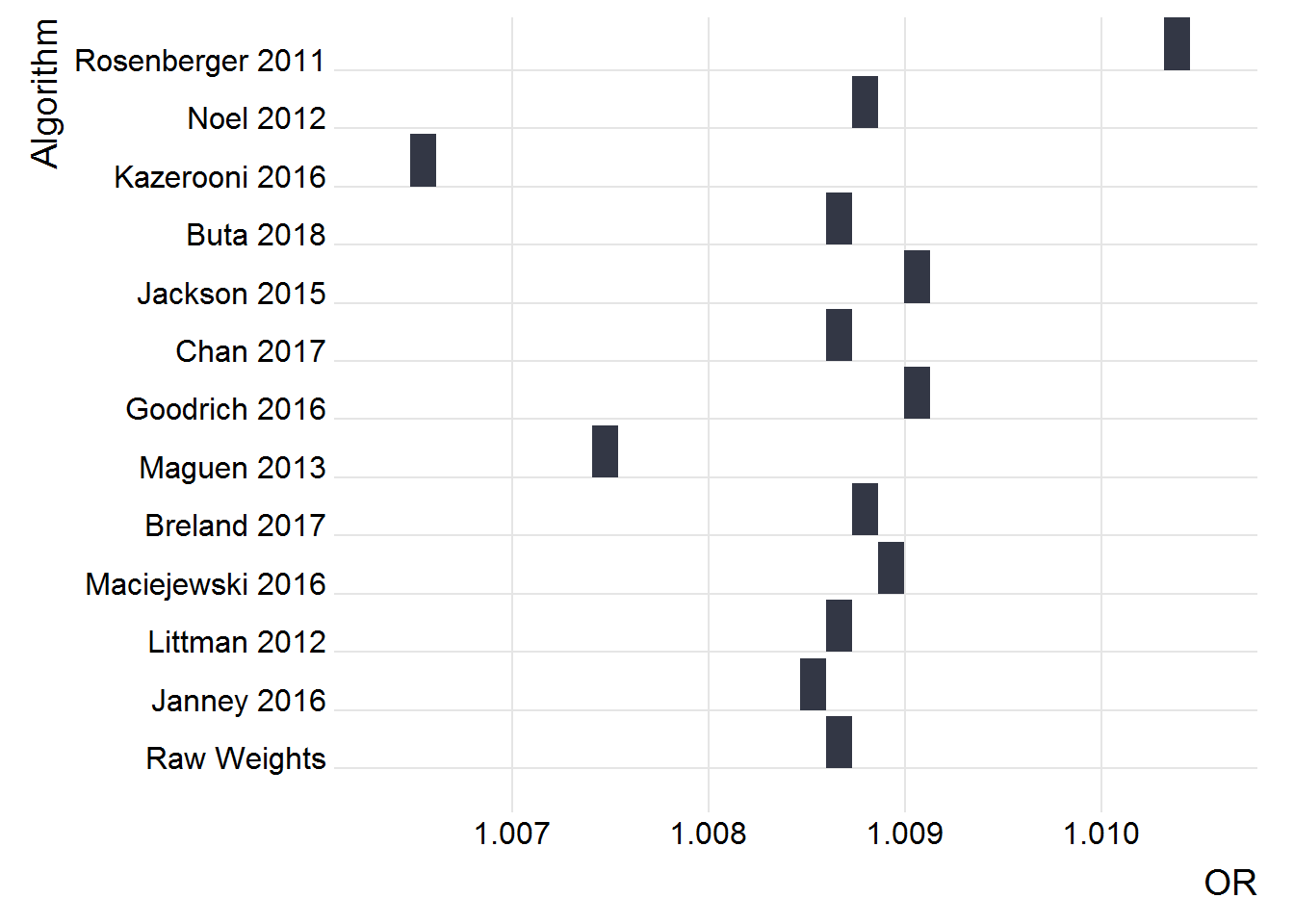
Impact of Algorithm choice
PCP 2008 Cohort
plot_rdf_ridges_by_dchoice(
eval.df %>% filter(Cohort == "PCP 2008"),
est = "OR",
dchoice = "Algorithm",
hist = TRUE,
scale = 0.9,
fill = "#333745",
color = NA
)
General PCP Cohort, 2016
plot_rdf_ridges_by_dchoice(
eval.df %>% filter(Cohort == "PCP 2016"),
est = "OR",
dchoice = "Algorithm",
hist = TRUE,
scale = 0.9,
fill = "#333745",
color = NA
)