Chapter 18 Appendix I: Full Analysis Code Examples
This section includes full blocks of code showing how to build examples of basic MPM types and conduct basic analyses using lefko3.
18.0.1 Basic ahistorical stage-based (Lefkovitch), raw (empirical) model
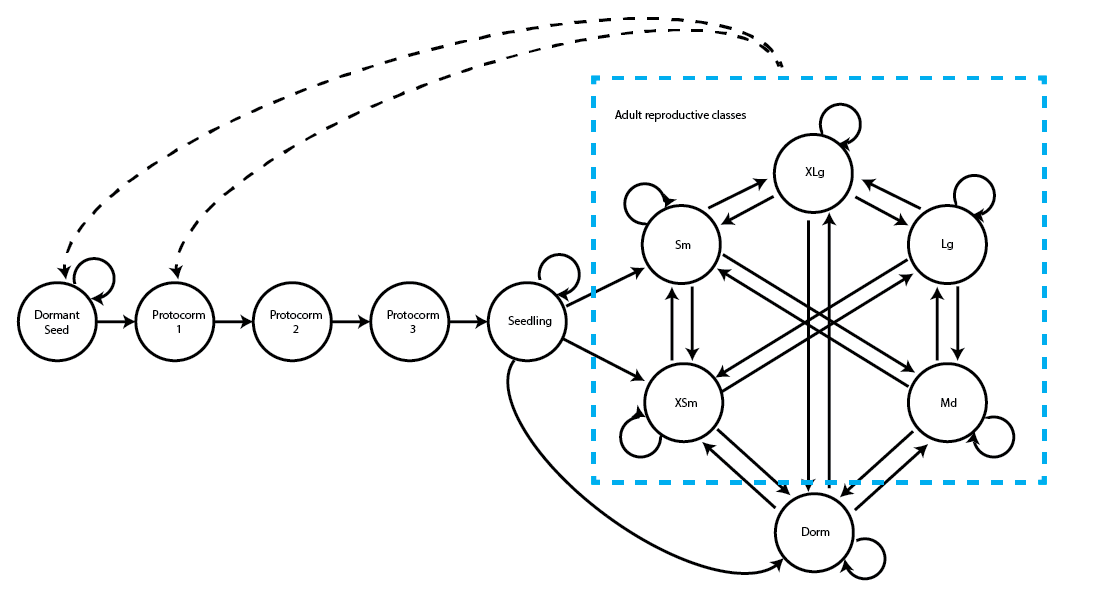
This life history model includes 11 stages:
1) dormant seed
2) first-year protocorm
3) second-year protocorm
4) third-year protocorm
5) seedling
6) dormant adult (no sprouts)
7) extra-small adult (1 sprout)
8) small adult (2-3 sprouts)
9) medium adult (4-5 sprouts)
10) large adult (6-7 sprouts)
11) extra-large adult (8+ sprouts)
The stageframe for this life history model can be programmed as follows.
data(cypdata)
sizevector <- c(0, 0, 0, 0, 0, 0, 1, 3, 6, 11, 19.5)
stagevector <- c("SD", "P1", "P2", "P3", "SL", "Dorm", "XSm", "Sm", "Md", "Lg",
"XLg")
repvector <- c(0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1)
obsvector <- c(0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1)
matvector <- c(0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1)
immvector <- c(0, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0)
propvector <- c(1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0)
indataset <- c(0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1)
binvec <- c(0, 0, 0, 0, 0, 0.5, 0.5, 1.5, 1.5, 3.5, 5)
comments <- c("Dormant seed", "1st yr protocorm", "2nd yr protocorm",
"3rd yr protocorm", "Seedling", "Dormant adult",
"Extra small adult (1 shoot)", "Small adult (2-4 shoots)",
"Medium adult (5-7 shoots)", "Large adult (8-14 shoots)",
"Extra large adult (>14 shoots)")
cyp_lefk_raw_frame <- sf_create(sizes = sizevector, stagenames = stagevector,
repstatus = repvector, obsstatus = obsvector, matstatus = matvector,
propstatus = propvector, immstatus = immvector, indataset = indataset,
binhalfwidth = binvec, comments = comments)Now we will create the MPM, first developing the standardized dataset and the supplemental data table.
cyp_lefk_raw_data <- verticalize3(data = cypdata, noyears = 6, firstyear = 2004,
patchidcol = "patch", individcol = "plantid", blocksize = 4,
sizeacol = "Inf2.04", sizebcol = "Inf.04", sizeccol = "Veg.04",
repstracol = "Inf.04", repstrbcol = "Inf2.04", fecacol = "Pod.04",
stageassign = cyp_lefk_raw_frame, stagesize = "sizeadded", NAas0 = TRUE,
NRasRep = TRUE, age_offset = 4)
seeds_per_fruit <- 5000
sl_mult <- 0.7
cyp_lefk_raw_supp <- supplemental(stage3 = c("SD", "P1", "P2", "P3", "SL", "SL",
"Dorm", "XSm", "Sm", "SD", "P1"),
stage2 = c("SD", "SD", "P1", "P2", "P3", "SL", "SL", "SL", "SL", "rep","rep"),
eststage3 = c(NA, NA, NA, NA, NA, NA, "Dorm", "XSm", "Sm", NA, NA),
eststage2 = c(NA, NA, NA, NA, NA, NA, "XSm", "XSm", "XSm", NA, NA),
givenrate = c(0.08, 0.10, 0.10, 0.10, 0.05, 0.05, NA, NA, NA, NA, NA),
multiplier = c(NA, NA, NA, NA, NA, NA, sl_mult, sl_mult, sl_mult,
0.5 * seeds_per_fruit, 0.5 * seeds_per_fruit),
type =c(1, 1, 1, 1, 1, 1, 1, 1, 1, 3, 3),
stageframe = cyp_lefk_raw_frame, historical = FALSE)
> Warning: NA values in argument multiplier will be treated as 1 values.
cyp_lefk_raw_mpm <- rlefko2(data = cyp_lefk_raw_data, year = "all",
patch = "all", stageframe = cyp_lefk_raw_frame, supplement = cyp_lefk_raw_supp,
stages = c("stage3", "stage2"), size = c("size3added", "size2added"),
yearcol = "year2", patchcol = "patchid", indivcol = "individ")
summary(cyp_lefk_raw_mpm)
>
> This ahistorical lefkoMat object contains 15 matrices.
>
> Each matrix is square with 11 rows and columns, and a total of 121 elements.
> A total of 266 survival transitions were estimated, with 17.733 per matrix.
> A total of 70 fecundity transitions were estimated, with 4.667 per matrix.
> This lefkoMat object covers 1 population, 3 patches, and 5 time steps.
>
> The dataset contains a total of 74 unique individuals and 320 unique transitions.
>
> Survival probability sum check (each matrix represented by column in order):
> [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10] [,11] [,12]
> Min. 0.000 0.000 0.000 0.000 0.000 0.000 0.050 0.050 0.000 0.050 0.000 0.000
> 1st Qu. 0.075 0.025 0.075 0.025 0.075 0.075 0.140 0.140 0.100 0.140 0.100 0.100
> Median 0.180 0.100 0.180 0.100 0.180 0.180 0.909 0.778 0.686 0.857 0.750 0.575
> Mean 0.457 0.361 0.471 0.328 0.417 0.464 0.631 0.611 0.530 0.631 0.562 0.523
> 3rd Qu. 0.955 0.769 1.000 0.592 0.781 1.000 1.000 1.000 0.955 1.000 1.000 1.000
> Max. 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000
> [,13] [,14] [,15]
> Min. 0.000 0.000 0.000
> 1st Qu. 0.075 0.075 0.100
> Median 0.180 0.180 0.750
> Mean 0.432 0.450 0.562
> 3rd Qu. 0.875 1.000 1.000
> Max. 1.000 1.000 1.000
>
>
> Stages without connections leading to the rest of the life cycle found in matrices: 1 2 3 4 5 6 9 11 12 13 14 15Some basic analyses.
lambda3(cyp_lefk_raw_mpm)
> pop patch year2 lambda
> 1 1 A 2004 1.0830875
> 2 1 A 2005 1.0000000
> 3 1 A 2006 1.0000000
> 4 1 A 2007 0.5000000
> 5 1 A 2008 0.8917561
> 6 1 B 2004 1.0214506
> 7 1 B 2005 1.1285250
> 8 1 B 2006 1.1815829
> 9 1 B 2007 0.9604335
> 10 1 B 2008 1.0893093
> 11 1 C 2004 1.1124223
> 12 1 C 2005 0.8333333
> 13 1 C 2006 1.1543634
> 14 1 C 2007 0.8619361
> 15 1 C 2008 1.078635218.0.2 Complex ahistorical stage-based (Lefkovitch), function-based model
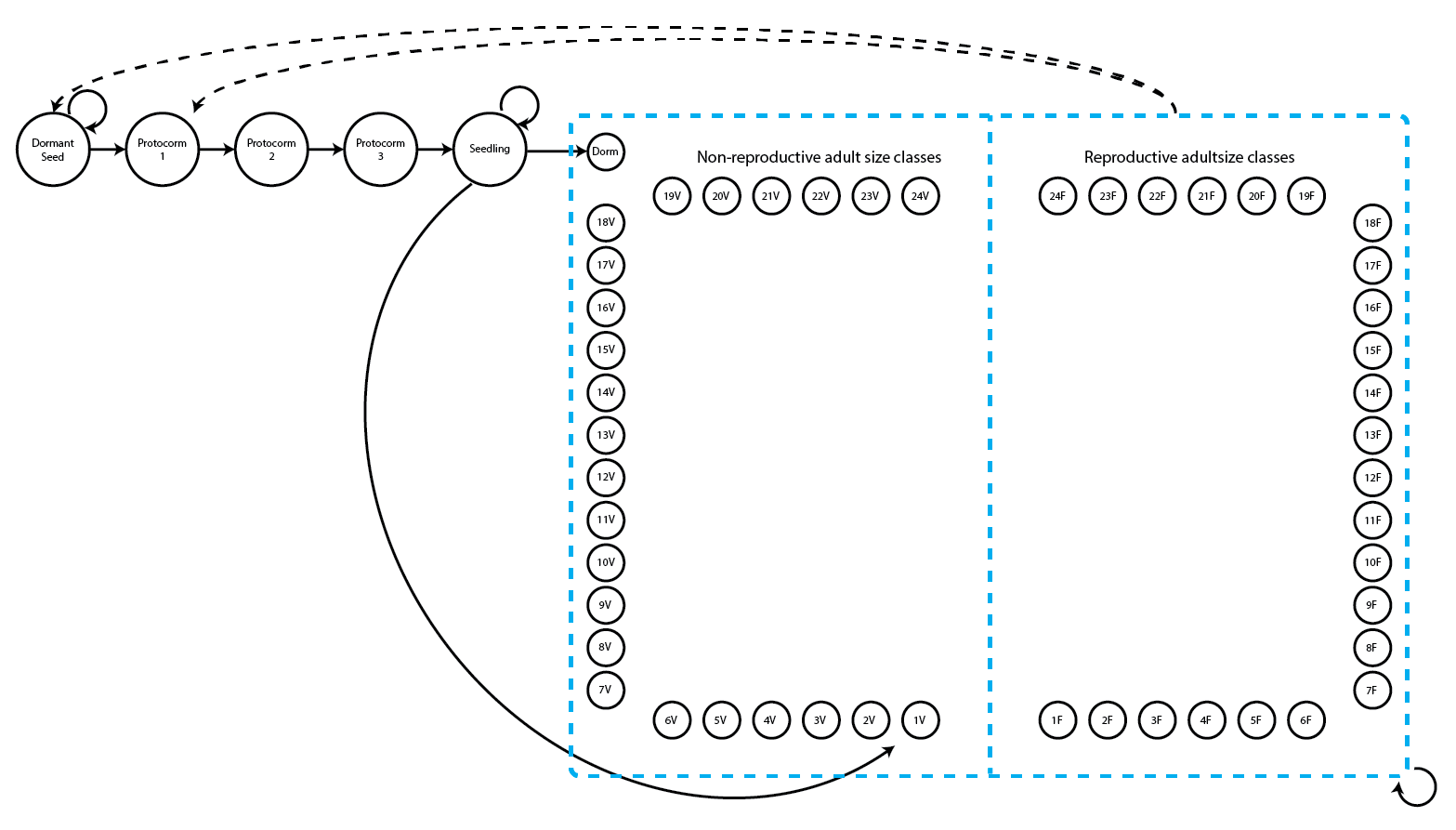
This life history model includes 54 stages:
1) dormant seed
2) first-year protocorm (juvenile)
3) second-year protocorm (juvenile)
4) third-year protocorm (juvenile)
5) seedling (juvenile)
6) dormant adult
7-30) non-flowering but sprouting adults with 1-24 sprouts (1 stage per # sprouts)
31-54) flowering adults with 1-24 sprouts (1 stage per # sprouts)
The stageframe for this life history model can be programmed as follows.
