Example
## BCOA BYDV.totalpos.BCOA BCOA.totaltested year long lat
## 1 12 2 10 2014 -95.16269 37.86238
## 2 1 0 1 2014 -95.28463 38.29669
## 3 2 0 2 2014 -95.33038 39.59482
## 4 0 NA 0 2014 -95.32098 39.50696
## 5 8 0 8 2014 -98.55469 38.48455
## 6 1 0 1 2014 -98.84792 38.32772
# Get shapefiles of KS
map.kansas <- map("state", "kansas", fill = TRUE, plot = FALSE)
map.crs <- CRS("+proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0")
sf.kansas <- map2SpatialPolygons(map.kansas, IDs = map.kansas$names, proj4string = map.crs)
# Make SpatialPoints data frame
pts.sample <- data.frame(long = df1$long, lat = df1$lat, count = df1$BCOA, BYVD.pos = df1$BYDV.totalpos.BCOA,
BYVD.tot = df1$BCOA.totaltested, BYVD.prop = df1$BYDV.totalpos.BCOA/df1$BCOA.totaltested)
coordinates(pts.sample) = ~long + lat
proj4string(pts.sample) <- CRS("+proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0")
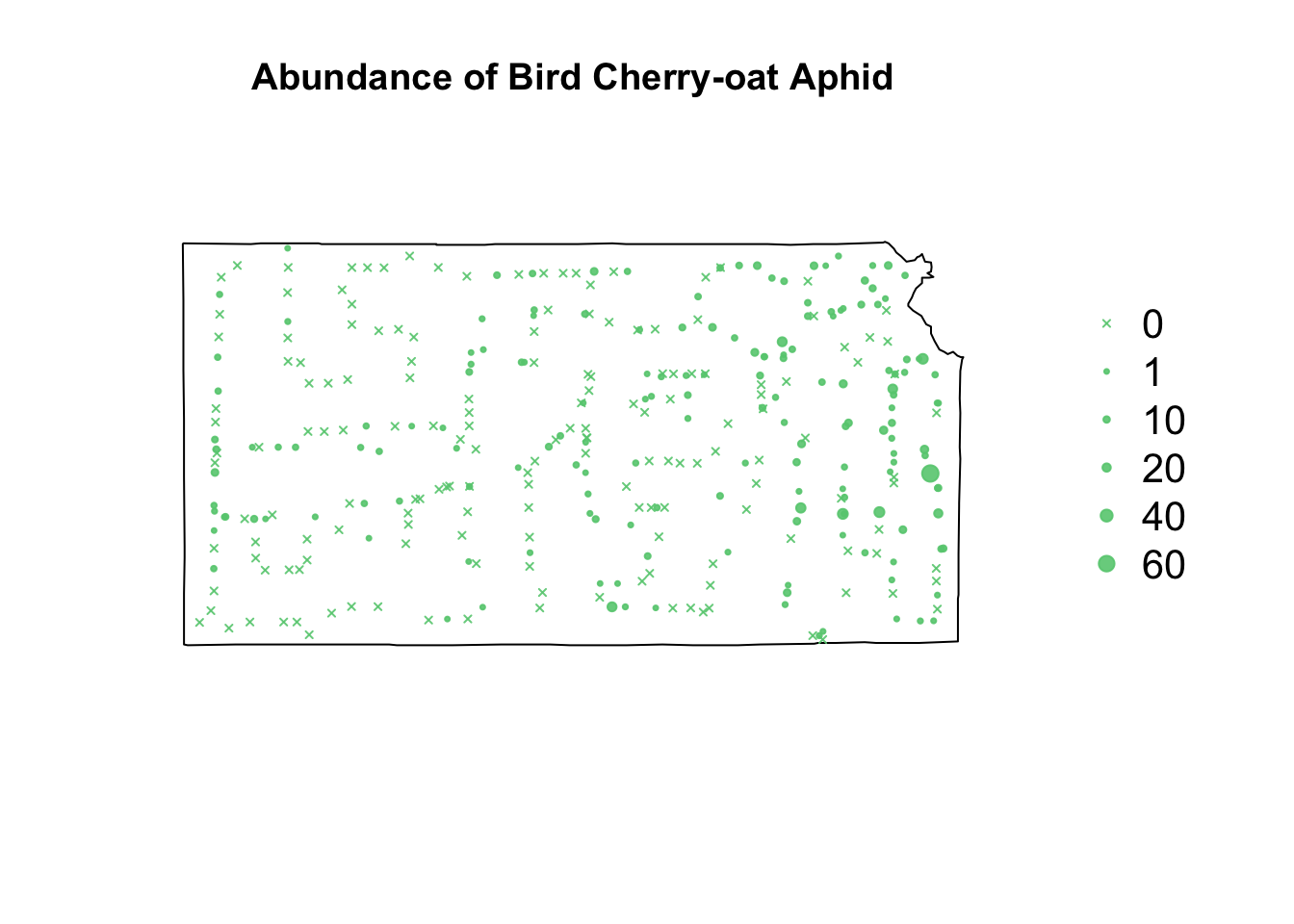
# Plot counts of Bird cherry-oat aphid
par(mar = c(5.1, 4.1, 4.1, 8.1), xpd = TRUE)
