# states
n_states = 30
i = 1:n_states
# target distribution - proportional to
pi_i = i ^ 2 * exp(-0.5 * i) # notice: not probabilities
n_steps = 10000
i_sim = rep(NA, n_steps)
i_sim[1] = 3 # initialize
for (n in 2:n_steps){
current = i_sim[n - 1]
# propose next state
proposed = sample(c(current + 1, current - 1), size = 1, prob = c(0.5, 0.5))
# compute acceptance probability
if (!(proposed %in% i)){ # to correct for proposing moves outside of boundaries
proposed = current
}
a = min(1, pi_i[proposed] / pi_i[current])
# simulate the next state
i_sim[n] = sample(c(proposed, current), size = 1, prob = c(a, 1 - a))
}Metropolis-Hastings Algorithm
Island hopping
First few steps
# display the first few steps
data.frame(step = 1:n_steps, i_sim) |> head(10) |> kbl() |> kable_styling() | step | i_sim |
|---|---|
| 1 | 3 |
| 2 | 2 |
| 3 | 2 |
| 4 | 3 |
| 5 | 2 |
| 6 | 2 |
| 7 | 3 |
| 8 | 3 |
| 9 | 4 |
| 10 | 5 |
First 100 steps
# trace plot
plot(1:100, i_sim[1:100], type = "o",
ylim = range(i), xlab = "Day", ylab = "Island")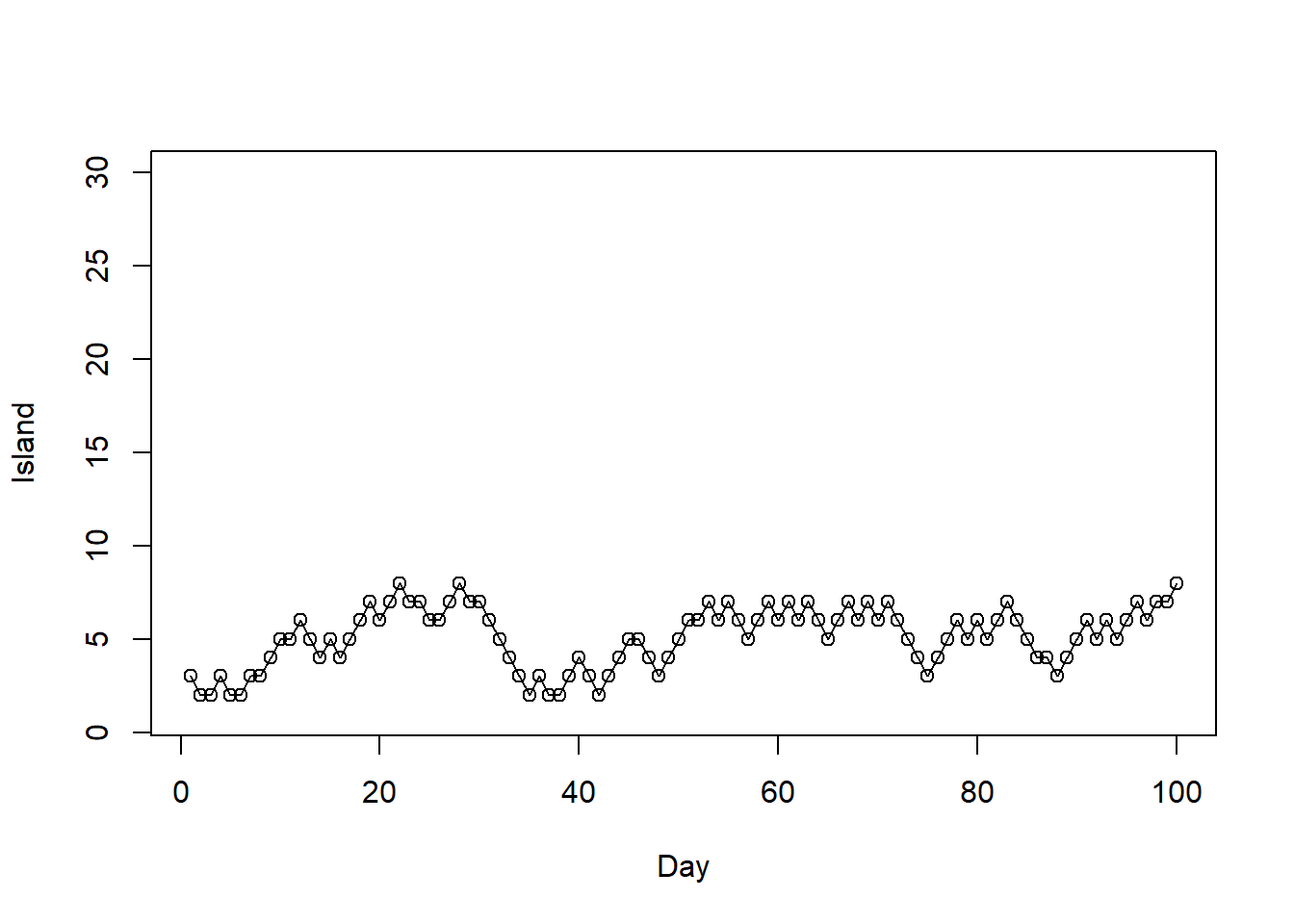
Relative frequencies of first 100 steps
# simulated relative frequencies
plot(table(i_sim[1:100]) / 100,
xlab = "Island", ylab = "Proportion of days")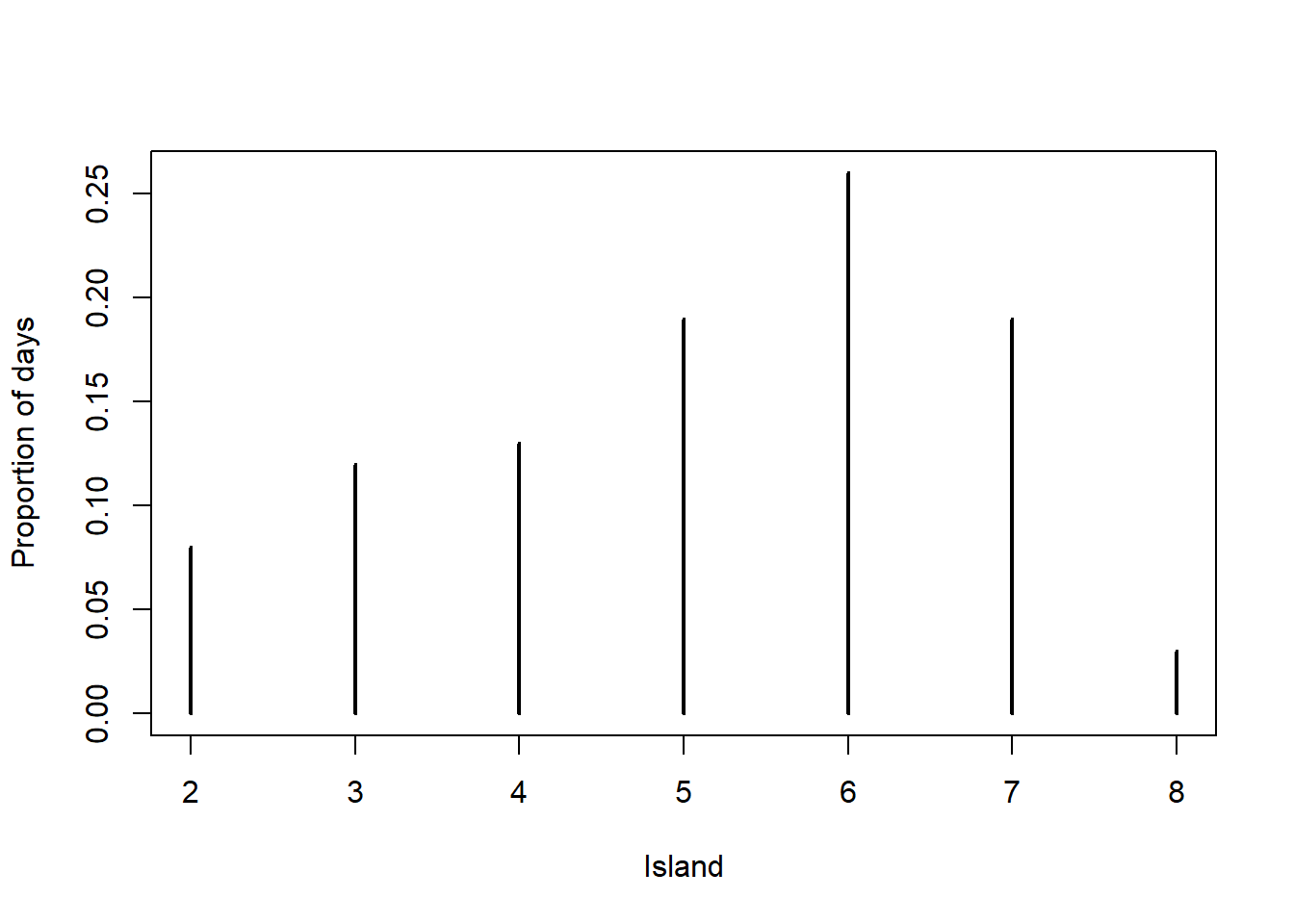
