16 October 8
16.1 Announcements
- Assignment 4 will be posted on Friday
- Remember that project proposal is due on Friday
16.2 Trajectories
Most of the statistical methods we are covering can be found in this book or this paper.
- So far we have considered examples where the location and time of the observations are fixed and can be chosen
- Example: weather stations
- There are many examples were the location (or time) can not be chosen and may be the random variable
- Example: earthquakes, tornadoes, movement of animals or people.
- Trajectories are a time series of the spatial location of an object (or animal).
- We can usually pick the object and the time that we obtain its spatial location (i.e., time is fixed)
- The location is a random variable in most cases, but time can also be a random variable.
- Other example include PIT tag sensors and capture recapture data.
- Examples of trajectories
- Course website homepage that shows data from Hooten et al (2018) which is available from USGS.
library(maps) library(maptools) library(lubridate) library(plotKML) # Load sandhill crane data # https://www.sciencebase.gov/catalog/item/59a472d7e4b077f005673486 url <- "https://www.dropbox.com/s/akhfy5hwa5d8oon/sacr_locations.csv?dl=1" df.sacr <- read.csv(url) head(df.sacr)## Crane Time X Y ## 1 100840 9/1/2013 0:59 -2663.988 3273.347 ## 2 100840 9/1/2013 12:56 -2665.651 3273.454 ## 3 100840 9/1/2013 18:54 -2666.122 3271.090 ## 4 100840 9/2/2013 6:50 -2663.497 3270.656 ## 5 100840 9/2/2013 12:50 -2665.664 3271.351 ## 6 100840 9/3/2013 0:48 -2663.884 3272.595# Change date/time to class 'POSIXct' df.sacr$Time <- mdy_hm(df.sacr$Time, tz = "America/Chicago") pt.sacr <- SpatialPointsDataFrame(coords = df.sacr[, 3:4], data = data.frame(crane = df.sacr$Crane, time = df.sacr$Time), proj4string = CRS("+proj=aeqd +lat_0=51 +lon_0=-110 +ellps=GRS80 +units=km +no_defs")) # Get map of World map.world <- map("world", fill = TRUE, plot = FALSE) #projection = 'azequalarea', plot = FALSE)orientation = c(51,-101,0) sf.world <- map2SpatialPolygons(map.world, IDs = map.world$names) map.crs <- CRS("+proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0") # Plot data for all individuals on world map plot(sf.world, xlim = c(-200, -101), ylim = c(29.3, 68.5)) plot(spTransform(pt.sacr, map.crs), add = TRUE)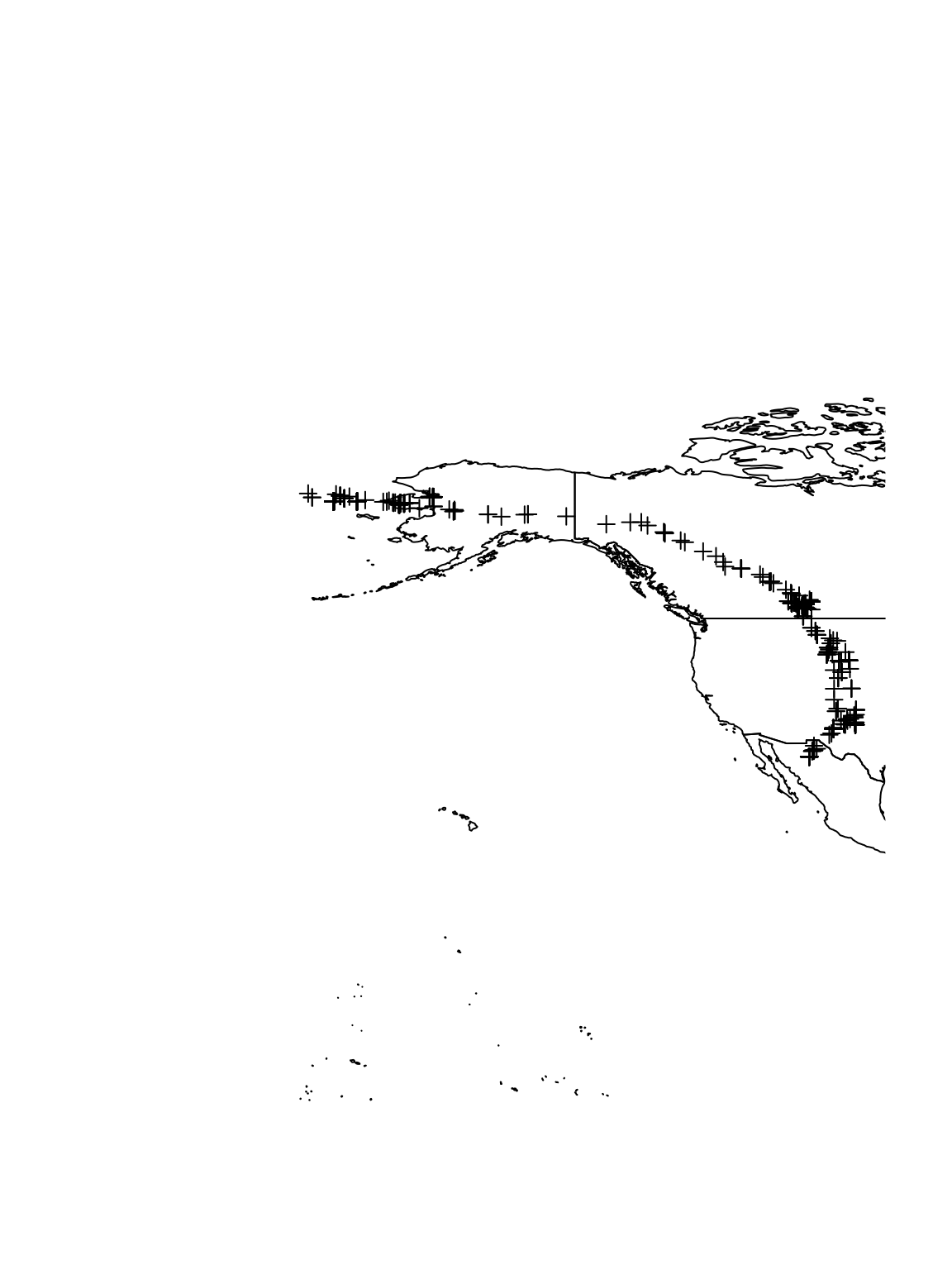
# Plot data for crane 100840 on world map plot(sf.world, xlim = c(-200, -101), ylim = c(29.3, 68.5)) plot(spTransform(pt.sacr, map.crs)[which(df.sacr$Crane == 100840), ], add = TRUE)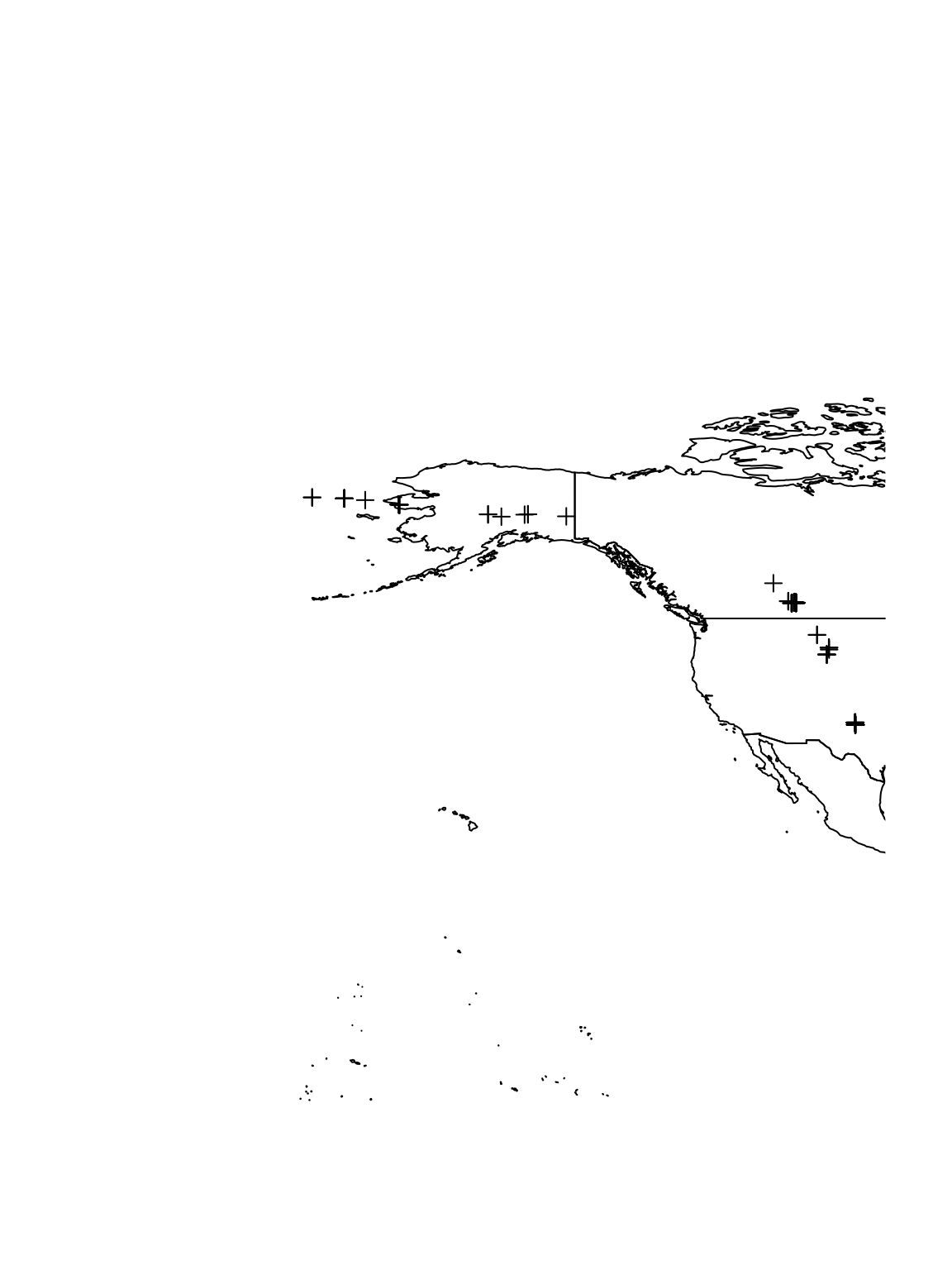
# Plot time series of locations par(mfrow = c(2, 1)) plot(df.sacr[which(df.sacr$Crane == 100840), 2], df.sacr[which(df.sacr$Crane == 100840), 3], xlab = "Date", ylab = "Location") plot(df.sacr[which(df.sacr$Crane == 100840), 2], df.sacr[which(df.sacr$Crane == 100840), 4], , xlab = "Date", ylab = "Location")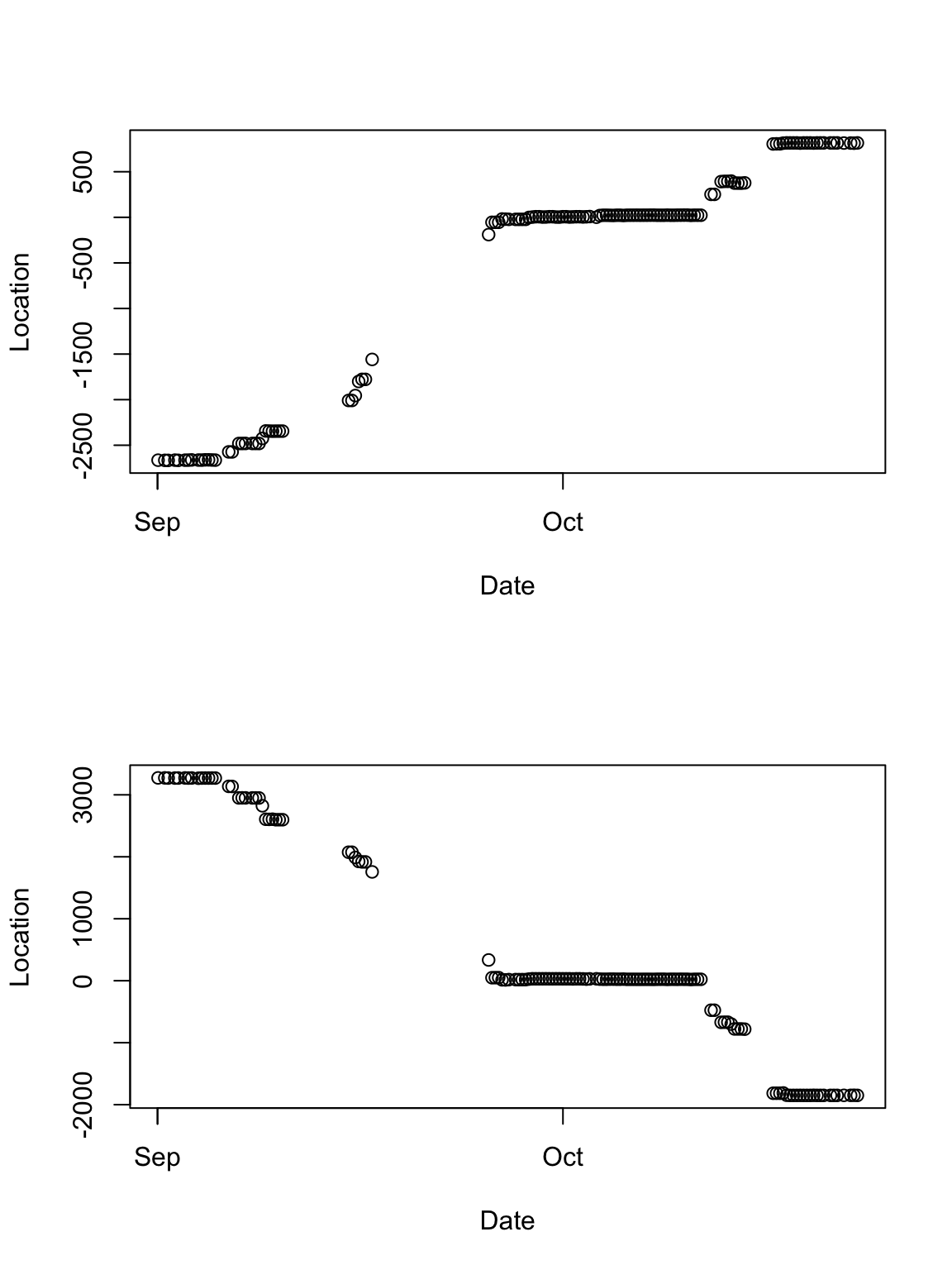
- Sports data
- Data from wearable gps devices