data(cypdata)
stagevector <- c("SD", "P1", "P2", "P3", "SL", "Dorm", "1V", "2V", "3V", "4V",
"5V", "6V", "7V", "8V", "9V", "10V", "11V", "12V", "13V", "14V", "15V", "16V",
"17V", "18V", "19V", "20V", "21V", "22V", "23V", "24V", "1F", "2F", "3F",
"4F", "5F", "6F", "7F", "8F", "9F", "10F", "11F", "12F", "13F", "14F", "15F",
"16F", "17F", "18F", "19F", "20F", "21F", "22F", "23F", "24F")
indataset <- c(0, 0, 0, 0, 0, rep(1, 49))
sizevector <- c(0, 0, 0, 0, 0, seq(from = 0, t = 24), seq(from = 1, to = 24))
repvector <- c(0, 0, 0, 0, 0, rep(0, 25), rep(1, 24))
obsvector <- c(0, 0, 0, 0, 0, 0, rep(1, 48))
matvector <- c(0, 0, 0, 0, 0, rep(1, 49))
immvector <- c(0, 1, 1, 1, 1, rep(0, 49))
propvector <- c(1, rep(0, 53))
comments <- c("Dormant seed", "Yr1 protocorm", "Yr2 protocorm", "Yr3 protocorm",
"Seedling", "Veg dorm", "Veg adult 1 stem", "Veg adult 2 stems",
"Veg adult 3 stems", "Veg adult 4 stems", "Veg adult 5 stems",
"Veg adult 6 stems", "Veg adult 7 stems", "Veg adult 8 stems",
"Veg adult 9 stems", "Veg adult 10 stems", "Veg adult 11 stems",
"Veg adult 12 stems", "Veg adult 13 stems", "Veg adult 14 stems",
"Veg adult 15 stems", "Veg adult 16 stems", "Veg adult 17 stems",
"Veg adult 18 stems", "Veg adult 19 stems", "Veg adult 20 stems",
"Veg adult 21 stems", "Veg adult 22 stems", "Veg adult 23 stems",
"Veg adult 24 stems", "Flo adult 1 stem", "Flo adult 2 stems",
"Flo adult 3 stems", "Flo adult 4 stems", "Flo adult 5 stems",
"Flo adult 6 stems", "Flo adult 7 stems", "Flo adult 8 stems",
"Flo adult 9 stems", "Flo adult 10 stems", "Flo adult 11 stems",
"Flo adult 12 stems", "Flo adult 13 stems", "Flo adult 14 stems",
"Flo adult 15 stems", "Flo adult 16 stems", "Flo adult 17 stems",
"Flo adult 18 stems", "Flo adult 19 stems", "Flo adult 20 stems",
"Flo adult 21 stems", "Flo adult 22 stems", "Flo adult 23 stems",
"Flo adult 24 stems")
cyp_lefk_fb_frame <- sf_create(sizes = sizevector, stagenames = stagevector,
repstatus = repvector, obsstatus = obsvector, matstatus = matvector,
propstatus = propvector, immstatus = immvector, indataset = indataset,
comments = comments)Now we will create the MPM, first developing the standardized dataset, the supplemental data table, and the vital rate models.
cyp_lefk_fb_data <- verticalize3(data = cypdata, noyears = 6, firstyear = 2004,
patchidcol = "patch", individcol = "plantid", blocksize = 4,
sizeacol = "Inf2.04", sizebcol = "Inf.04", sizeccol = "Veg.04",
repstracol = "Inf.04", repstrbcol = "Inf2.04", fecacol = "Pod.04",
stageassign = cyp_lefk_fb_frame, stagesize = "sizeadded", NAas0 = TRUE,
age_offset = 4)
seeds_per_fruit <- 5000
sl_mult <- 0.7
cyp_lefk_fb_supp <- supplemental(stage3 = c("SD", "P1", "P2", "P3", "SL", "SL",
"Dorm", "1V", "2V", "3V", "SD", "P1"),
stage2 = c("SD", "SD", "P1", "P2", "P3", "SL", "SL", "SL", "SL", "SL", "rep",
"rep"),
eststage3 = c(NA, NA, NA, NA, NA, NA, "Dorm", "1V", "2V", "3V", NA, NA),
eststage2 = c(NA, NA, NA, NA, NA, NA, "Dorm", "Dorm", "Dorm", "Dorm", NA, NA),
givenrate = c(0.08, 0.1, 0.1, 0.1, 0.05, 0.05, NA, NA, NA, NA, NA, NA),
multiplier = c(NA, NA, NA, NA, NA, NA, sl_mult, sl_mult, sl_mult, sl_mult,
0.5 * seeds_per_fruit, 0.5 * seeds_per_fruit),
type = c("S", "S", "S", "S", "S", "S", "S", "S", "S", "S", "R", "R"),
stageframe = cyp_lefk_fb_frame, historical = FALSE)
> Warning: NA values in argument multiplier will be treated as 1 values.
cyp_lefk_fb_vital <- modelsearch(cyp_lefk_fb_data, historical = FALSE,
suite = "main", approach = "mixed",
vitalrates = c("surv", "obs", "size", "repst", "fec"), patch = "patchid",
sizedist = "negbin", size.trunc = TRUE, fecdist = "poisson", fec.zero = TRUE,
size = c("size3added", "size2added", "size1added"), quiet = "partial")
>
> Developing global model of survival probability...
>
> Global model of survival probability developed. Proceeding with model dredge...
>
> Developing global model of observation probability...
>
> Global model of observation probability developed. Proceeding with model dredge...
>
> Developing global model of primary size...
>
> Global model of primary size developed. Proceeding with model dredge...
>
> Developing global model of reproduction probability...
>
> Global model of reproduction probability developed. Proceeding with model dredge...
>
> Developing global model of fecundity...
>
> Global model of fecundity developed. Proceeding with model dredge...
>
> Finished selecting best-fit models.
cyp_lefk_fb_mpm <- flefko2(stageframe = cyp_lefk_fb_frame,
supplement = cyp_lefk_fb_supp, modelsuite = cyp_lefk_fb_vital,
data = cyp_lefk_fb_data)
summary(cyp_lefk_fb_mpm)
>
> This ahistorical lefkoMat object contains 15 matrices.
>
> Each matrix is square with 54 rows and columns, and a total of 2916 elements.
> A total of 36165 survival transitions were estimated, with 2411 per matrix.
> A total of 720 fecundity transitions were estimated, with 48 per matrix.
> This lefkoMat object covers 1 population, 3 patches, and 5 time steps.
>
> The dataset contains a total of 74 unique individuals and 320 unique transitions.
>
> Vital rate modeling quality control:
>
> Survival estimated with 74 individuals and 320 individual transitions.
> Observation estimated with 70 individuals and 303 individual transitions.
> Primary size estimated with 70 individuals and 288 individual transitions.
> Secondary size transition not estimated.
> Tertiary size transition not estimated.
> Reproductive status estimated with 70 individuals and 288 individual transitions.
> Fecundity estimated with 51 individuals and 118 individual transitions.
> Juvenile survival not estimated.
> Juvenile observation probability not estimated.
> Juvenile primary size transition not estimated.
> Juvenile secondary size transition not estimated.
> Juvenile tertiary size transition not estimated.
> Juvenile reproduction probability not estimated.
> Juvenile maturity transition probability not estimated.
>
> Survival probability sum check (each matrix represented by column in order):
> [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10] [,11] [,12]
> Min. 0.050 0.050 0.050 0.050 0.050 0.050 0.050 0.050 0.050 0.050 0.050 0.050
> 1st Qu. 0.991 0.991 0.991 0.991 0.991 0.991 0.991 0.991 0.991 0.991 0.991 0.991
> Median 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000
> Mean 0.916 0.917 0.917 0.917 0.914 0.916 0.917 0.917 0.917 0.914 0.914 0.916
> 3rd Qu. 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000
> Max. 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000
> [,13] [,14] [,15]
> Min. 0.050 0.050 0.050
> 1st Qu. 0.991 0.991 0.991
> Median 1.000 1.000 1.000
> Mean 0.916 0.916 0.913
> 3rd Qu. 1.000 1.000 1.000
> Max. 1.000 1.000 1.000
>
>
> All matrices have no stage discontinuities.Now some basic analyses.
lambda3(cyp_lefk_fb_mpm)
> pop patch year2 lambda
> 1 1 A 2004 1.0302681
> 2 1 A 2005 1.0400000
> 3 1 A 2006 1.0369609
> 4 1 A 2007 0.9872717
> 5 1 A 2008 1.0164516
> 6 1 B 2004 1.0723217
> 7 1 B 2005 1.0791790
> 8 1 B 2006 1.0770056
> 9 1 B 2007 1.0148237
> 10 1 B 2008 1.0605714
> 11 1 C 2004 1.0442739
> 12 1 C 2005 1.0674346
> 13 1 C 2006 1.0603017
> 14 1 C 2007 1.0006138
> 15 1 C 2008 1.023001018.0.3 Basic historical stage-based (Lefkovitch), raw (empirical) model

This life history model includes 11 stages:
1) dormant seed
2) first-year protocorm
3) second-year protocorm
4) third-year protocorm
5) seedling
6) dormant adult (no sprouts)
7) extra-small adult (1 sprout)
8) small adult (2-3 sprouts)
9) medium adult (4-5 sprouts)
10) large adult (6-7 sprouts)
11) extra-large adult (8+ sprouts)
The stageframe for this life history model can be programmed as follows.
data(cypdata)
sizevector <- c(0, 0, 0, 0, 0, 0, 1, 3, 6, 11, 19.5)
stagevector <- c("SD", "P1", "P2", "P3", "SL", "Dorm", "XSm", "Sm", "Md", "Lg",
"XLg")
repvector <- c(0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1)
obsvector <- c(0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1)
matvector <- c(0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1)
immvector <- c(0, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0)
propvector <- c(1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0)
indataset <- c(0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1)
binvec <- c(0, 0, 0, 0, 0, 0.5, 0.5, 1.5, 1.5, 3.5, 5)
comments <- c("Dormant seed", "1st yr protocorm", "2nd yr protocorm",
"3rd yr protocorm", "Seedling", "Dormant adult",
"Extra small adult (1 shoot)", "Small adult (2-4 shoots)",
"Medium adult (5-7 shoots)", "Large adult (8-14 shoots)",
"Extra large adult (>14 shoots)")
cyp_lefk_raw_frame <- sf_create(sizes = sizevector, stagenames = stagevector,
repstatus = repvector, obsstatus = obsvector, matstatus = matvector,
propstatus = propvector, immstatus = immvector, indataset = indataset,
binhalfwidth = binvec, comments = comments)Now we will create the MPM, first developing the standardized dataset and the supplemental data table.