First 10000 steps
# trace plot
plot(1:n_steps, i_sim, type = "l",
ylim = range(i), xlab = "Day", ylab = "Island")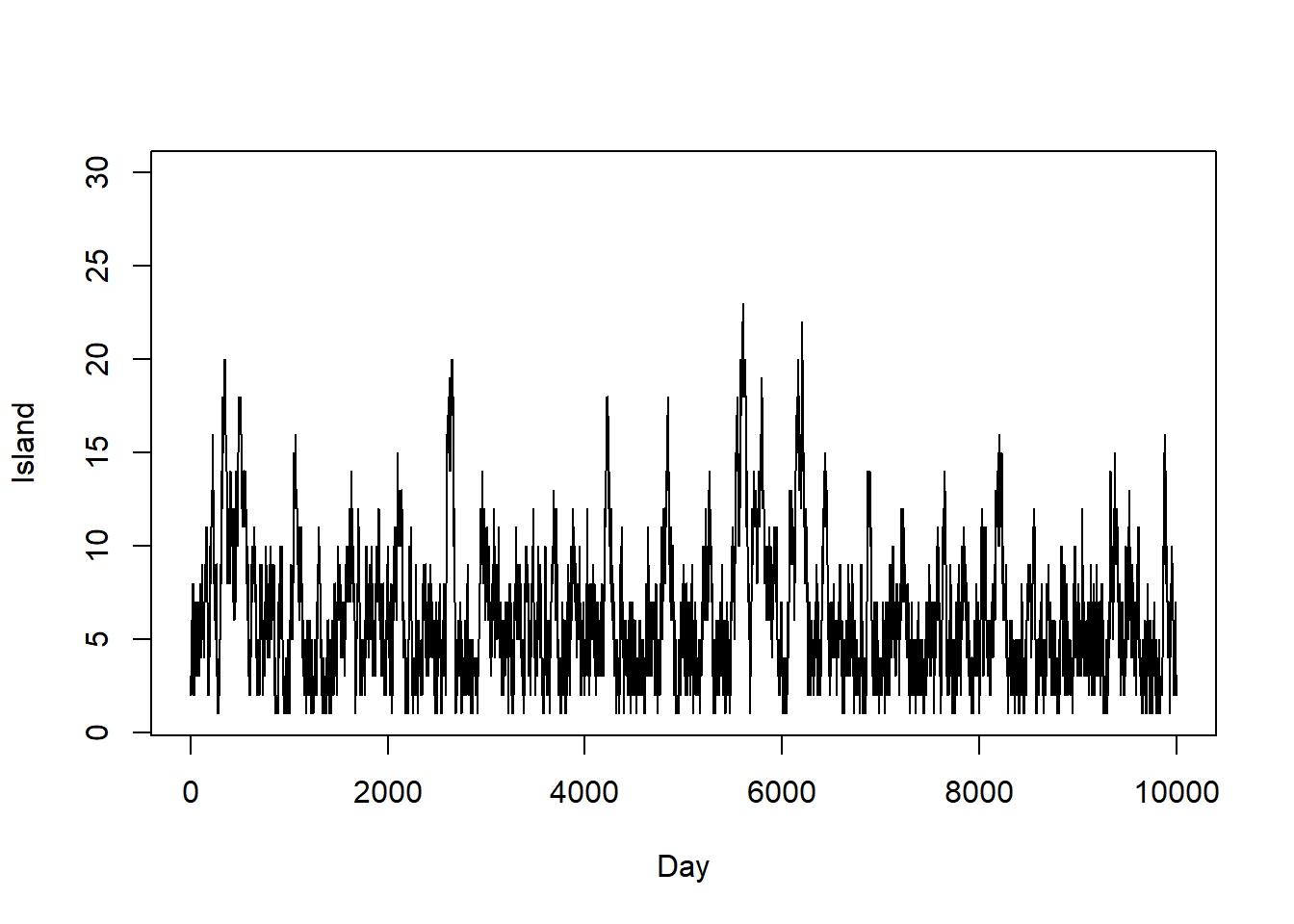
Relative frequencies of first 10000 steps
# simulated relative frequencies
plot(table(i_sim) / n_steps,
xlab = "Island", ylab = "Proportion of days")
# target distribution
points(i, pi_i / sum(pi_i), type = "o",
col = "seagreen")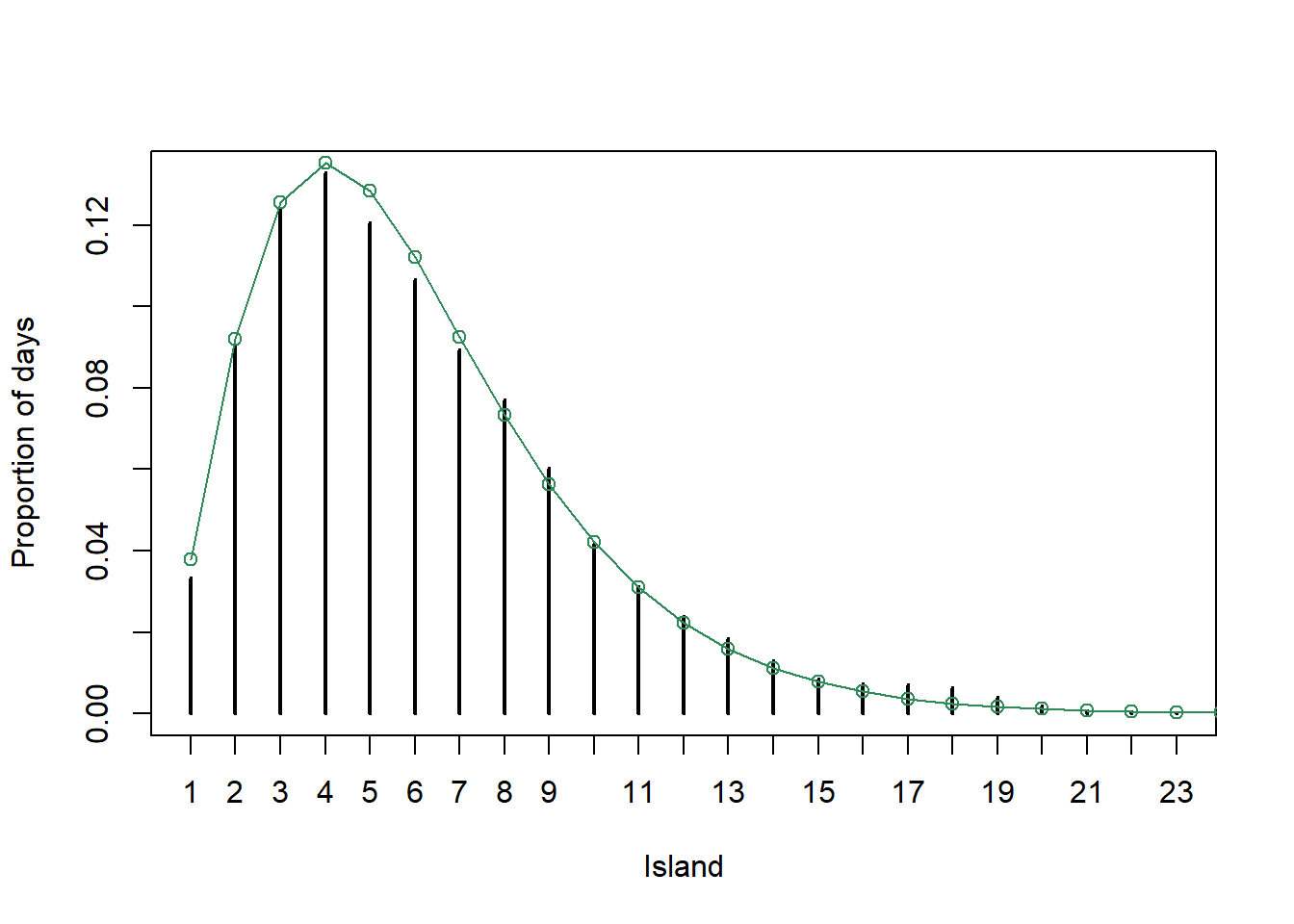
Explicit M-H transition matrix
P = rbind(
c(0.5, 0.5, 0),
c(0.25, 0.25, 0.5),
c(0, 1 / 3, 2 / 3)
)
compute_stationary_distribution(P) [,1] [,2] [,3]
[1,] 0.1666667 0.3333333 0.5Zipf
# parameters of Zipf
alpha = 1
