Resampling: Evaluating models across resamples
Learning outcomes/objective: Learn…
- …why resampling is helpful.
- …how to use resampling to evaluate accuracy and avoid overfitting.
Sources: James et al. (2013) (Ch. 5); Fit a model with resampling;
1 Retake: Simple setup to build predictive model
- A simple setup to built a predictive model might look as follows (validation set approach):
- Randomly split data into one training dataset and one test dataset
- Randomly split the training dataset into one analysis and one assessment (validation/hold-out) dataset
- Train model based on analysis data
- Evaluate model based on the assessment dataset
- Repeat (3.) and (4.) until happy with the model
- This could involved different changes, e.g., parameter tuning, feature selection, up-/down-sampling of imbalanced data prior to training
- Finally, test the model’s accuracy using test dataset (that the model has not seen yet)
- Q: What could be disadvantages of this approach (think of data set sizes/splitting!)? Can you explain the problem of overfitting when we are just doing a single split (as visualzed here)?
- Q: James et al. (2013) use Figure 5.1 [p.177] to explain the validation set approach. Please inspect the figure and explain it in a few words.
- We could also use this approach if we don’t have enough observations for a proper test dataset
Insights
- Disadvantages
- Estimate of test error rate can be highly variable, depending on which observations included in training set/validation set