cyp_lefk_raw_data <- verticalize3(data = cypdata, noyears = 6, firstyear = 2004,
patchidcol = "patch", individcol = "plantid", blocksize = 4,
sizeacol = "Inf2.04", sizebcol = "Inf.04", sizeccol = "Veg.04",
repstracol = "Inf.04", repstrbcol = "Inf2.04", fecacol = "Pod.04",
stageassign = cyp_lefk_raw_frame, stagesize = "sizeadded", NAas0 = TRUE,
NRasRep = TRUE, age_offset = 4)
seeds_per_fruit <- 5000
sl_mult <- 0.7
cyp_lefk_raw_hsupp <- supplemental(stage3 = c("SD", "SD", "P1", "P1", "P2", "P2",
"P3", "SL", "SL", "SL", "Dorm", "XSm", "Sm", "Dorm", "XSm", "Sm", "mat",
"mat", "mat", "SD", "P1"),
stage2 = c("SD", "SD", "SD", "SD", "P1", "P1", "P2", "P3", "SL", "SL", "SL",
"SL", "SL", "SL", "SL", "SL", "Dorm", "XSm", "Sm", "rep", "rep"),
stage1 = c("SD", "rep", "SD", "rep", "SD", "rep", "P1", "P2", "P3", "SL",
"SL", "SL", "SL", "P3", "P3", "P3", "SL", "SL", "SL", "mat", "mat"),
eststage3 = c(NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, "Dorm", "XSm", "Sm",
"Dorm", "XSm", "Sm", "mat", "mat", "mat", NA, NA),
eststage2 = c(NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, "XSm", "XSm", "XSm",
"XSm", "XSm", "XSm", "Dorm", "XSm", "Sm", NA, NA),
eststage1 = c(NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, "XSm", "XSm", "XSm",
"XSm", "XSm", "XSm", "XSm", "XSm", "XSm", NA, NA),
givenrate = c(0.08, 0.10, 0.10, 0.10, 0.10, 0.10, 0.10, 0.05, 0.05, 0.05, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA),
multiplier = c(NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, sl_mult, sl_mult,
sl_mult, sl_mult, sl_mult, sl_mult, 1, 1, 1, 0.5 * seeds_per_fruit,
0.5 * seeds_per_fruit),
type =c(1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 3, 3),
stageframe = cyp_lefk_raw_frame, historical = TRUE)
> Warning: NA values in argument multiplier will be treated as 1 values.
cyp_lefk_raw_hmpm <- rlefko3(data = cyp_lefk_raw_data,
stageframe = cyp_lefk_raw_frame, supplement = cyp_lefk_raw_hsupp,
year = "all", patch = "all", stages = c("stage3", "stage2", "stage1"),
size = c("size3added", "size2added", "size1added"),
yearcol = "year2", patchcol = "patchid", indivcol = "individ")
summary(cyp_lefk_raw_hmpm)
>
> This historical lefkoMat object contains 12 matrices.
>
> Each matrix is square with 121 rows and columns, and a total of 14641 elements.
> A total of 516 survival transitions were estimated, with 43 per matrix.
> A total of 70 fecundity transitions were estimated, with 5.833 per matrix.
> This lefkoMat object covers 1 population, 3 patches, and 4 time steps.
>
> The dataset contains a total of 74 unique individuals and 320 unique transitions.
>
> Survival probability sum check (each matrix represented by column in order):
> [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10] [,11]
> Min. 0.000 0.0000 0.0000 0.000 0.000 0.000 0.00 0.000 0.000 0.0000 0.000
> 1st Qu. 0.000 0.0000 0.0000 0.000 0.000 0.000 0.00 0.000 0.000 0.0000 0.000
> Median 0.000 0.0000 0.0000 0.000 0.000 0.000 0.00 0.000 0.000 0.0000 0.000
> Mean 0.107 0.0945 0.0851 0.101 0.158 0.158 0.14 0.169 0.119 0.0851 0.119
> 3rd Qu. 0.000 0.0000 0.0000 0.000 0.100 0.100 0.05 0.100 0.000 0.0000 0.000
> Max. 1.000 1.0000 1.0000 1.000 1.000 1.000 1.00 1.000 1.000 1.0000 1.000
> [,12]
> Min. 0.000
> 1st Qu. 0.000
> Median 0.000
> Mean 0.144
> 3rd Qu. 0.000
> Max. 1.000
>
>
> Stage-pairs without connections leading to the rest of the life cycle found in matrices: 1 2 3 4 5 6 7 8 9 10 11 12Some basic analyses.
18.0.4 Complex historical stage-based (Lefkovitch), function-based model

This life history model includes 54 stages:
1) dormant seed
2) first-year protocorm (juvenile)
3) second-year protocorm (juvenile)
4) third-year protocorm (juvenile)
5) seedling (juvenile)
6) dormant adult
7-30) non-flowering but sprouting adults with 1-24 sprouts (1 stage per # sprouts)
31-54) flowering adults with 1-24 sprouts (1 stage per # sprouts)
The stageframe for this life history model can be programmed as follows.
data(cypdata)
stagevector <- c("SD", "P1", "P2", "P3", "SL", "Dorm", "1V", "2V", "3V", "4V",
"5V", "6V", "7V", "8V", "9V", "10V", "11V", "12V", "13V", "14V", "15V", "16V",
"17V", "18V", "19V", "20V", "21V", "22V", "23V", "24V", "1F", "2F", "3F",
"4F", "5F", "6F", "7F", "8F", "9F", "10F", "11F", "12F", "13F", "14F", "15F",
"16F", "17F", "18F", "19F", "20F", "21F", "22F", "23F", "24F")
indataset <- c(0, 0, 0, 0, 0, rep(1, 49))
sizevector <- c(0, 0, 0, 0, 0, seq(from = 0, t = 24), seq(from = 1, to = 24))
repvector <- c(0, 0, 0, 0, 0, rep(0, 25), rep(1, 24))
obsvector <- c(0, 0, 0, 0, 0, 0, rep(1, 48))
matvector <- c(0, 0, 0, 0, 0, rep(1, 49))
immvector <- c(0, 1, 1, 1, 1, rep(0, 49))
propvector <- c(1, rep(0, 53))
comments <- c("Dormant seed", "Yr1 protocorm", "Yr2 protocorm", "Yr3 protocorm",
"Seedling", "Veg dorm", "Veg adult 1 stem", "Veg adult 2 stems",
"Veg adult 3 stems", "Veg adult 4 stems", "Veg adult 5 stems",
"Veg adult 6 stems", "Veg adult 7 stems", "Veg adult 8 stems",
"Veg adult 9 stems", "Veg adult 10 stems", "Veg adult 11 stems",
"Veg adult 12 stems", "Veg adult 13 stems", "Veg adult 14 stems",
"Veg adult 15 stems", "Veg adult 16 stems", "Veg adult 17 stems",
"Veg adult 18 stems", "Veg adult 19 stems", "Veg adult 20 stems",
"Veg adult 21 stems", "Veg adult 22 stems", "Veg adult 23 stems",
"Veg adult 24 stems", "Flo adult 1 stem", "Flo adult 2 stems",
"Flo adult 3 stems", "Flo adult 4 stems", "Flo adult 5 stems",
"Flo adult 6 stems", "Flo adult 7 stems", "Flo adult 8 stems",
"Flo adult 9 stems", "Flo adult 10 stems", "Flo adult 11 stems",
"Flo adult 12 stems", "Flo adult 13 stems", "Flo adult 14 stems",
"Flo adult 15 stems", "Flo adult 16 stems", "Flo adult 17 stems",
"Flo adult 18 stems", "Flo adult 19 stems", "Flo adult 20 stems",
"Flo adult 21 stems", "Flo adult 22 stems", "Flo adult 23 stems",
"Flo adult 24 stems")
cyp_lefk_fb_frame <- sf_create(sizes = sizevector, stagenames = stagevector,
repstatus = repvector, obsstatus = obsvector, matstatus = matvector,
propstatus = propvector, immstatus = immvector, indataset = indataset,
comments = comments)Now we will create the MPM, first developing the standardized dataset, the supplemental data table, and the vital rate models.
cyp_lefk_fb_data <- verticalize3(data = cypdata, noyears = 6, firstyear = 2004,
patchidcol = "patch", individcol = "plantid", blocksize = 4,
sizeacol = "Inf2.04", sizebcol = "Inf.04", sizeccol = "Veg.04",
repstracol = "Inf.04", repstrbcol = "Inf2.04", fecacol = "Pod.04",
stageassign = cyp_lefk_fb_frame, stagesize = "sizeadded", NAas0 = TRUE,
age_offset = 4)
seeds_per_fruit <- 5000
sl_mult <- 0.7
cyp_lefk_fb_hsupp <- supplemental(stage3 = c("SD", "SD", "P1", "P1", "P2", "P3",
"SL", "SL", "SL", "Dorm", "1V", "2V", "3V", "Dorm", "1V", "2V", "3V", "mat",
"mat", "mat", "mat", "SD", "P1"),
stage2 = c("SD", "SD", "SD", "SD", "P1", "P2", "P3", "SL", "SL", "SL", "SL",
"SL", "SL", "SL", "SL", "SL", "SL", "Dorm", "1V", "2V", "3V", "rep", "rep"),
stage1 = c("SD", "rep", "SD", "rep", "SD", "P1", "P2", "P3", "SL", "P3", "P3",
"P3", "P3", "SL", "SL", "SL", "SL", "SL", "SL", "SL", "SL", "mat", "mat"),
eststage3 = c(NA, NA, NA, NA, NA, NA, NA, NA, NA, "Dorm", "1V", "2V", "3V",
"Dorm", "1V", "2V", "3V", "mat", "mat", "mat", "mat", NA, NA),
eststage2 = c(NA, NA, NA, NA, NA, NA, NA, NA, NA, "Dorm", "Dorm", "Dorm",
"Dorm", "Dorm", "Dorm", "Dorm", "Dorm", "Dorm", "1V", "2V", "3V", NA, NA),
eststage1 = c(NA, NA, NA, NA, NA, NA, NA, NA, NA, "Dorm", "Dorm", "Dorm",
"Dorm", "Dorm", "Dorm", "Dorm", "Dorm", "1V", "1V", "1V", "1V", NA, NA),
givenrate = c(0.08, 0.08, 0.1, 0.1, 0.1, 0.1, 0.1, 0.05, 0.05, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA),
multiplier = c(NA, NA, NA, NA, NA, NA, NA, NA, NA, sl_mult, sl_mult,
sl_mult, sl_mult, sl_mult, sl_mult, sl_mult, sl_mult, 1, 1, 1, 1,
0.5 * seeds_per_fruit, 0.5 * seeds_per_fruit),
type = c("S", "S", "S", "S", "S", "S", "S", "S", "S", "S", "S", "S", "S", "S",
"S", "S", "S", "S", "S", "S", "S", "R", "R"),
type_t12 = c("S", "F", "S", "F", "S", "S", "S", "S", "S", "S", "S", "S", "S",
"S", "S", "S", "S", "S", "S", "S", "S", "S", "S"),
stageframe = cyp_lefk_fb_frame, historical = TRUE)
> Warning: NA values in argument multiplier will be treated as 1 values.
cyp_lefk_fb_vital <- modelsearch(cyp_lefk_fb_data, historical = TRUE, approach = "mixed",
vitalrates = c("surv", "obs", "size", "repst", "fec"), patch = "patchid",
sizedist = "negbin", size.trunc = TRUE, fecdist = "poisson", fec.zero = TRUE,
suite = "main", size = c("size3added", "size2added", "size1added"),
quiet = "partial")
>
> Developing global model of survival probability...