- Strava example
library(plotKML)
library(lubridate)
library(rgdal)
url <- "https://www.dropbox.com/s/ulbn9m3uyz6d0a5/Abilene_Running.gpx?dl=1"
ogrListLayers(url)## [1] "waypoints" "routes" "tracks" "route_points"
## [5] "track_points"
## attr(,"driver")
## [1] "GPX"
## attr(,"nlayers")
## [1] 5## OGR data source with driver: GPX
## Source: "https://www.dropbox.com/s/ulbn9m3uyz6d0a5/Abilene_Running.gpx?dl=1", layer: "track_points"
## with 2095 features
## It has 27 fields## track_fid track_seg_id track_seg_point_id ele time
## 1 0 0 0 350.3 2018/04/07 12:00:07+00
## 2 0 0 1 350.3 2018/04/07 12:00:08+00
## 3 0 0 2 350.3 2018/04/07 12:00:11+00
## 4 0 0 3 350.0 2018/04/07 12:00:15+00
## 5 0 0 4 350.0 2018/04/07 12:00:20+00
## 6 0 0 5 350.0 2018/04/07 12:00:24+00
## magvar geoidheight name cmt desc src link1_href link1_text link1_type
## 1 NA NA <NA> <NA> <NA> <NA> <NA> <NA> <NA>
## 2 NA NA <NA> <NA> <NA> <NA> <NA> <NA> <NA>
## 3 NA NA <NA> <NA> <NA> <NA> <NA> <NA> <NA>
## 4 NA NA <NA> <NA> <NA> <NA> <NA> <NA> <NA>
## 5 NA NA <NA> <NA> <NA> <NA> <NA> <NA> <NA>
## 6 NA NA <NA> <NA> <NA> <NA> <NA> <NA> <NA>
## link2_href link2_text link2_type sym type fix sat hdop vdop pdop
## 1 <NA> <NA> <NA> <NA> <NA> <NA> NA NA NA NA
## 2 <NA> <NA> <NA> <NA> <NA> <NA> NA NA NA NA
## 3 <NA> <NA> <NA> <NA> <NA> <NA> NA NA NA NA
## 4 <NA> <NA> <NA> <NA> <NA> <NA> NA NA NA NA
## 5 <NA> <NA> <NA> <NA> <NA> <NA> NA NA NA NA
## 6 <NA> <NA> <NA> <NA> <NA> <NA> NA NA NA NA
## ageofdgpsdata dgpsid
## 1 NA NA
## 2 NA NA
## 3 NA NA
## 4 NA NA
## 5 NA NA
## 6 NA NA
## gpxtpx_TrackPointExtension
## 1 <gpxtpx:hr>143</gpxtpx:hr> <gpxtpx:cad>0</gpxtpx:cad>
## 2 <gpxtpx:hr>142</gpxtpx:hr> <gpxtpx:cad>0</gpxtpx:cad>
## 3 <gpxtpx:hr>139</gpxtpx:hr> <gpxtpx:cad>60</gpxtpx:cad>
## 4 <gpxtpx:hr>136</gpxtpx:hr> <gpxtpx:cad>60</gpxtpx:cad>
## 5 <gpxtpx:hr>134</gpxtpx:hr> <gpxtpx:cad>99</gpxtpx:cad>
## 6 <gpxtpx:hr>136</gpxtpx:hr> <gpxtpx:cad>99</gpxtpx:cad>
## coords.x1 coords.x2 optional
## 1 -97.21403 