M = 10
states = 1:M
# proposal chain is random walk (with reflection at boundaries)
pup = 0.5
Q = matrix(rep(0, M * M), nrow = M)
Q[1, 1] = 1 - pup
Q[1, 2] = pup
Q[M, M] = pup
Q[M, M - 1] = 1 - pup
for (i in 2:(M - 1)){
Q[i, i - 1] = 1 - pup
Q[i, i + 1] = pup
}
# Metropolis-Hastings chain
N = 10000
Xn = rep(NA, N)
Xn[1] = 1
for (n in 2:N){
# current state
i = Xn[n - 1]
# proposed state
j = sample(states, 1 , prob = Q[i, ])
# acceptance probability
aij = min(1, (1 / j) ^ alpha / (1 / i) ^ alpha)
# next state - accept j or stay at i
Xn[n] = sample(c(i, j), 1, prob = c(1 - aij, aij))
}# trace plot
plot(1:N, Xn, type = "l",
ylim = range(states), xlab = "n", ylab = "X_n")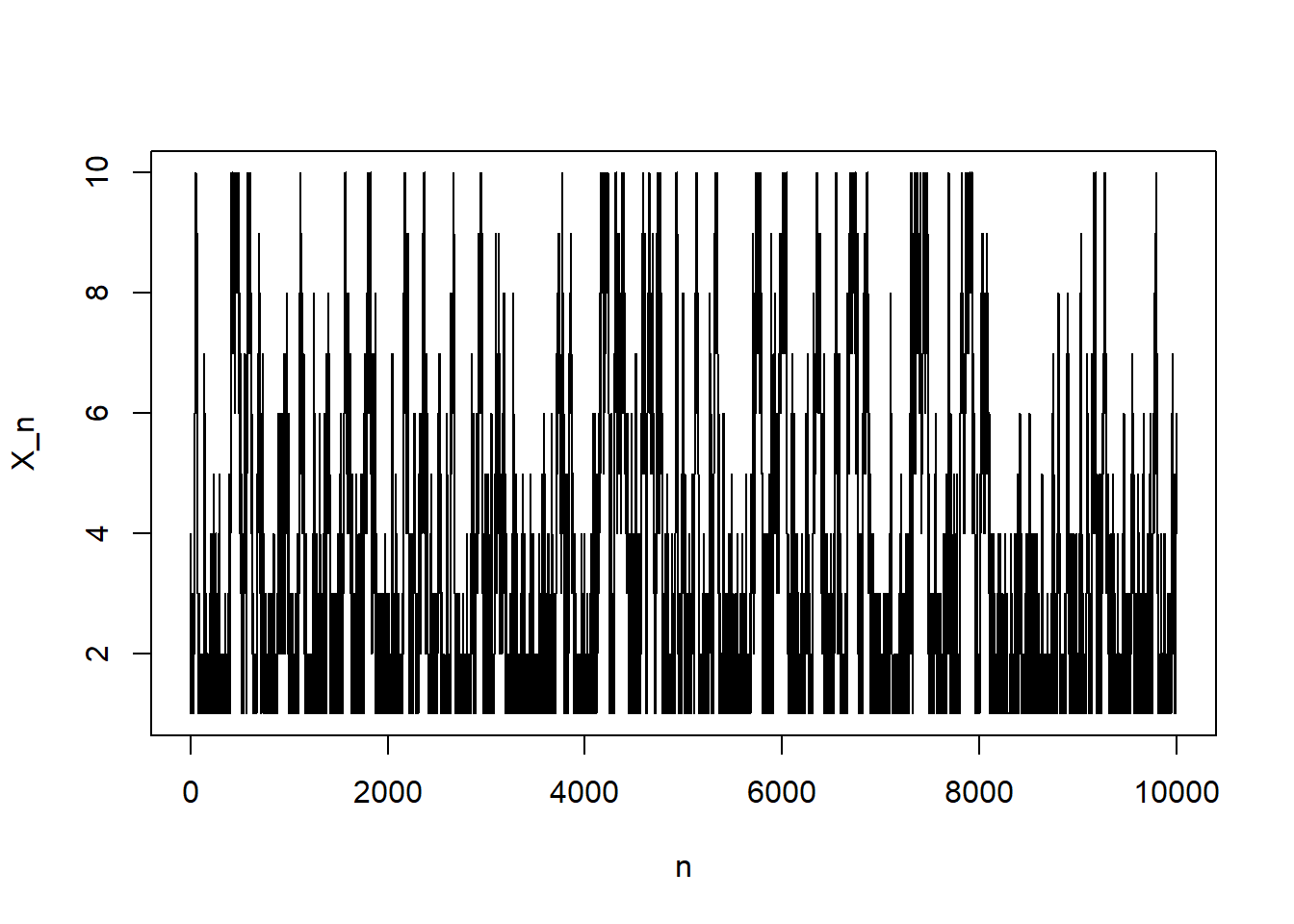
# simulated relative frequencies
plot(table(Xn) / N,
xlab = "x", ylab = "Relative frequency")
# target Zipf distribution
pi_x = (1 / states) ^ alpha
pi_x = pi_x / sum(pi_x)
points(states, pi_x, type = "o",
col = "seagreen")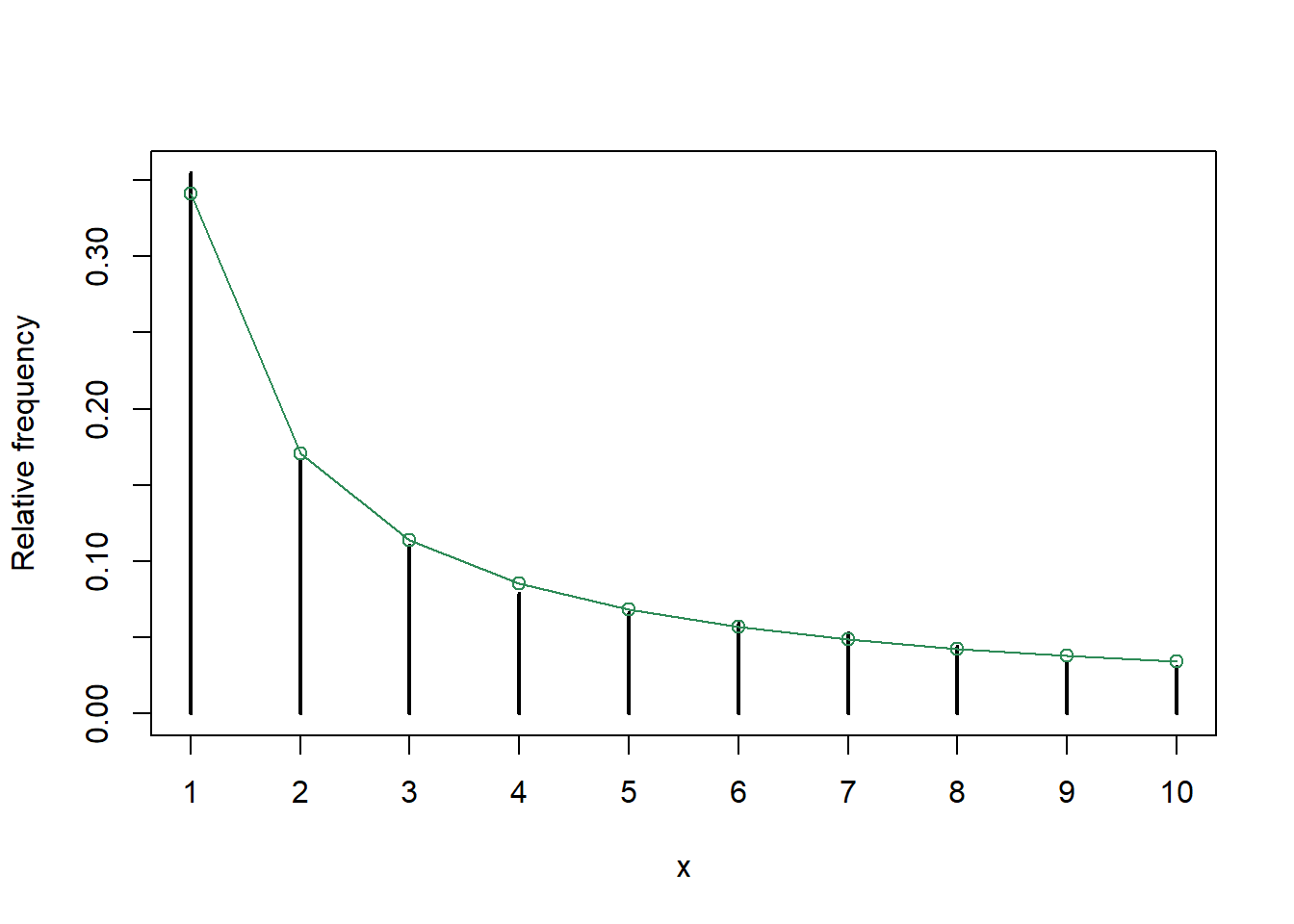
Gamma
Normal proposals
pi_x = function(u) {
if (u > 0) {
u ^ (7.2 - 1) * exp(-3 * u)
} else {
0
}
}
sigma = 0.5
N = 10000
x_n = rep(NA, N)
x_n[1] = 4
for (i in 2:N){
current = x_n[i-1]
proposed = rnorm(1, current, sigma)
a = min(1, pi_x(proposed) / pi_x(current))