>
> Global model of survival probability developed. Proceeding with model dredge...
>
> Developing global model of observation probability...
>
> Global model of observation probability developed. Proceeding with model dredge...
>
> Developing global model of primary size...
>
> Global model of primary size developed. Proceeding with model dredge...
>
> Developing global model of reproduction probability...
>
> Global model of reproduction probability developed. Proceeding with model dredge...
>
> Developing global model of fecundity...
>
> Global model of fecundity developed. Proceeding with model dredge...
>
> Finished selecting best-fit models.
cyp_lefk_fb_hmpm <- flefko3(stageframe = cyp_lefk_fb_frame, supplement = cyp_lefk_fb_hsupp,
modelsuite = cyp_lefk_fb_vital, data = cyp_lefk_fb_data, sparse_output = TRUE)
summary(cyp_lefk_fb_hmpm)
>
> This historical lefkoMat object contains 15 matrices.
>
> Each matrix is square with 2916 rows and columns, and a total of 8503056 elements.
> A total of 1768620 survival transitions were estimated, with 117908 per matrix.
> A total of 35280 fecundity transitions were estimated, with 2352 per matrix.
> This lefkoMat object covers 1 population, 3 patches, and 5 time steps.
>
> The dataset contains a total of 74 unique individuals and 320 unique transitions.
>
> Vital rate modeling quality control:
>
> Survival estimated with 74 individuals and 320 individual transitions.
> Observation estimated with 70 individuals and 303 individual transitions.
> Primary size estimated with 70 individuals and 288 individual transitions.
> Secondary size transition not estimated.
> Tertiary size transition not estimated.
> Reproductive status estimated with 70 individuals and 288 individual transitions.
> Fecundity estimated with 51 individuals and 118 individual transitions.
> Juvenile survival not estimated.
> Juvenile observation probability not estimated.
> Juvenile primary size transition not estimated.
> Juvenile secondary size transition not estimated.
> Juvenile tertiary size transition not estimated.
> Juvenile reproduction probability not estimated.
> Juvenile maturity transition probability not estimated.
>
> Survival probability sum check (each matrix represented by column in order):
> [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10] [,11] [,12]
> Min. 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000
> 1st Qu. 0.965 0.965 0.965 0.965 0.965 0.964 0.965 0.965 0.965 0.964 0.964 0.965
> Median 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000
> Mean 0.820 0.820 0.820 0.820 0.820 0.820 0.820 0.820 0.820 0.820 0.820 0.820
> 3rd Qu. 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000
> Max. 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000
> [,13] [,14] [,15]
> Min. 0.000 0.000 0.000
> 1st Qu. 0.965 0.964 0.964
> Median 1.000 1.000 1.000
> Mean 0.820 0.820 0.820
> 3rd Qu. 1.000 1.000 1.000
> Max. 1.000 1.000 1.000
>
>
> Stage-pairs without connections leading to them found in matrices: 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15Now some basic analyses.
lambda3(cyp_lefk_fb_hmpm)
> pop patch year2 lambda
> 1 1 A 2004 0.9790556
> 2 1 A 2005 0.9751305
> 3 1 A 2006 0.9779635
> 4 1 A 2007 0.9631103
> 5 1 A 2008 0.9772806
> 6 1 B 2004 0.9836386
> 7 1 B 2005 0.9778615
> 8 1 B 2006 0.9812224
> 9 1 B 2007 0.9673247
> 10 1 B 2008 0.9836171
> 11 1 C 2004 0.9844678
> 12 1 C 2005 0.9841072
> 13 1 C 2006 0.9861658
> 14 1 C 2007 0.9695788
> 15 1 C 2008 0.980886418.0.5 Age-based (Leslie) raw (empirical) model
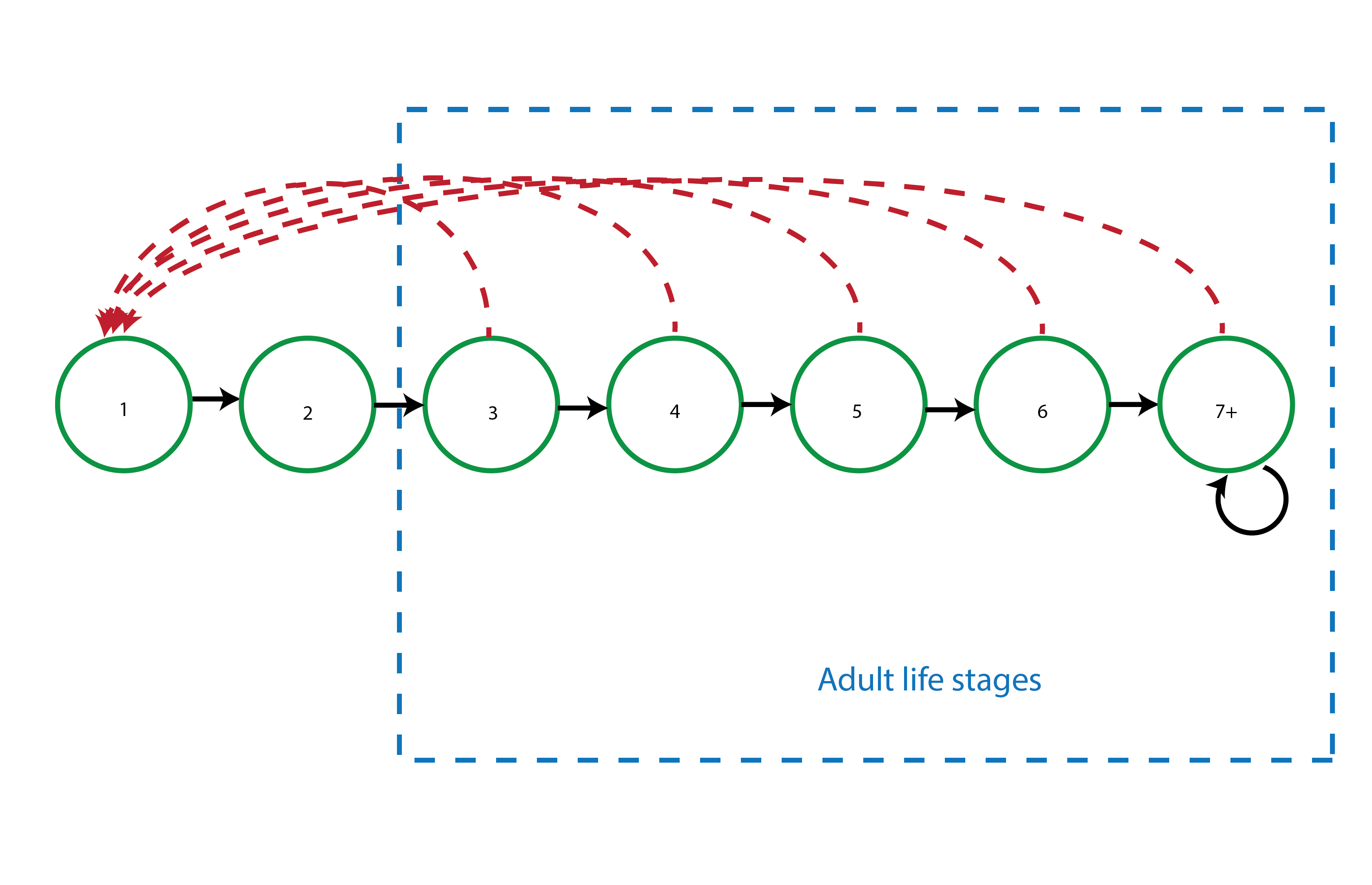
This life history model includes 7 main ages, assuming the possibility of survival past age 7. This model does not include a dormant seed stage, instead assuming that all seeds either germinate immediately or die.
This life history model does not require a stageframe to be programmed. Instead, we will standardize the dataset, develop a supplemental data table, and create the MPM.
data(cypdata)
cyp_lesl_data <- verticalize3(data = cypdata, noyears = 6, firstyear = 2004,
patchidcol = "patch", individcol = "plantid", blocksize = 4,
sizeacol = "Inf2.04", sizebcol = "Inf.04", sizeccol = "Veg.04",
repstracol = "Inf.04", repstrbcol = "Inf2.04", fecacol = "Pod.04",
stagesize = "sizeadded", NAas0 = TRUE, age_offset = 2)
germination <- 0.08
protocorm_to_seedling <- 0.10
seeding_to_adult <- 0.20
seeds_per_fruit <- 8000
cyp_lesl_supp <- supplemental(historical = FALSE, stagebased = FALSE,
agebased = TRUE, age2 = c(1, 2), type = c(1, 1),
givenrate = c(protocorm_to_seedling, seeding_to_adult))
cyp_lesl_raw_mpm <- rleslie(data = cyp_lesl_data, last_age = 7, fecage_min = 3,
fecmod = (germination * seeds_per_fruit), supplement = cyp_lesl_supp)Now some basic analyses.
lambda3(cyp_lesl_raw_mpm)
> Warning: A matrix with an eigenvalue of 0 has been detected.
> Warning: A matrix with an eigenvalue of 0 has been detected.
> pop patch year2 lambda
> 1 1 1 2004 1.367981
> 2 1 1 2005 1.602825
> 3 1 1 2006 0.000000
> 4 1 1 2007 0.000000
> 5 1 1 2008 0.962963
cyp_lesl_raw_meanmpm <- lmean(cyp_lesl_raw_mpm)
lambda3(cyp_lesl_raw_meanmpm)
> pop patch lambda
> 1 1 1 1.4262418.0.6 Age-based (Leslie) function-based model

This life history model includes 7 main ages, assuming the possibility of survival past age 7. This model does not include a dormant seed stage, instead assuming that all seeds either germinate immediately or die.
This life history model does not require a stageframe to be programmed. Instead, we will standardize the dataset, create the vital rate models, and develop a supplemental data table.
data(cypdata)
cyp_lesl_data <- verticalize3(data = cypdata, noyears = 6, firstyear = 2004,
patchidcol = "patch", individcol = "plantid", blocksize = 4,
sizeacol = "Inf2.04", sizebcol = "Inf.04", sizeccol = "Veg.04",
repstracol = "Inf.04", repstrbcol = "Inf2.04", fecacol = "Pod.04",
stagesize = "sizeadded", NAas0 = TRUE, age_offset = 2)
cyp_lesl_vital <- modelsearch(cyp_lesl_data, historical = FALSE,
approach = "mixed", suite = "cons", bestfit = "AICc&k", age = "obsage",
vitalrates = c("surv", "fec"), fecdist = "poisson", indiv = "individ",
year = "year2", year.as.random = TRUE, patch.as.random = TRUE,
show.model.tables = TRUE, fec.zero = TRUE, global.only = TRUE,
test.age = TRUE, quiet = "partial")
>
> Developing global model of survival probability...
>
> Global model of survival probability developed. Proceeding with model dredge...