- Only subset of observations in training set used to fit model.. but stat. methods perform worse when trained on fewer observations.. (ideally use more observations!)
2 Splitting the data and overfitting
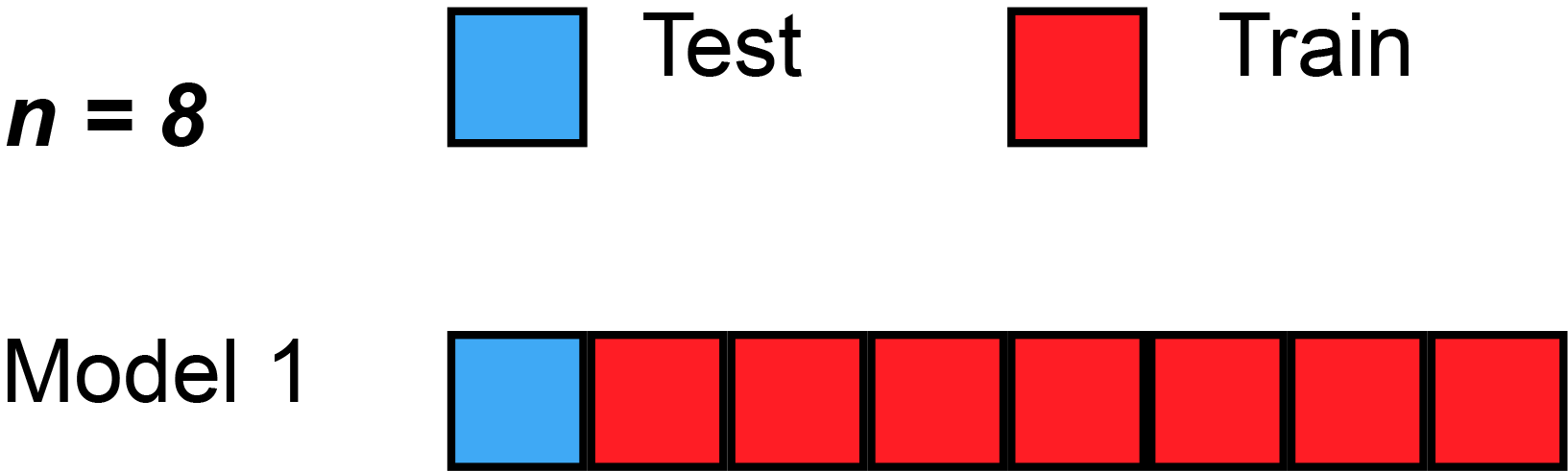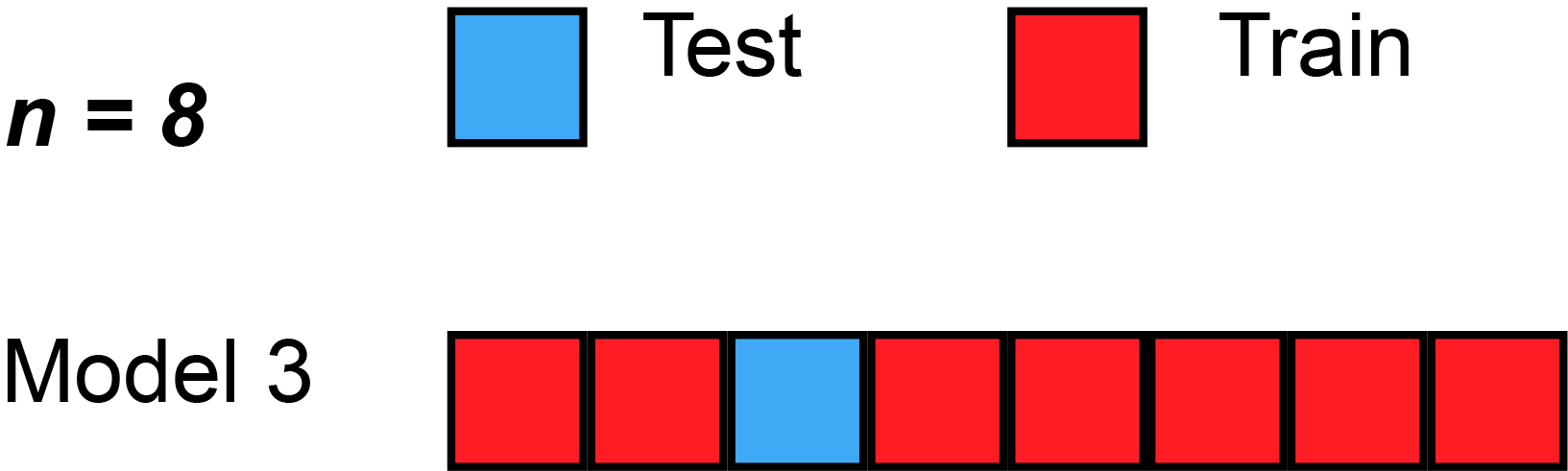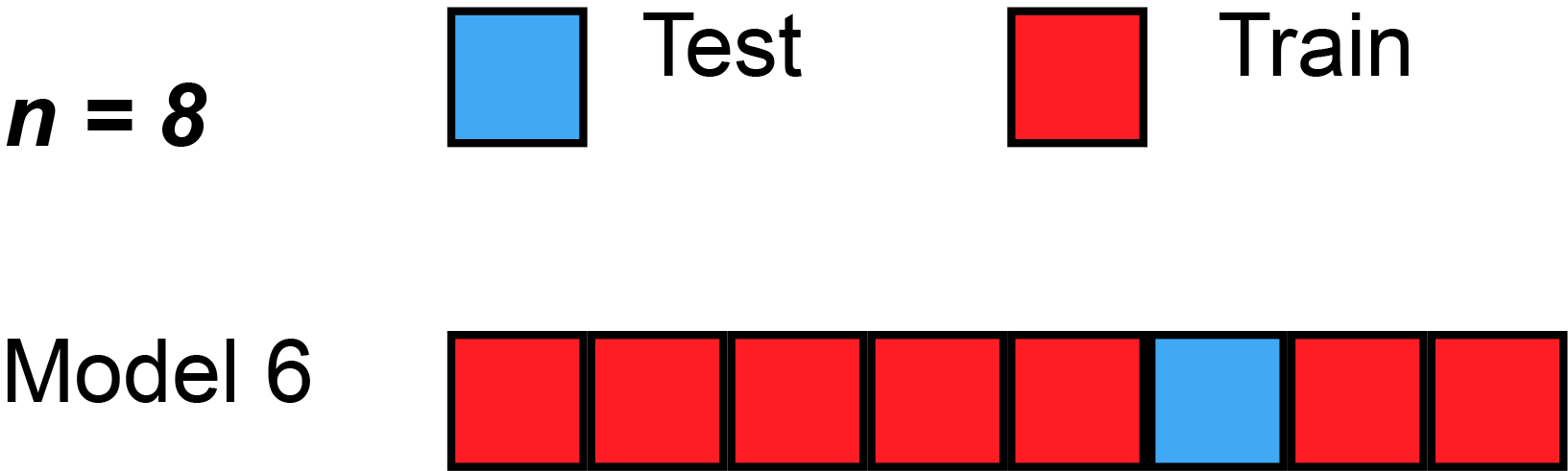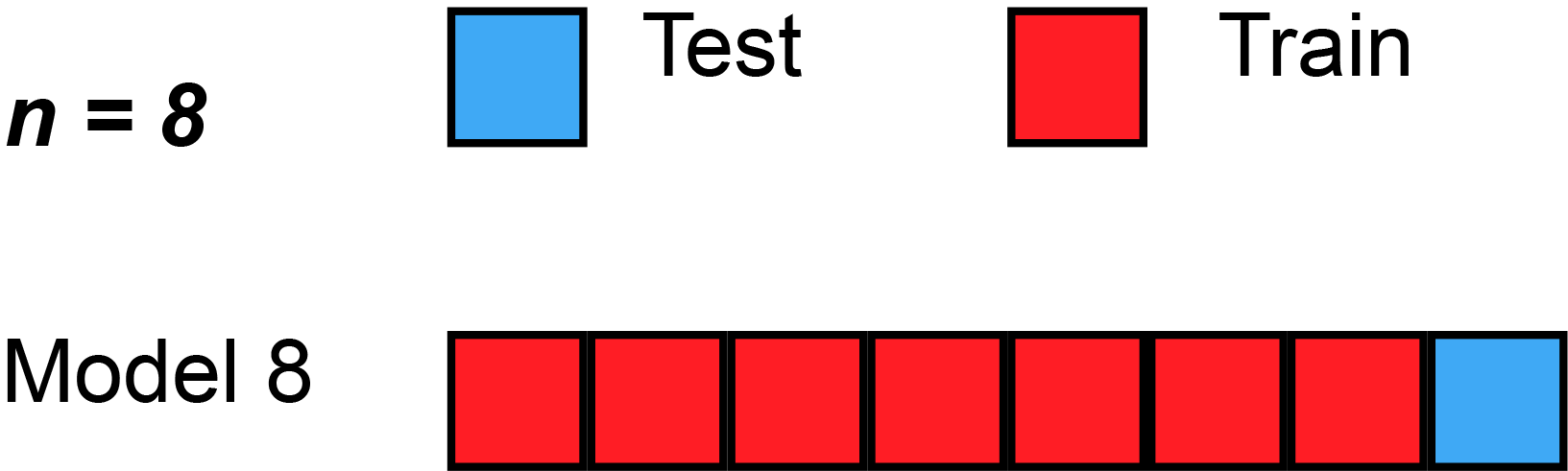
- To avoid overfitting we can estimate our model based on different resamples as shown in Figure 1
- Datasets obtained from the initial split are called training and test data
- Datasets obtained from further splits to the training data are called analysis and assessment datasets
- Often such further splits are called folds and we resample for each fold
3 Resampling methods (1)
- Resampling methods (James et al. 2013, Ch. 5)
- Repeatedly draw samples from training (analysis) set and refit model of interest on each sample to obtain additional information about the fitted model, e.g., the variability of a logistic regression fit
- Aim: Obtain information that would not be available from fitting model only once using the original training sample
- Model assessment: Process of evaluating a model’s performance
- Model selection: Process of selecting the proper level of flexibility for a model
- Common resampling methods: Cross-validation (CV) and bootstrap
- Cross-validation used to (1) estimate the test error associated with a given statistical learning method in order to evaluate its performance, or (2) to select the appropriate level of flexibility
- Bootstrap most commonly used in model selection to provide a measure of accuracy of a parameter estimate or of a given statistical learning method
- Here we discuss cross-validation!
4 Resampling methods (2): Cross-validation
- Recap: Training error rate vs. test error rate (James et al. 2013, chap. 2.2.3)
- “test error is the average error that results from using a statistical learning method to predict the response on a new observation – that is, a measurement that was not used in training the method” (James et al. 2013, 176)
- Objective: Estimate this test error
- i.e., prediction errors our model would make for new data, e.g., for prisoners that were not used to train the model
- In absence of large designated test set a number of techniques can be used to estimate test error rate using the available training data (James et al. 2013, 176)
- Cross-validation (CV) (James et al. 2013, chap. 5.1)
- Class of methods to estimate test error rate by holding out a subset of the training observations from the fitting process, and then applying the statistical learning method to those held out observations
- James et al. (2013) discuss CV for regression approach [Ch. 5.1.1-5.1.4] and classification approach [Ch. 5.1.5]
5 Resampling methods (3): Leave-one-out cross-validation (LOOCV)
- Q: James et al. (2013) use Figure 5.3 [p.179] to explain the Leave-One-Out Cross-Validation (LOOCV). Please inspect the figure and explain it in a few words.