x_n[i] = sample(c(current, proposed), 1, prob = c(1 - a, a))
}plot(1:N, x_n , type="l", xlab = "n", ylab = "X_n")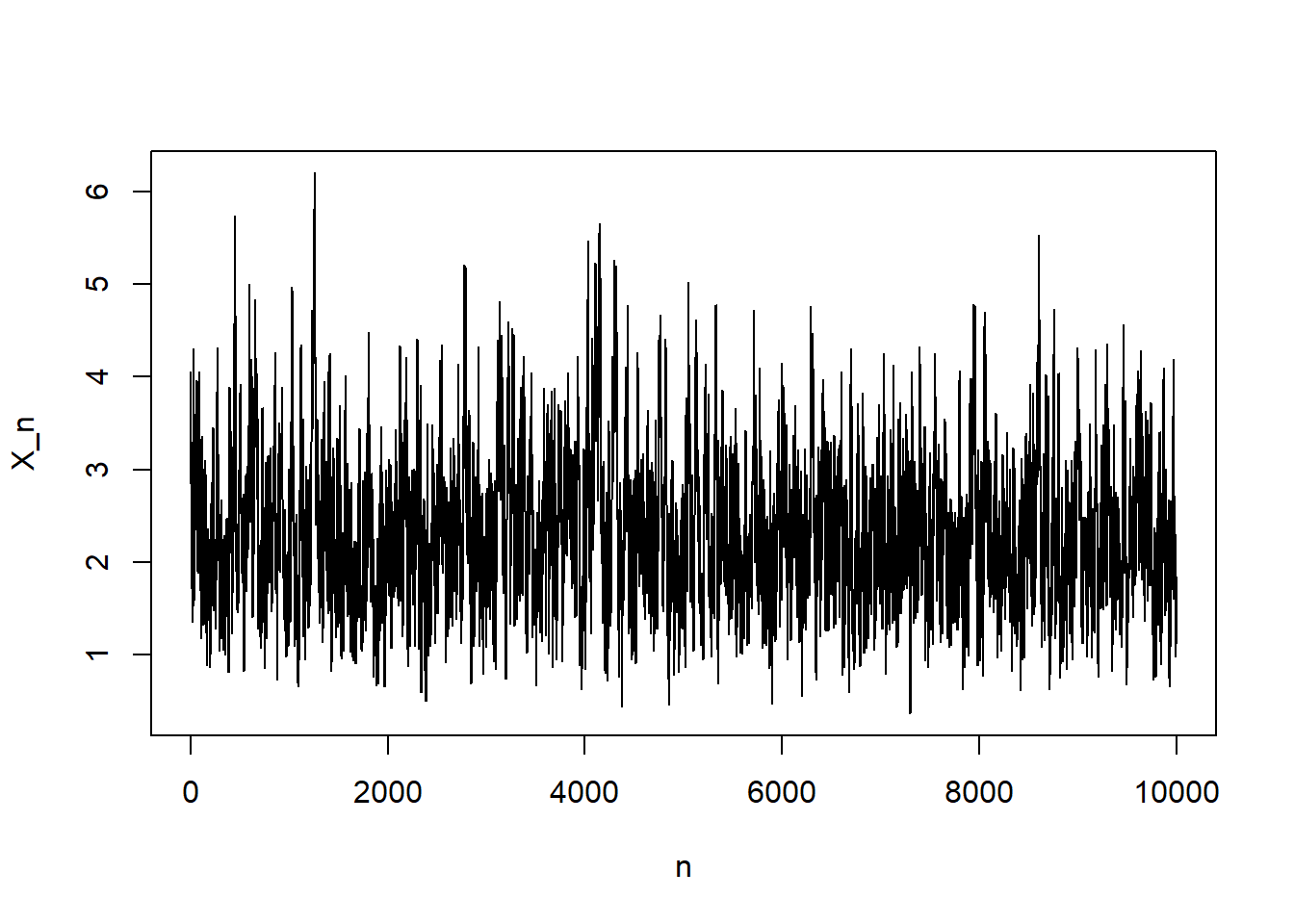
hist(x_n, freq = FALSE)
curve(dgamma(x, shape = 7.2, rate = 3), add = TRUE, col = "seagreen")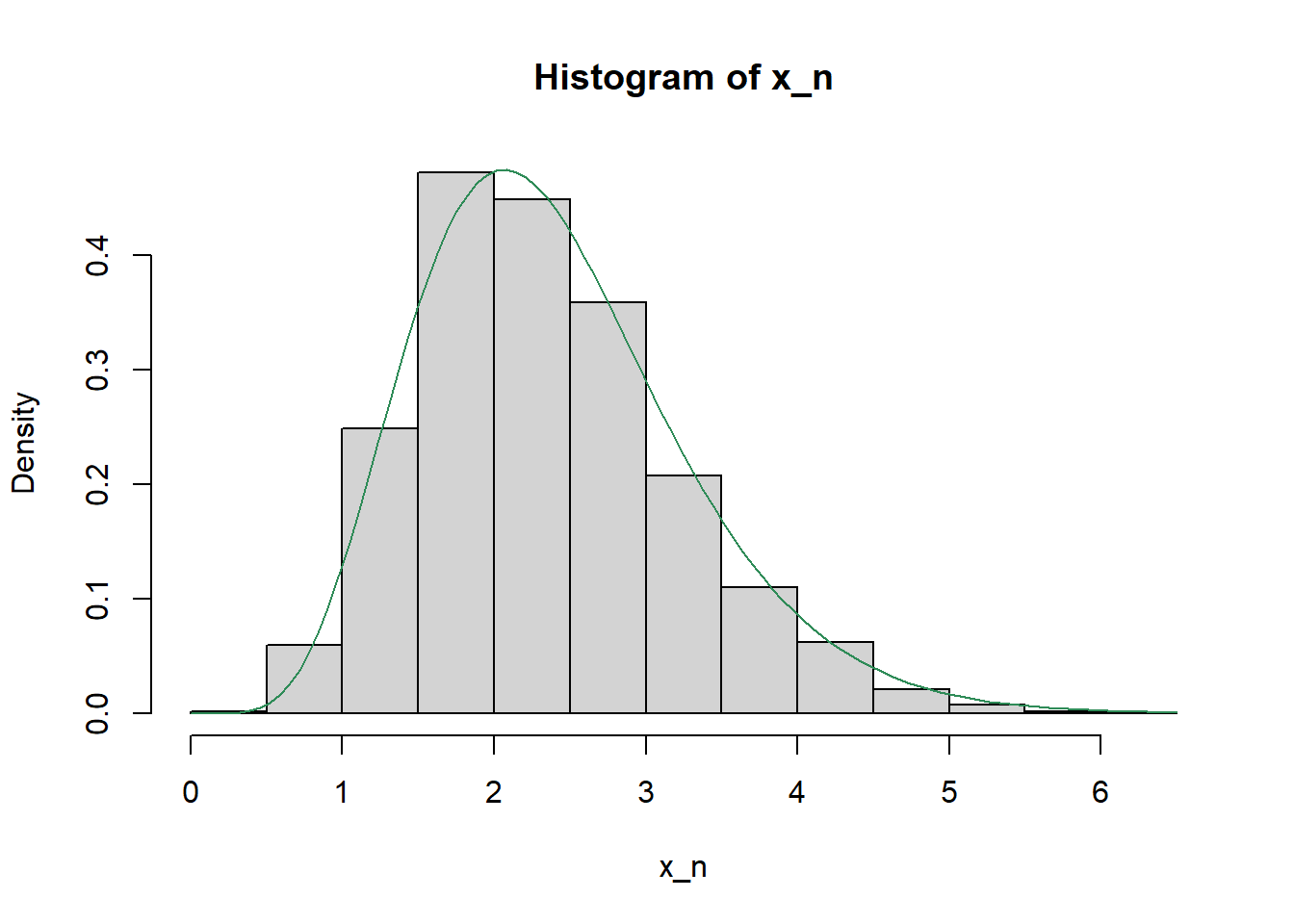
Moving Uniform proposals
delta = 0.5
N = 10000
x_n = rep(NA, N)
x_n[1] = 4
for (i in 2:N){
current = x_n[i-1]
proposed = runif(1, current - delta, current + delta)
a = min(1, pi_x(proposed) / pi_x(current))
x_n[i] = sample(c(current, proposed), 1, prob = c(1 - a, a))
}plot(1:N, x_n , type="l", xlab = "n", ylab = "X_n")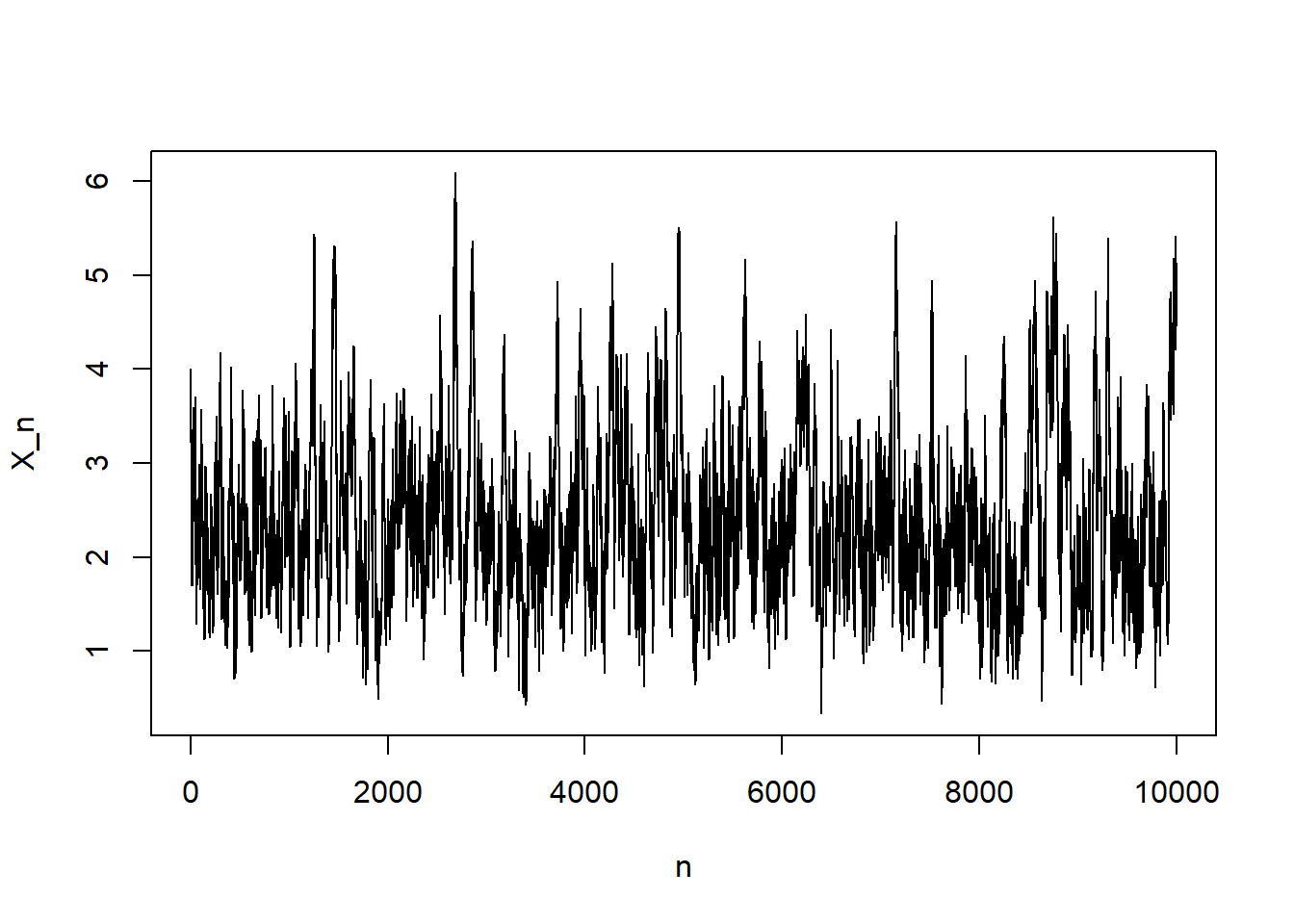