>
> Developing global model of fecundity...
>
> Global model of fecundity developed. Proceeding with model dredge...
germination <- 0.08
protocorm_to_seedling <- 0.10
seeding_to_adult <- 0.20
seeds_per_fruit <- 8000
cyp_lesl_supp <- supplemental(historical = FALSE, stagebased = FALSE,
agebased = TRUE, age2 = c(1, 2), type = c(1, 1),
givenrate = c(protocorm_to_seedling, seeding_to_adult))Now we will create the MPM.
cyp_lesl_fb_mpm <- fleslie(data = cyp_lesl_data, modelsuite = cyp_lesl_vital,
last_age = 7, fecage_min = 3, fecmod = (germination * seeds_per_fruit),
supplement = cyp_lesl_supp)
summary(cyp_lesl_fb_mpm)
>
> This ahistorical lefkoMat object contains 5 matrices.
>
> Each matrix is square with 7 rows and columns, and a total of 49 elements.
> A total of 35 survival transitions were estimated, with 7 per matrix.
> A total of 25 fecundity transitions were estimated, with 5 per matrix.
> This lefkoMat object covers 1 population, 1 patch, and 5 time steps.
>
> The dataset contains a total of 74 unique individuals and 320 unique transitions.
>
> Vital rate modeling quality control:
>
> Survival estimated with 74 individuals and 320 individual transitions.
> Observation probability not estimated.
> Primary size transition not estimated.
> Secondary size transition not estimated.
> Tertiary size transition not estimated.
> Reproduction probability not estimated.
> Fecundity estimated with 74 individuals and 320 individual transitions.
> Juvenile survival not estimated.
> Juvenile observation probability not estimated.
> Juvenile primary size transition not estimated.
> Juvenile secondary size transition not estimated.
> Juvenile tertiary size transition not estimated.
> Juvenile reproduction probability not estimated.
> Juvenile maturity transition probability not estimated.
>
> Survival probability sum check (each matrix represented by column in order):
> [,1] [,2] [,3] [,4] [,5]
> Min. 0.100 0.100 0.100 0.100 0.100
> 1st Qu. 0.573 0.573 0.573 0.573 0.573
> Median 0.946 0.946 0.946 0.946 0.946
> Mean 0.719 0.719 0.719 0.719 0.719
> 3rd Qu. 0.947 0.947 0.947 0.947 0.947
> Max. 0.949 0.949 0.949 0.949 0.949
>
>
> All matrices have no stage discontinuities.Finally some analyses.
18.0.7 Hybrid age-based (Leslie) model with dormant propagule
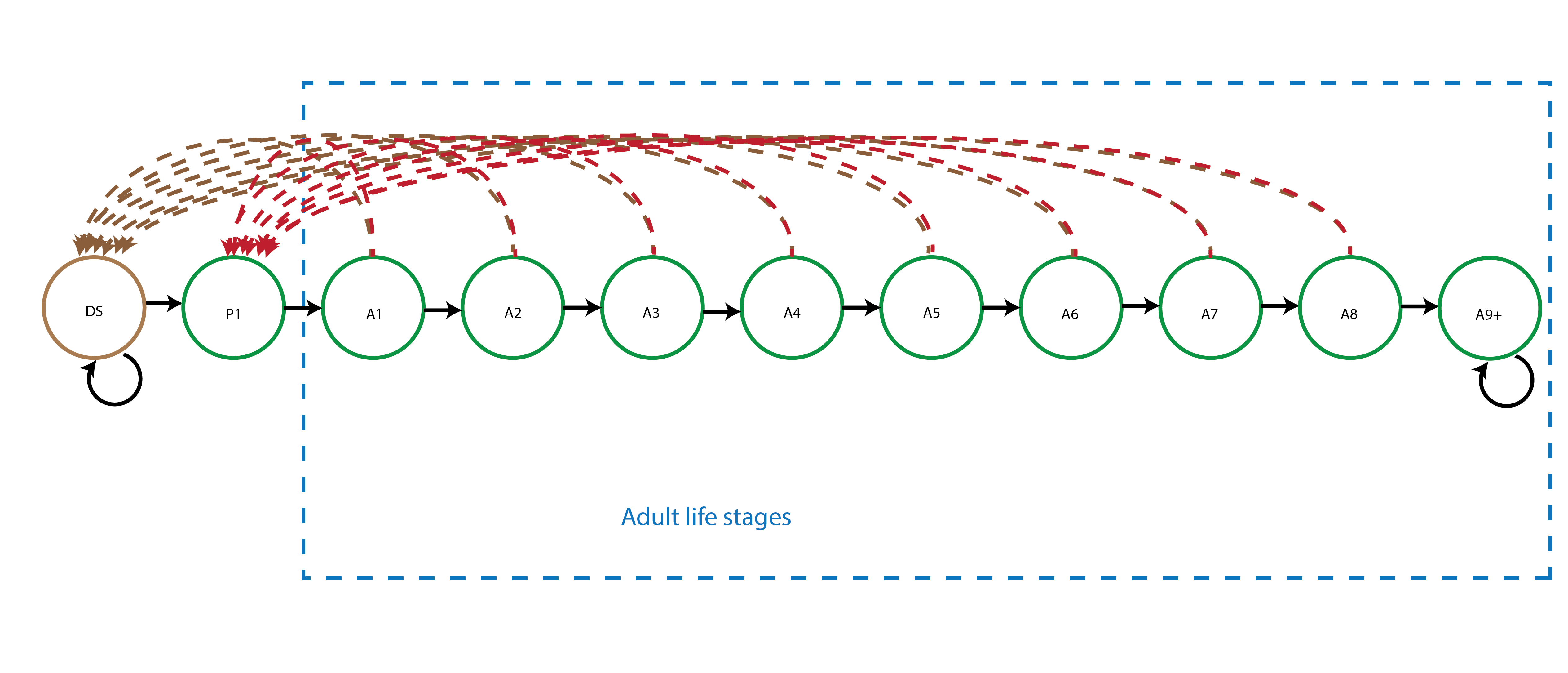
This life history model includes 7 (st)ages after germination, including 2 juvenile stages (protocorm and seedling) and 5 adult ages, plus a dormant seed stage.
We will use a Leslie model as the backbone for this. Leslie MPMs do not require stageframes to be built, but include auto-generated stageframes within the associated lefkoMat object that is produced. The first part of this exercise is the same as in the previous Leslie exercise.
data(cypdata)
cyp_lesl_data <- verticalize3(data = cypdata, noyears = 6, firstyear = 2004,
patchidcol = "patch", individcol = "plantid", blocksize = 4,
sizeacol = "Inf2.04", sizebcol = "Inf.04", sizeccol = "Veg.04",
repstracol = "Inf.04", repstrbcol = "Inf2.04", fecacol = "Pod.04",
stagesize = "sizeadded", NAas0 = TRUE, age_offset = 2)
cyp_lesl_vital <- modelsearch(cyp_lesl_data, historical = FALSE,
approach = "mixed", suite = "cons", bestfit = "AICc&k", age = "obsage",
vitalrates = c("surv", "fec"), fecdist = "poisson", indiv = "individ",
year = "year2", year.as.random = TRUE, patch.as.random = TRUE,
show.model.tables = TRUE, fec.zero = TRUE, global.only = TRUE,
test.age = TRUE, quiet = "partial")
>
> Developing global model of survival probability...
>
> Global model of survival probability developed. Proceeding with model dredge...
>
> Developing global model of fecundity...
>
> Global model of fecundity developed. Proceeding with model dredge...
germination <- 0.08
protocorm_to_seedling <- 0.10
seeding_to_adult <- 0.20
seeds_per_fruit <- 8000
cyp_lesl_supp <- supplemental(historical = FALSE, stagebased = FALSE,
agebased = TRUE, age2 = c(1, 2), type = c(1, 1),
givenrate = c(protocorm_to_seedling, seeding_to_adult))Now we will create the Leslie MPM.
cyp_lesl_fb_mpm <- fleslie(data = cyp_lesl_data, modelsuite = cyp_lesl_vital,
last_age = 7, fecage_min = 3, fecmod = (germination * seeds_per_fruit),
supplement = cyp_lesl_supp)
summary(cyp_lesl_fb_mpm)
>
> This ahistorical lefkoMat object contains 5 matrices.
>
> Each matrix is square with 7 rows and columns, and a total of 49 elements.
> A total of 35 survival transitions were estimated, with 7 per matrix.
> A total of 25 fecundity transitions were estimated, with 5 per matrix.
> This lefkoMat object covers 1 population, 1 patch, and 5 time steps.
>
> The dataset contains a total of 74 unique individuals and 320 unique transitions.
>
> Vital rate modeling quality control:
>
> Survival estimated with 74 individuals and 320 individual transitions.
> Observation probability not estimated.
> Primary size transition not estimated.
> Secondary size transition not estimated.
> Tertiary size transition not estimated.
> Reproduction probability not estimated.
> Fecundity estimated with 74 individuals and 320 individual transitions.
> Juvenile survival not estimated.
> Juvenile observation probability not estimated.
> Juvenile primary size transition not estimated.
> Juvenile secondary size transition not estimated.
> Juvenile tertiary size transition not estimated.
> Juvenile reproduction probability not estimated.
> Juvenile maturity transition probability not estimated.
>
> Survival probability sum check (each matrix represented by column in order):
> [,1] [,2] [,3] [,4] [,5]
> Min. 0.100 0.100 0.100 0.100 0.100
> 1st Qu. 0.573 0.573 0.573 0.573 0.573
> Median 0.946 0.946 0.946 0.946 0.946
> Mean 0.719 0.719 0.719 0.719 0.719
> 3rd Qu. 0.947 0.947 0.947 0.947 0.947
> Max. 0.949 0.949 0.949 0.949 0.949
>
>
> All matrices have no stage discontinuities.Next we will create the age-hybrid MPM, and edit the structure to include the proper stage description and the proper transition values for the new stage.
cyp_hybrid <- add_stage(cyp_lesl_fb_mpm, add_before = 1,
stage_name = "DormantSeed")
cyp_hybrid$ahstages$propstatus[1] <- 1 # Dormant seeds are propagules
cyp_hybrid <- edit_lM(cyp_hybrid,
stage3 = c("DormantSeed", "Age1", "Age1", "DormantSeed"),
stage2 = c("DormantSeed", "DormantSeed", "rep", "rep"),
eststage3 = c(NA, NA, NA, "Age1"),
eststage2 = c(NA, NA, NA, "rep"),
givenrate = c(0.5, 0.2, NA, NA),
multiplier = c(NA, NA, 0.5, 1),
type = c(1, 1, 3, 2))
summary(cyp_hybrid)
>
> This ahistorical lefkoMat object contains 5 matrices.
>
> Each matrix is square with 8 rows and columns, and a total of 64 elements.
> A total of 45 survival transitions were estimated, with 9 per matrix.
> A total of 25 fecundity transitions were estimated, with 5 per matrix.
> This lefkoMat object covers 1 population, 1 patch, and 5 time steps.
>
> The dataset contains a total of 74 unique individuals and 320 unique transitions.
>
> Vital rate modeling quality control:
>
> Survival estimated with 74 individuals and 320 individual transitions.
> Observation probability not estimated.
> Primary size transition not estimated.
> Secondary size transition not estimated.
> Tertiary size transition not estimated.
> Reproduction probability not estimated.
> Fecundity estimated with 74 individuals and 320 individual transitions.
> Juvenile survival not estimated.
> Juvenile observation probability not estimated.
> Juvenile primary size transition not estimated.