38.91273 TRUE
## 2 -97.21403 38.91273 TRUE
## 3 -97.21403 38.91270 TRUE
## 4 -97.21404 38.91255 TRUE
## 5 -97.21406 38.91235 TRUE
## 6 -97.21406 38.91215 TRUE# Change date/time to class 'POSIXct'
pt.marathon$time <- ymd_hms(pt.marathon$time, "%Y-%m-%d%H:%M:%S", tz = "America/Chicago")[-2096]
# Drop all extra variables
pt.marathon <- pt.marathon[, 5]
# Plot locations
plot(pt.marathon, pch = 20, cex = 0.1)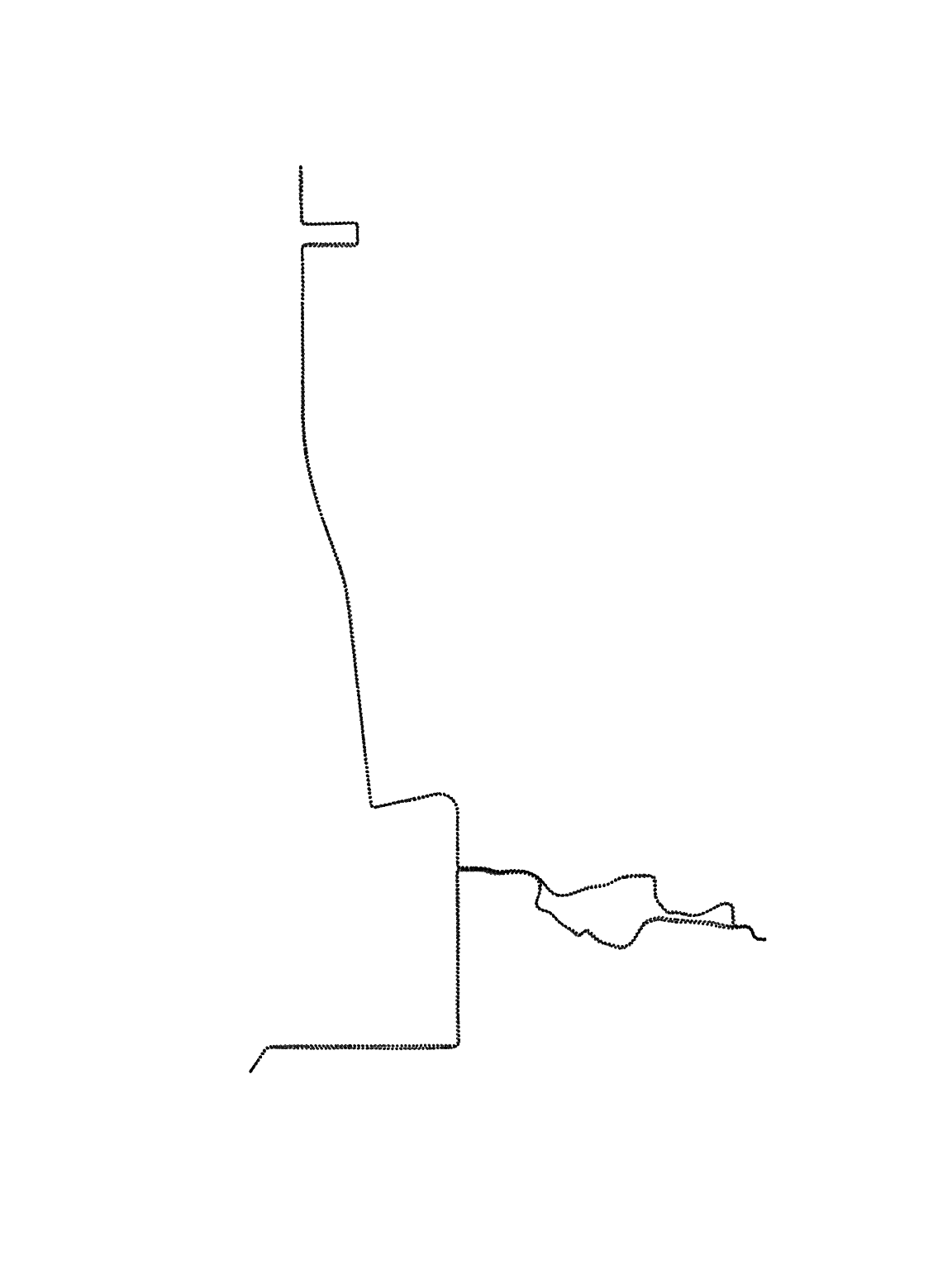
# Plot time series of spatial locations
par(mfrow = c(2, 1))
plot(data.frame(pt.marathon)[, 1], data.frame(pt.marathon)[, 2], pch = "*",
xlab = "Time", ylab = "Location")
plot(data.frame(pt.marathon)[, 1], data.frame(pt.marathon)[, 3], pch = "*",
xlab = "Time", ylab = "Location")
16.3 Descriptive spatio-temporal models for trajectories
- Descriptive vs. mechanistic spatio-temporal statistical models for trajectories
- What are the goals?
- Prediction
- Inference
- Mechanistic understanding
- Functional movement model Buderman et al. 2018