hist(x_n, freq = FALSE)
curve(dgamma(x, shape = 7.2, rate = 3), add = TRUE, col = "seagreen")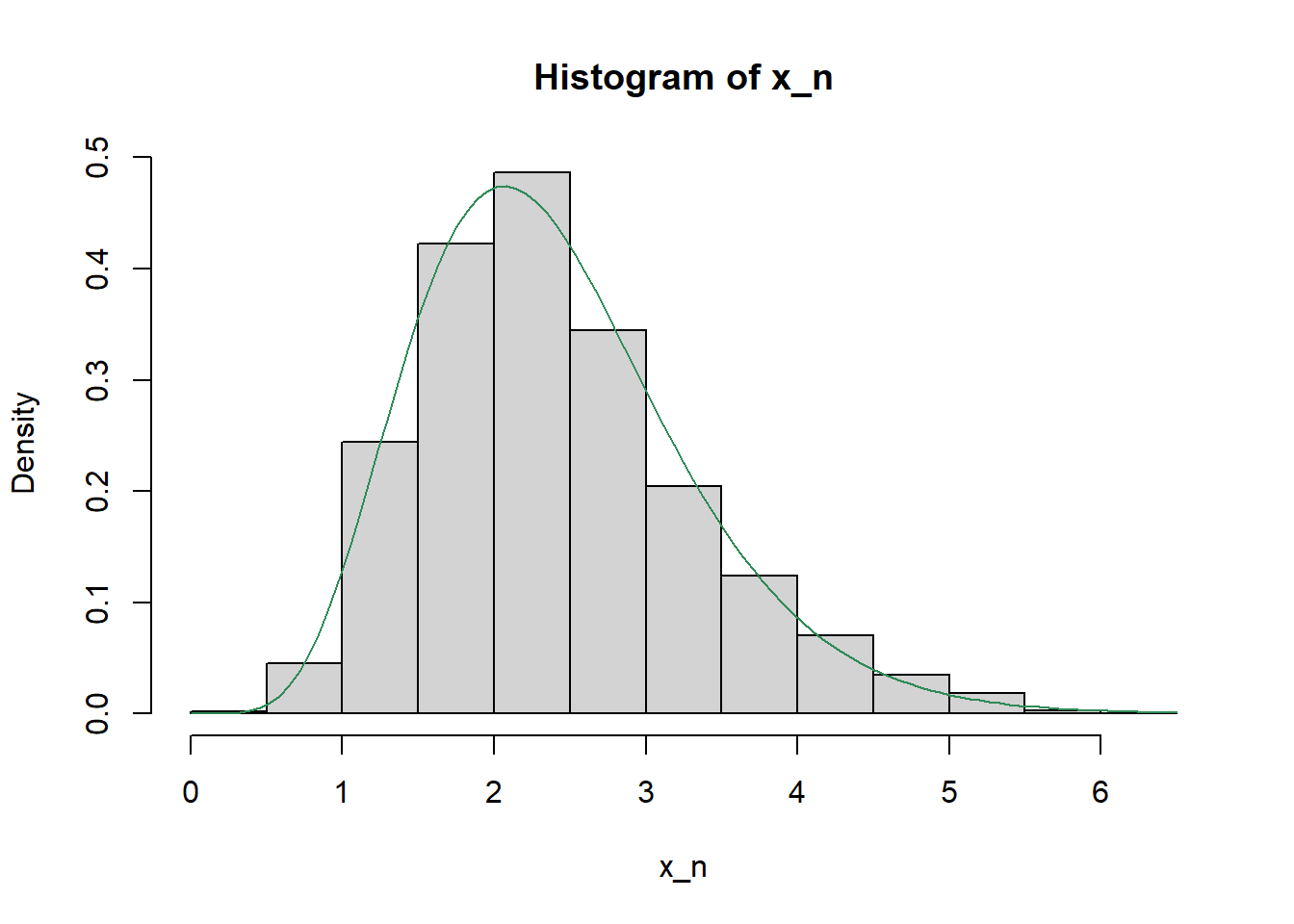
Multivariable
Normal proposals
pi_xy = function(x, y) {
if ((x > 0) & (y > 0)) {
exp(-(x + y + x * y))
} else {
0
}
}sigma = 0.5
N = 10000
x_n = rep(NA, N)
y_n = rep(NA, N)
x_n[1] = 1
y_n[1] = 1
for (i in 2:N){
current_x = x_n[i-1]
current_y = y_n[i-1]
proposed_x = rnorm(1, current_x, sigma)
proposed_y = rnorm(1, current_y, sigma)
a = min(1, pi_xy(proposed_x, proposed_y) / pi_xy(current_x, current_y))
action = sample(c("reject", "accept"), 1, prob = c(1 - a, a))