> Juvenile secondary size transition not estimated.
> Juvenile tertiary size transition not estimated.
> Juvenile reproduction probability not estimated.
> Juvenile maturity transition probability not estimated.
>
> Survival probability sum check (each matrix represented by column in order):
> [,1] [,2] [,3] [,4] [,5]
> Min. 0.100 0.100 0.100 0.100 0.100
> 1st Qu. 0.575 0.575 0.575 0.575 0.575
> Median 0.946 0.946 0.946 0.946 0.946
> Mean 0.717 0.717 0.717 0.717 0.717
> 3rd Qu. 0.947 0.947 0.947 0.947 0.947
> Max. 0.949 0.949 0.949 0.949 0.949
>
>
> All matrices have no stage discontinuities.Finally some analyses.
18.0.8 Simple raw (empirical) age-by-stage model
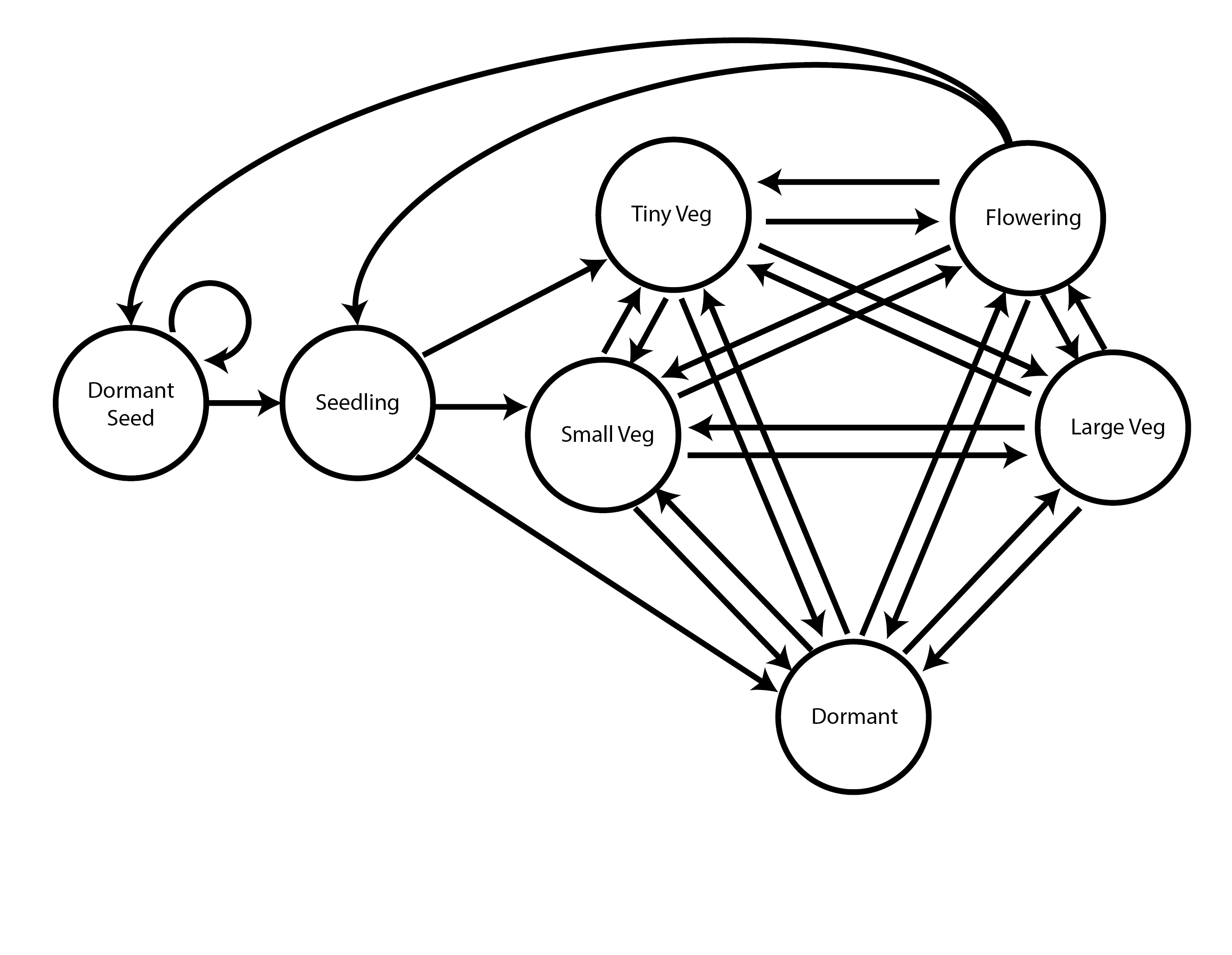
This life history model assumes 7 stages, including 4 aboveground adult stages of various sizes and 1 belowground adult stage.
The stageframe can be programmed as follows.
data(lathyrus)
sizevector <- c(0, 100, 13, 127, 3730, 3800, 0)
stagevector <- c("Sd", "Sdl", "VSm", "Sm", "VLa", "Flo", "Dorm")
repvector <- c(0, 0, 0, 0, 0, 1, 0)
obsvector <- c(0, 1, 1, 1, 1, 1, 0)
matvector <- c(0, 0, 1, 1, 1, 1, 1)
immvector <- c(1, 1, 0, 0, 0, 0, 0)
propvector <- c(1, 0, 0, 0, 0, 0, 0)
indataset <- c(0, 1, 1, 1, 1, 1, 1)
binvec <- c(0, 100, 11, 103, 3500, 3800, 0.5)
lathframe_raw <- sf_create(sizes = sizevector, stagenames = stagevector,
repstatus = repvector, obsstatus = obsvector, matstatus = matvector,
immstatus = immvector, indataset = indataset, binhalfwidth = binvec,
propstatus = propvector)Next we will standardize the dataset, input the supplemental data, and create the MPM.
lathyrus$indiv_id <- paste(lathyrus$SUBPLOT, lathyrus$GENET)
lathvert_raw <- verticalize3(lathyrus, noyears = 4, firstyear = 1988,
patchidcol = "SUBPLOT", individcol = "indiv_id", blocksize = 9,
juvcol = "Seedling1988", sizeacol = "Volume88", repstracol = "FCODE88",
fecacol = "Intactseed88", deadacol = "Dead1988", nonobsacol = "Dormant1988",
stageassign = lathframe_raw, stagesize = "sizea", censorcol = "Missing1988",
censorkeep = NA, censorRepeat = TRUE, censor = TRUE)
lathvert_raw_small <- subset(lathvert_raw, firstseen > 1988)
lathsupp2 <- supplemental(stage3 = c("Sd", "Sdl", "Sd", "Sdl"),
stage2 = c("Sd", "Sd", "rep", "rep"), givenrate = c(0.345, 0.054, NA, NA),
multiplier = c(NA, NA, 0.345, 0.054), type = c(1, 1, 3, 3),
stageframe = lathframe_raw, historical = FALSE, stagebased = TRUE,
agebased = TRUE)
> Warning: NA values in argument multiplier will be treated as 1 values.
lathmat2p_raw <- arlefko2(data = lathvert_raw_small, stageframe = lathframe_raw,
supplement = lathsupp2, stages = c("stage3", "stage2", "stage1"),
patch = "all", patchcol = "patchid", yearcol = "year2", agecol = "obsage",
indivcol = "individ")
summary(lathmat2p_raw)
>
> This ahistorical lefkoMat object contains 12 matrices.
>
> Each matrix is square with 21 rows and columns, and a total of 441 elements.
> A total of 209 survival transitions were estimated, with 17.417 per matrix.
> A total of 12 fecundity transitions were estimated, with 1 per matrix.
> This lefkoMat object covers 1 population, 6 patches, and 2 time steps.
>
> The dataset contains a total of 345 unique individuals and 531 unique transitions.
>
> Survival probability sum check (each matrix represented by column in order):
> [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10] [,11] [,12]
> Min. 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000
> 1st Qu. 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000
> Median 0.000 0.000 0.000 0.399 0.000 0.399 0.000 0.399 0.000 0.399 0.000 0.000
> Mean 0.281 0.349 0.261 0.385 0.274 0.463 0.268 0.423 0.223 0.388 0.234 0.279
> 3rd Qu. 0.399 0.778 0.399 0.750 0.399 1.000 0.399 0.778 0.399 0.818 0.399 0.399
> Max. 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.000
>
>
> Age-stages without connections leading to the rest of the life cycle found in matrices: 1 2 3 4 5 6 7 8 9 10 11 12Finally, some basic analyses.
18.0.9 Complex function-based age-by-stage model
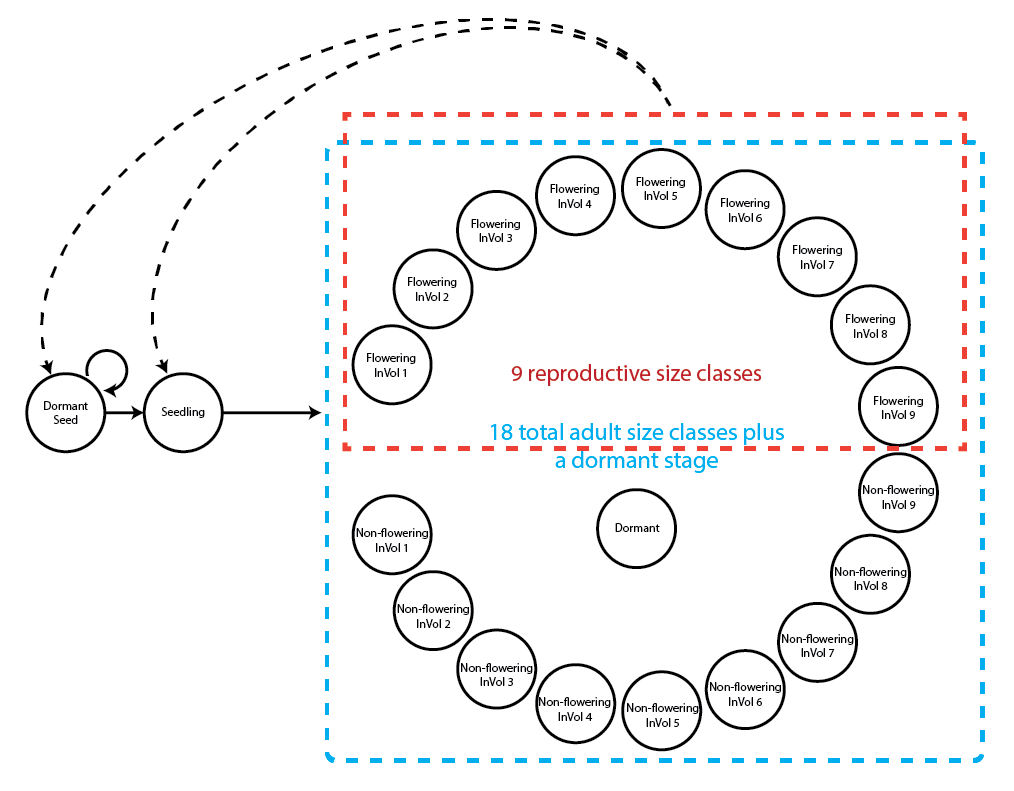
This life history model includes 21 life history stages, of which 19 are adult size-based stages.