- Q: What is shown in Figure 2?
- Data set repeatedly split into two parts, whereby a single observation \((x_{1},y_{1})\) is used for assessment (validation) set
- Remaining observations \({(x_{2},y_{2}),...,(x_{n},y_{n})}\) make up the analysis (training) set
- Statistical learning method is fit on the \(n−1\) training observations and prediction is made for excluded observation
- Repeated for all splits
- Q: What might be advantages/disadvantages? Bias? Reproducibility? Computational intensity?
Insights
- Advantages over validation set approach
- Bias: Less bias because training data is bigger (Why?)
- Reproducibility: Performing LOOCV repeatedly will always produces the same results (Why? No randomness in training/validation set splits)
- Computational intensity: Disadvantage: Can be computationally intensive (Why?) [estimate it n times]
6 Resampling methods (4): Leave-one-out cross-validation (LOOCV)
- In classification setting LOOCV error rate takes the form (James et al. 2013, 184):
\[CV_{(n)} = \frac{1}{n}\sum_{i=1}^{n}Err_{i}\] where \(Err_{i}=I(y_{i}\neq\hat{y}_{i})\), i.e., is 1 when left-out \(y_{i}\) was wrongly predicted
Linear regression: Use MSE rather than number of misclassified observations to quantify test error
The k-fold CV error rate (see next slide) and validation set error rates are defined analogously
7 Resampling methods (5): k-Fold Cross-Validation
- James et al. (2013) use Figure 5.5 [p.179] to explain the k-Fold Cross-Validation. Please inspect the figure and explain it in a few words.
- Q: What is shown in Figure 3?
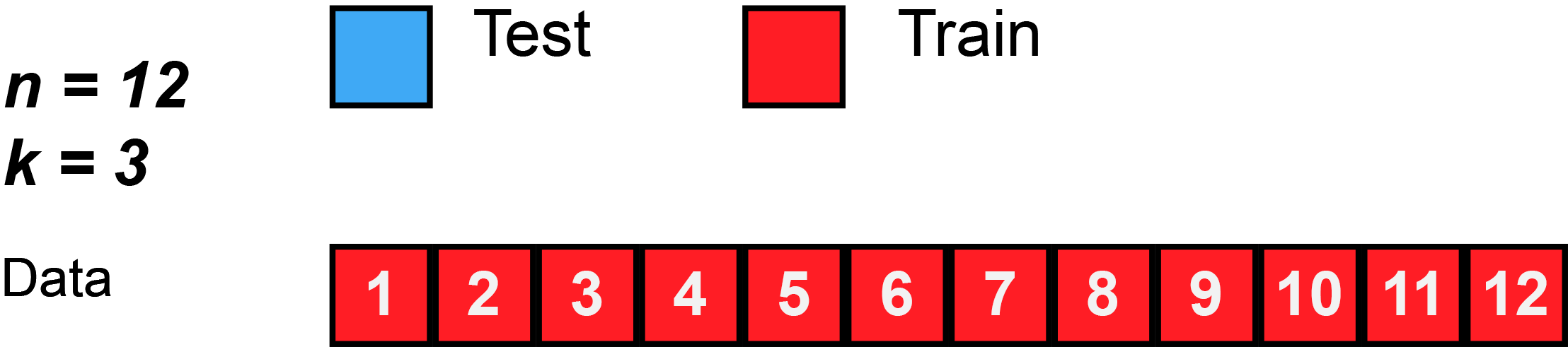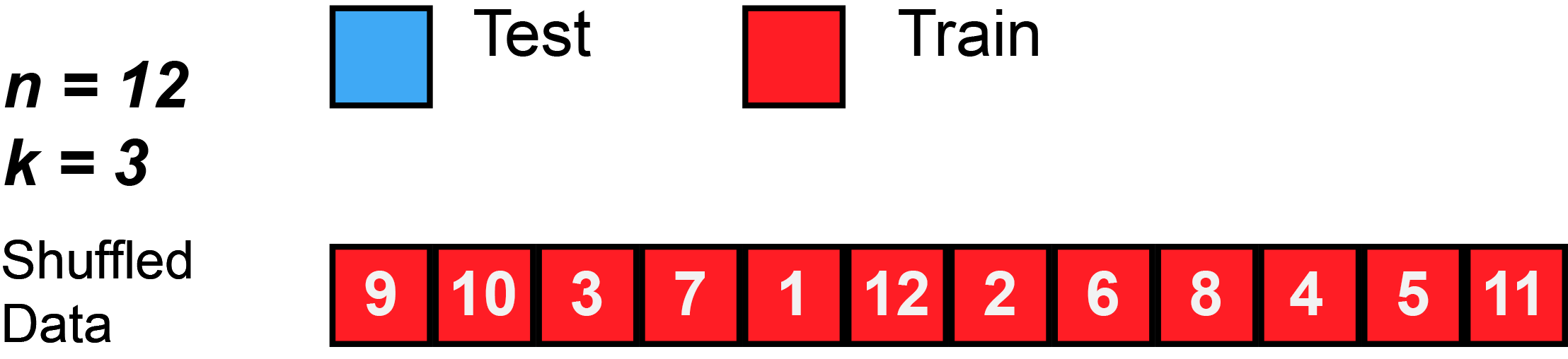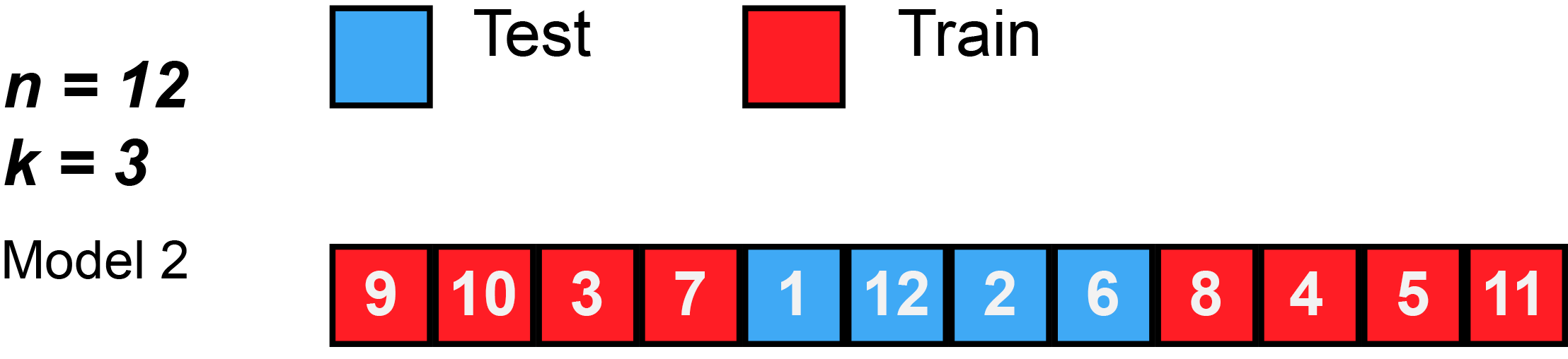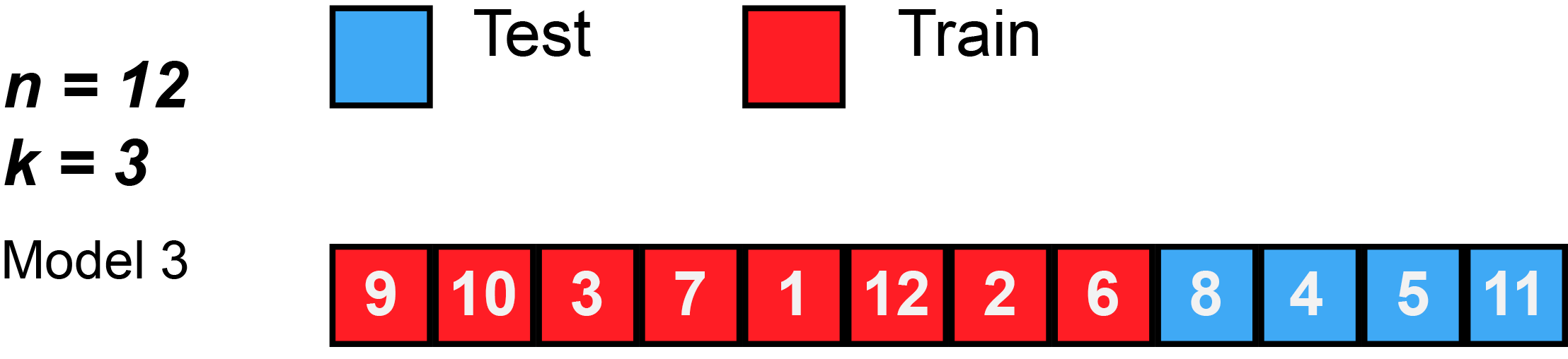
k-Fold Cross-Validation is special case of LOOCV where \(k\) is set to equal \(n\)
Error rate: \(CV_{(k)} = \frac{1}{k}\sum_{i=1}^{k}Err_{i}\)
Advantages
- Q: What is the advantage of using \(k = 5\) or \(k = 10\) rather than \(k = n\)? [computation!]