if (action == "accept"){
x_n[i] = proposed_x
y_n[i] = proposed_y
} else {
x_n[i] = current_x
y_n[i] = current_y
}
}
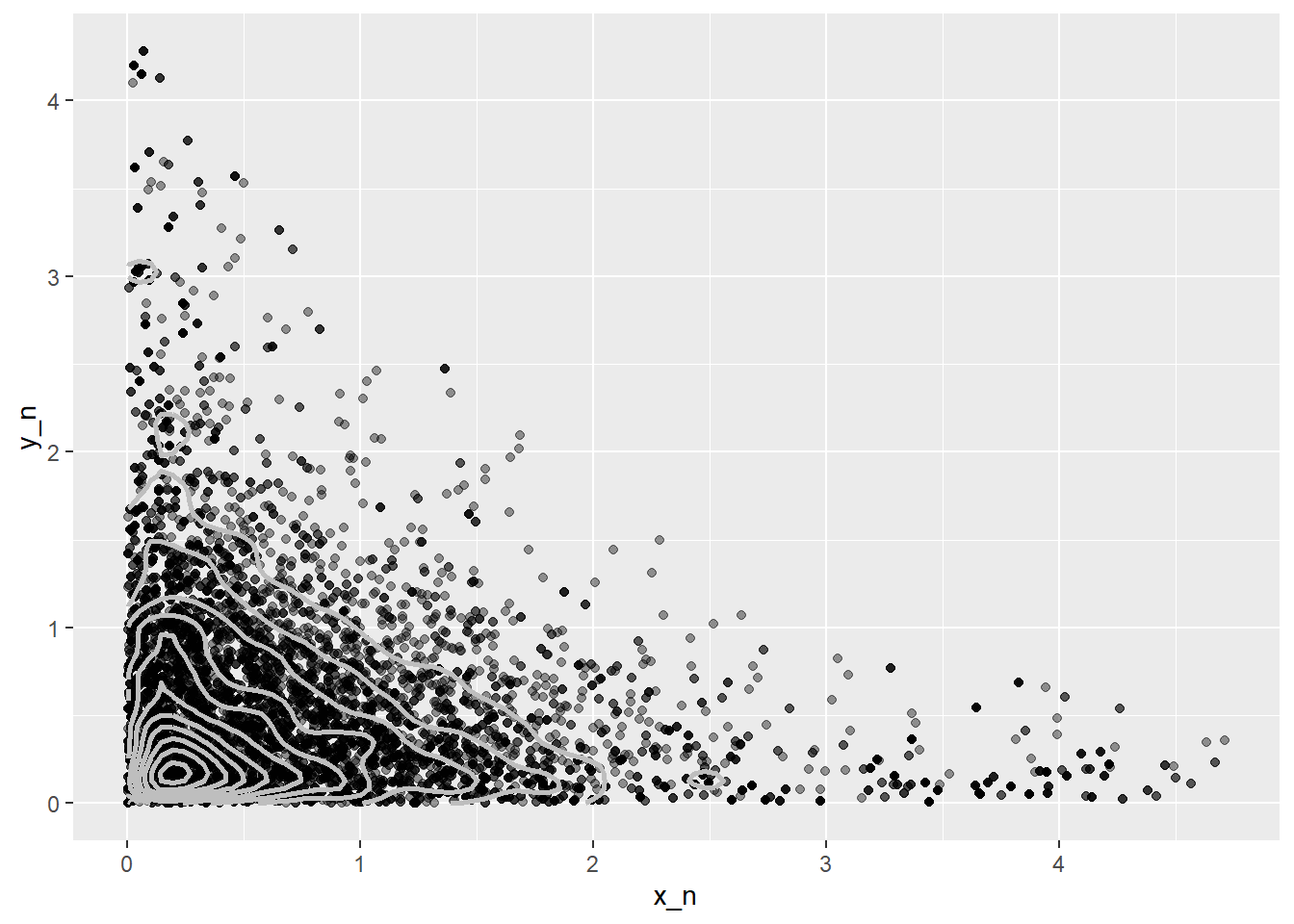
plot(x_n, y_n)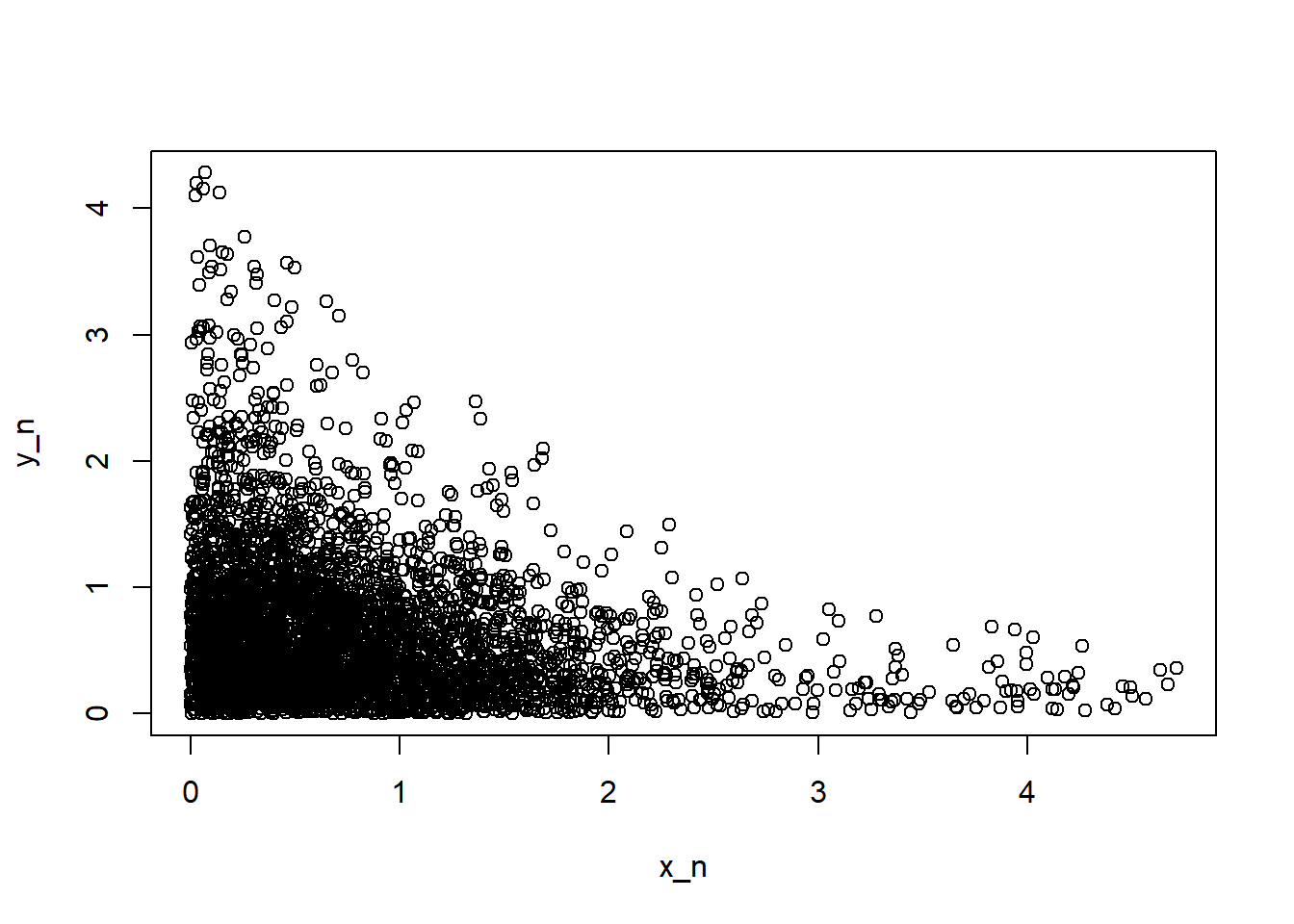
# Trace plot of first 100 steps
xy = data.frame(x_n, y_n)
ggplot(xy[1:100, ] |>
mutate(label = 1:100),
aes(x_n, y_n)) +
geom_path() +
geom_point(size = 2) +
geom_text(aes(label = label, x = x_n + 0.1, y = y_n + 0.01)) +
labs(title = "Trace plot of first 100 steps")
ggplot(xy, aes(x_n, y_n)) +
geom_point(alpha = 0.4) +
stat_density_2d(color = "grey", size = 1)Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.