Here we program the stageframe.
data(lathyrus)
sizevector <- c(0, 4.6, 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 1, 2, 3, 4, 5, 6, 7, 8, 9)
stagevector <- c("Sd", "Sdl", "Dorm", "Sz1nr", "Sz2nr", "Sz3nr", "Sz4nr",
"Sz5nr","Sz6nr", "Sz7nr", "Sz8nr", "Sz9nr", "Sz1r", "Sz2r", "Sz3r", "Sz4r",
"Sz5r", "Sz6r", "Sz7r", "Sz8r", "Sz9r")
repvector <- c(0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1)
obsvector <- c(0, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1)
matvector <- c(0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1)
immvector <- c(1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0)
propvector <- c(1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0)
indataset <- c(0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1)
binvec <- c(0, 4.6, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5,
0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5)
minima <- c(1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2)
maxima <- c(NA, 1, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA)
comments <- c("Dormant seed", "Seedling", "Dormant", "Size 1 Veg", "Size 2 Veg",
"Size 3 Veg", "Size 4 Veg", "Size 5 Veg", "Size 6 Veg", "Size 7 Veg",
"Size 8 Veg", "Size 9 Veg", "Size 1 Flo", "Size 2 Flo", "Size 3 Flo",
"Size 4 Flo", "Size 5 Flo", "Size 6 Flo", "Size 7 Flo", "Size 8 Flo",
"Size 9 Flo")
lathframeln <- sf_create(sizes = sizevector, stagenames = stagevector,
repstatus = repvector, obsstatus = obsvector, propstatus = propvector,
immstatus = immvector, matstatus = matvector, indataset = indataset,
binhalfwidth = binvec, minage = minima, maxage = maxima, comments = comments)Next we will standardize the dataset, input the supplemental data, create the vital rate models, and create our MPM.
lathyrus$indiv_id <- paste(lathyrus$SUBPLOT, lathyrus$GENET)
lathvertln <- verticalize3(lathyrus, noyears = 4, firstyear = 1988,
patchidcol = "SUBPLOT", individcol = "indiv_id", blocksize = 9,
juvcol = "Seedling1988", sizeacol = "lnVol88", repstracol = "FCODE88",
fecacol = "Intactseed88", deadacol = "Dead1988", nonobsacol = "Dormant1988",
stageassign = lathframeln, stagesize = "sizea", censorcol = "Missing1988",
censorkeep = NA, censorRepeat = TRUE, NAas0 = TRUE, censor = TRUE)
lathvertln_small <- subset(lathvertln, firstseen > 1988)
lathvertln$feca3 <- round(lathvertln$feca3)
lathvertln$feca2 <- round(lathvertln$feca2)
lathvertln$feca1 <- round(lathvertln$feca1)
lathsupp2 <- supplemental(stage3 = c("Sd", "Sdl", "Sd", "Sdl"),
stage2 = c("Sd", "Sd", "rep", "rep"), givenrate = c(0.345, 0.054, NA, NA),
multiplier = c(NA, NA, 0.345, 0.054), type = c(1, 1, 3, 3),
stageframe = lathframeln, historical = FALSE, stagebased = TRUE,
agebased = TRUE)
> Warning: NA values in argument multiplier will be treated as 1 values.
lathmodelsln2 <- modelsearch(lathvertln_small, historical = FALSE,
approach = "mixed", suite = "main", bestfit = "AICc&k", juvestimate = "Sdl",
vitalrates = c("surv", "obs", "size", "repst", "fec"), sizedist = "gaussian",
fecdist = "negbin", indiv = "individ", year = "year2", age = "obsage",
year.as.random = TRUE, patch.as.random = TRUE, show.model.tables = TRUE,
fec.zero = TRUE, test.age = TRUE, quiet = "partial")
>
> Developing global model of survival probability...
>
> Global model of survival probability developed. Proceeding with model dredge...
>
> Developing global model of observation probability...
>
> Global model of observation probability developed. Proceeding with model dredge...
>
> Developing global model of primary size...
>
> Global model of primary size developed. Proceeding with model dredge...
>
> Developing global model of reproduction probability...
>
> Global model of reproduction probability developed. Proceeding with model dredge...
>
> Developing global model of fecundity...
>
> Global model of fecundity developed. Proceeding with model dredge...
>
> Developing global model of juvenile survival probability...
>
> Global model of juvenile survival probability developed. Proceeding with model dredge...
> Warning: Juvenile maturity status in time t+1 appears to be constant (1). Setting to constant.
>
> Developing global model of juvenile observation probability...
>
> Global model of juvenile observation probability developed. Proceeding with model dredge...
>
> Developing global model of juvenile primary size...
>
> Global model estimation failed. Dropping individual identity term.
>
> Redeveloping global model of juvenile primary size...
>
> Global model of juvenile primary size developed. Proceeding with model dredge...
> Warning: Juvenile reproductive status in time t+1 appears to be constant, and so will be set to constant.
>
> Finished selecting best-fit models.
lathmat2age <- aflefko2(year = "all", stageframe = lathframeln,
modelsuite = lathmodelsln2, data = lathvertln_small, supplement = lathsupp2,
continue = TRUE, reduce = FALSE)
summary(lathmat2age)
>
> This ahistorical lefkoMat object contains 2 matrices.
>
> Each matrix is square with 63 rows and columns, and a total of 3969 elements.
> A total of 1434 survival transitions were estimated, with 717 per matrix.
> A total of 72 fecundity transitions were estimated, with 36 per matrix.
> This lefkoMat object covers 1 population, 1 patch, and 2 time steps.
>
> The dataset contains a total of 345 unique individuals and 531 unique transitions.
>
> Vital rate modeling quality control:
>
> Survival estimated with 247 individuals and 347 individual transitions.
> Observation estimated with 203 individuals and 294 individual transitions.
> Primary size estimated with 191 individuals and 266 individual transitions.
> Secondary size transition not estimated.
> Tertiary size transition not estimated.
> Reproductive status estimated with 191 individuals and 266 individual transitions.
> Fecundity estimated with 16 individuals and 19 individual transitions.
> Juvenile survival estimated with 184 individuals and 184 individual transitions.
> Juvenile observation estimated with 137 individuals and 137 individual transitions.
> Juvenile primary size estimated with 130 individuals and 130 individual transitions.
> Juvenile secondary size transition not estimated.
> Juvenile tertiary size transition not estimated.
> Juvenile reproduction probability not estimated.
> Juvenile maturity transition probability not estimated.
>
> Survival probability sum check (each matrix represented by column in order):
> [,1] [,2]
> Min. 0.000 0.000
> 1st Qu. 0.000 0.000
> Median 0.185 0.185
> Mean 0.352 0.365
> 3rd Qu. 0.861 0.955
> Max. 1.000 1.000
>
>
> Age-stages without connections leading to the rest of the life cycle found in matrices: 1 2Now some basic analyses.
18.0.10 Ahistorical discretized integral projection model (IPM)
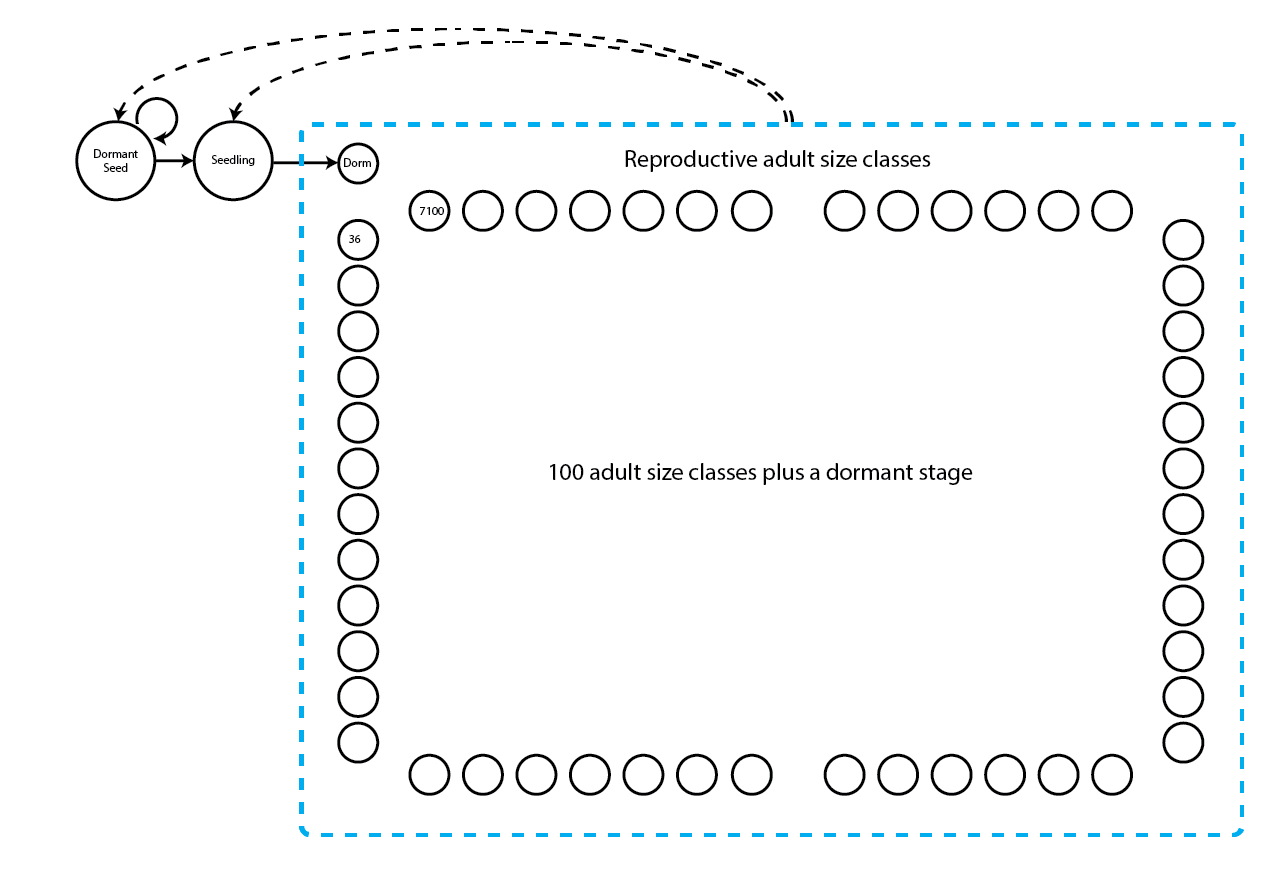
This life history model includes 103 stages, of which 100 are adult, size-classified IPM bins.