plot(sf.kansas, main = "Abundance of Bird Cherry-oat Aphid")
points(pts.sample[, 1], col = rgb(0.4, 0.8, 0.5, 0.9), pch = ifelse(pts.sample$count >
0, 20, 4), cex = pts.sample$count/50 + 0.5)
legend("right", inset = c(-0.25, 0), legend = c(0, 1, 10, 20, 40, 60), bty = "n",
text.col = "black", pch = c(4, 20, 20, 20, 20, 20), cex = 1.3, pt.cex = c(0,
1, 10, 20, 40, 60)/50 + 0.5, col = rgb(0.4, 0.8, 0.5, 0.9))
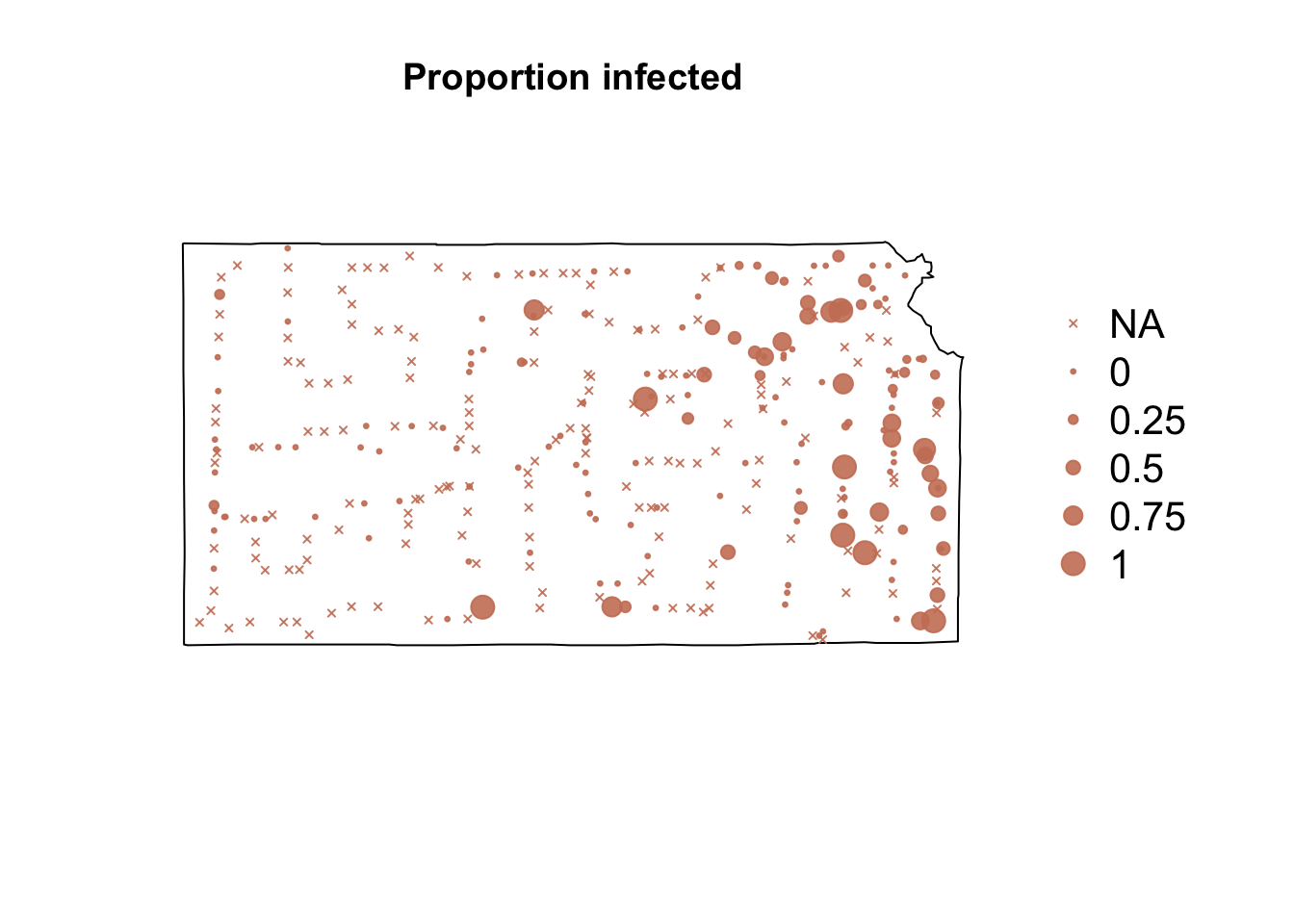
# Plot proportion of number of Bird cherry-oat aphid infected with BYVD
par(mar = c(5.1, 4.1, 4.1, 8.1), xpd = TRUE)
plot(sf.kansas, main = "Proportion infected")
points(pts.sample[, 4], col = rgb(0.8, 0.5, 0.4, 0.9), pch = ifelse(is.na(pts.sample$BYVD.prop) ==
FALSE, 20, 4), cex = ifelse(is.na(pts.sample$BYVD.prop) == FALSE, pts.sample$BYVD.prop,
0)/0.5 + 0.5)
legend("right", inset = c(-0.25, 0), legend = c("NA", 0, 0.25, 0.5, 0.75, 1),
bty = "n", text.col = "black", pch = c(4, 20, 20, 20, 20, 20), cex = 1.3,
pt.cex = c(0, 0, 0.25, 0.5, 0.75, 1)/0.5 + 0.5, col = rgb(0.8, 0.5, 0.4,
0.9))