Here we program the stageframe.
data(lathyrus)
sizevector <- c(0, 100, 0, 1, 9)
stagevector <- c("Sd", "Sdl", "Dorm", "ipm", "ipm")
repvector <- c(0, 0, 0, 1, 1)
obsvector <- c(0, 1, 0, 1, 1)
matvector <- c(0, 0, 1, 1, 1)
immvector <- c(1, 1, 0, 0, 0)
propvector <- c(1, 0, 0, 0, 0)
indataset <- c(0, 1, 1, 1, 1)
binvec <- c(0, 100, 0.5, 0.5, 0.5)
comments <- c("Dormant seed", "Seedling", "Dormant", "ipm adult stage",
"ipm adult stage")
lathframeipm <- sf_create(sizes = sizevector, stagenames = stagevector,
repstatus = repvector, obsstatus = obsvector, propstatus = propvector,
immstatus = immvector, matstatus = matvector, comments = comments,
indataset = indataset, binhalfwidth = binvec, ipmbins = 100, roundsize = 3)
> Warning: Values supplied in vectors sizemin, sizemax, and binhalfwidth will be overwritten for ipm bins.Now we will standardize the dataset, input the supplemental data, estimate the vital rate models, and create the IPM.
lathyrus$indiv_id <- paste(lathyrus$SUBPLOT, lathyrus$GENET)
lathvertipm <- verticalize3(lathyrus, noyears = 4, firstyear = 1988,
individcol = "indiv_id", blocksize = 9, juvcol = "Seedling1988",
sizeacol = "lnVol88", repstracol = "FCODE88", fecacol = "Intactseed88",
deadacol = "Dead1988", nonobsacol = "Dormant1988", stageassign = lathframeipm,
stagesize = "sizea", censorcol = "Missing1988", censorkeep = NA,
censorRepeat = TRUE, censor = TRUE, NAas0 = TRUE, NRasRep = TRUE)
> Warning: Some stages occurring in the dataset do not match any characteristics
> in the input stageframe.
lathvertipm$feca3 <- round(lathvertipm$feca3)
lathvertipm$feca2 <- round(lathvertipm$feca2)
lathvertipm$feca1 <- round(lathvertipm$feca1)
lathsupp2 <- supplemental(stage3 = c("Sd", "Sdl", "Sd", "Sdl"),
stage2 = c("Sd", "Sd", "rep", "rep"),
givenrate = c(0.345, 0.054, NA, NA),
multiplier = c(NA, NA, 0.345, 0.054),
type = c(1, 1, 3, 3), stageframe = lathframeipm, historical = FALSE)
> Warning: NA values in argument multiplier will be treated as 1 values.
lathmodels2ipm <- modelsearch(lathvertipm, historical = FALSE,
approach = "mixed", suite = "size", juvestimate = "Sdl",
vitalrates = c("surv", "obs", "size", "fec"), bestfit = "AICc&k",
sizedist = "gaussian", fecdist = "negbin", fec.zero = TRUE, indiv = "individ",
year = "year2", year.as.random = TRUE, juvsize = TRUE,
show.model.tables = TRUE, quiet = "partial")
>
> Developing global model of survival probability...
>
> Global model estimation failed. Dropping individual identity term.
>
> Redeveloping global model of survival probability...
>
> Global model of survival probability developed. Proceeding with model dredge...
>
> Developing global model of observation probability...
>
> Global model of observation probability developed. Proceeding with model dredge...
>
> Developing global model of primary size...
>
> Global model of primary size developed. Proceeding with model dredge...
>
> Developing global model of fecundity...
>
> Global model of fecundity developed. Proceeding with model dredge...
>
> Developing global model of juvenile survival probability...
>
> Global model of juvenile survival probability developed. Proceeding with model dredge...
> Warning: Juvenile maturity status in time t+1 appears to be constant (1). Setting to constant.
>
> Developing global model of juvenile observation probability...
>
> Global model of juvenile observation probability developed. Proceeding with model dredge...
>
> Developing global model of juvenile primary size...
>
> Global model estimation failed. Dropping individual identity term.
>
> Redeveloping global model of juvenile primary size...
>
> Global model of juvenile primary size developed. Proceeding with model dredge...
>
> Finished selecting best-fit models.
lathmat2ipm <- flefko2(stageframe = lathframeipm, modelsuite = lathmodels2ipm,
supplement = lathsupp2, data = lathvertipm, reduce = FALSE)
summary(lathmat2ipm)
>
> This ahistorical lefkoMat object contains 3 matrices.
>
> Each matrix is square with 103 rows and columns, and a total of 10609 elements.
> A total of 30608 survival transitions were estimated, with 10202.667 per matrix.
> A total of 600 fecundity transitions were estimated, with 200 per matrix.
> This lefkoMat object covers 1 population, 1 patch, and 3 time steps.
>
> The dataset contains a total of 1053 unique individuals and 2527 unique transitions.
>
> Vital rate modeling quality control:
>
> Survival estimated with 931 individuals and 2246 individual transitions.
> Observation estimated with 858 individuals and 2121 individual transitions.
> Primary size estimated with 845 individuals and 1916 individual transitions.
> Secondary size transition not estimated.
> Tertiary size transition not estimated.
> Reproduction probability not estimated.
> Fecundity estimated with 931 individuals and 2246 individual transitions.
> Juvenile survival estimated with 281 individuals and 281 individual transitions.
> Juvenile observation estimated with 210 individuals and 210 individual transitions.
> Juvenile primary size estimated with 193 individuals and 193 individual transitions.
> Juvenile secondary size transition not estimated.
> Juvenile tertiary size transition not estimated.
> Juvenile reproduction probability not estimated.
> Juvenile maturity transition probability not estimated.
>
> Survival probability sum check (each matrix represented by column in order):
> [,1] [,2] [,3]
> Min. 0.000 0.000 0.000
> 1st Qu. 0.922 0.921 0.921
> Median 0.951 0.951 0.951
> Mean 0.930 0.930 0.930
> 3rd Qu. 0.970 0.970 0.970
> Max. 0.982 0.982 0.982
>
>
> Stages without connections leading to the rest of the life cycle found in matrices: 1 2 3Now some basic analyses.
18.0.11 Historical discretized integral projection model (IPM)

This life history model includes 103 stages, of which 100 are adult, size-classified IPM bins.
Here we program the stageframe.
data(lathyrus)
sizevector <- c(0, 100, 0, 1, 9)
stagevector <- c("Sd", "Sdl", "Dorm", "ipm", "ipm")
repvector <- c(0, 0, 0, 1, 1)
obsvector <- c(0, 1, 0, 1, 1)
matvector <- c(0, 0, 1, 1, 1)
immvector <- c(1, 1, 0, 0, 0)
propvector <- c(1, 0, 0, 0, 0)
indataset <- c(0, 1, 1, 1, 1)
binvec <- c(0, 100, 0.5, 0.5, 0.5)
comments <- c("Dormant seed", "Seedling", "Dormant", "ipm adult stage",
"ipm adult stage")
lathframeipm <- sf_create(sizes = sizevector, stagenames = stagevector,
repstatus = repvector, obsstatus = obsvector, propstatus = propvector,
immstatus = immvector, matstatus = matvector, comments = comments,
indataset = indataset, binhalfwidth = binvec, ipmbins = 100, roundsize = 3)
> Warning: Values supplied in vectors sizemin, sizemax, and binhalfwidth will be overwritten for ipm bins.Now we will standardize the dataset, input the supplemental data, estimate the vital rate models, and create the IPM.
lathyrus$indiv_id <- paste(lathyrus$SUBPLOT, lathyrus$GENET)
lathvertipm <- verticalize3(lathyrus, noyears = 4, firstyear = 1988,
individcol = "indiv_id", blocksize = 9, juvcol = "Seedling1988",
sizeacol = "lnVol88", repstracol = "FCODE88", fecacol = "Intactseed88",
deadacol = "Dead1988", nonobsacol = "Dormant1988", stageassign = lathframeipm,
stagesize = "sizea", censorcol = "Missing1988", censorkeep = NA,
censorRepeat = TRUE, censor = TRUE, NAas0 = TRUE, NRasRep = TRUE)
> Warning: Some stages occurring in the dataset do not match any characteristics
> in the input stageframe.
lathvertipm$feca3 <- round(lathvertipm$feca3)
lathvertipm$feca2 <- round(lathvertipm$feca2)
lathvertipm$feca1 <- round(lathvertipm$feca1)
lathsupp3 <- supplemental(stage3 = c("Sd","Sd","Sdl","Sdl","npr","Sd","Sdl"),
stage2 = c("Sd", "Sd", "Sd", "Sd", "Sdl", "rep", "rep"),
stage1 = c("Sd", "rep", "Sd", "rep", "Sd", "mat", "mat"),
eststage3 = c(NA, NA, NA, NA, "npr", NA, NA),
eststage2 = c(NA, NA, NA, NA, "Sdl", NA, NA),
eststage1 = c(NA, NA, NA, NA, "Sdl", NA, NA),
givenrate = c(0.345, 0.345, 0.054, 0.054, NA, NA, NA),
multiplier = c(NA, NA, NA, NA, NA, 0.345, 0.054),
type = c(1, 1, 1, 1, 1, 3, 3), type_t12 = c(1, 2, 1, 2, 1, 1, 1),
stageframe = lathframeipm, historical = TRUE)
> Warning: NA values in argument multiplier will be treated as 1 values.
lathmodels3ipm <- modelsearch(lathvertipm, historical = TRUE, approach= "mixed",
suite = "size", vitalrates = c("surv", "obs", "size", "fec"),
juvestimate = "Sdl", bestfit = "AICc&k", sizedist = "Gaussian",
fecdist = "negbin", fec.zero = TRUE, indiv = "individ", year = "year2",
year.as.random = TRUE, juvsize = TRUE, show.model.tables = TRUE,
quiet = "partial")
>
> Developing global model of survival probability...
>
> Global model of survival probability developed. Proceeding with model dredge...
>
> Developing global model of observation probability...
>
> Global model of observation probability developed. Proceeding with model dredge...
>
> Developing global model of primary size...
>
> Global model of primary size developed. Proceeding with model dredge...
>
> Developing global model of fecundity...
>
> Global model of fecundity developed. Proceeding with model dredge...
>
> Developing global model of juvenile survival probability...
>
> Global model of juvenile survival probability developed. Proceeding with model dredge...
> Warning: Juvenile maturity status in time t+1 appears to be constant (1). Setting to constant.
>
> Developing global model of juvenile observation probability...
>
> Global model of juvenile observation probability developed. Proceeding with model dredge...
>
> Developing global model of juvenile primary size...
>
> Global model estimation failed. Dropping individual identity term.
>
> Redeveloping global model of juvenile primary size...
>
> Global model of juvenile primary size developed. Proceeding with model dredge...
>
> Finished selecting best-fit models.
lathmat3ipm <- flefko3(stageframe = lathframeipm, modelsuite = lathmodels3ipm,
supplement = lathsupp3, data = lathvertipm, reduce = FALSE,
sparse_output = TRUE)
summary(lathmat3ipm)
>
> This historical lefkoMat object contains 3 matrices.
>
> Each matrix is square with 10609 rows and columns, and a total of 112550881 elements.
> A total of 3121968 survival transitions were estimated, with 1040656 per matrix.
> A total of 60600 fecundity transitions were estimated, with 20200 per matrix.
> This lefkoMat object covers 1 population, 1 patch, and 3 time steps.
>
> The dataset contains a total of 1053 unique individuals and 2527 unique transitions.
>
> Vital rate modeling quality control:
>
> Survival estimated with 931 individuals and 2246 individual transitions.
> Observation estimated with 858 individuals and 2121 individual transitions.
> Primary size estimated with 845 individuals and 1916 individual transitions.
> Secondary size transition not estimated.
> Tertiary size transition not estimated.
> Reproduction probability not estimated.
> Fecundity estimated with 931 individuals and 2246 individual transitions.
> Juvenile survival estimated with 281 individuals and 281 individual transitions.
> Juvenile observation estimated with 210 individuals and 210 individual transitions.
> Juvenile primary size estimated with 193 individuals and 193 individual transitions.
> Juvenile secondary size transition not estimated.
> Juvenile tertiary size transition not estimated.
> Juvenile reproduction probability not estimated.
> Juvenile maturity transition probability not estimated.
>
> Survival probability sum check (each matrix represented by column in order):
> [,1] [,2] [,3]
> Min. 0.000 0.000 0.000
> 1st Qu. 0.996 1.000 1.000
> Median 1.000 1.000 1.000
> Mean 0.969 0.975 0.975
> 3rd Qu. 1.000 1.000 1.000
> Max. 1.000 1.000 1.000
>
>
> Stage-pairs without connections leading to them found in matrices: 1 2 3Now some basic analyses.