Bias-variance trade-off (James et al. 2013, Ch. 5.1.4)
- Q: k-Fold CV may give more accurate estimates of test error rate than LOOCV: Why?
Insights
Advantages: What is the advantage of using \(k = 5\) or \(k = 10\) rather than \(k = n\)? [computation!]
Bias-variance trade-off (James et al. 2013, Ch. 5.1.4)
- Q: k-Fold CV may give more accurate estimates of test error rate than LOOCV: Why?
- Datasets smaller than LOOCV, but larger than validation set approach
- LOOCV best from bias perspective but (most observations) but each model fit on identical observations (highly correlated)
- Mean of many highly correlated quantities has higher variance than does the mean of many quantities that are notas highly correlated → test error estimate from LOOCV higher variance than test error estimate from k-fold CV
- LOOCV best from bias perspective but (most observations) but each model fit on identical observations (highly correlated)
- Datasets smaller than LOOCV, but larger than validation set approach
- Recommendation use k-fold cross-validation using k = 5 or k = 10
- Q: k-Fold CV may give more accurate estimates of test error rate than LOOCV: Why?
8 Resampling methods (6): Some caveats
- Beware: Conceptual confusion
- Cross-validation four meanings in political science (Neunhoeffer and Sternberg 2019, 102)
- Validating new measures or instruments
- Resampling:
- Robustness check of coefficents across data subsets
- procedure to obtain estimate of true error (unseen data)
- procedure of model tuning (e.g., parameter tuning, feature selection, up-/down-sampling of imbalanced data prior to training)
- Cross-validation four meanings in political science (Neunhoeffer and Sternberg 2019, 102)
- Cross-validation for tuning vs. test error rate
- “A problematic use of cross-validation occurs when a single cross-validation procedure is used for model tuning and to estimate true error at the same time […]. Ignoring this can lead to serious misreporting of performance measures. If the goal of cross-validation is to obtain an estimate of true error, every step involved in training the model (including (hyper-) parameter tuning, feature selection or up-/down-sampling) has to be performed on each of the training folds of the cross-validation procedure. Hastie, Tibshirani, and Friedman (2011, 245) refer to this problem as the wrong way of doing cross-validation. We take down-sampling of imbalanced data as a simple example of this problem.” (Neunhoeffer and Sternberg 2019, 102)
9 Exercise: Resampling methods
- Let’s quickly review cross-validation
- Why/when do we normally use cross-validation (“resampling methods”)?
- Explain how k-fold cross-validation is implemented.
- What are the advantages and disadvantages of k-fold cross-validation relative to the validation set approach?
- What are the advantages and disadvantages of k-fold cross-validation relative to LOOCV?
10 Lab: Evaluate a model with resampling
# install.packages(pacman)
pacman::p_load(tidyverse,
tidymodels,
naniar,
knitr,
kableExtra)We first import the data into R:
# Build and evaluate model without resampling
data <- read_csv(sprintf("https://docs.google.com/uc?id=%s&export=download",
"1plUxvMIoieEcCZXkBpj4Bxw1rDwa27tj"))10.1 Build and evaluate model without resampling
Below we:
- Split the data
- Define the model (
lr_mod= “logistic regression model”) - Bundle the model/formula into workflow (
ml_wflow= “machine learning workflow) - Train our workflow/model using a single call to
fit - Extract the model coefficients
# Convert outcome to factor
data <- data %>%
mutate(is_recid = factor(is_recid))
# 1.
# Split the data into training and test data
data_split <- initial_split(data, prop = 0.80)
data_split # Inspect<Training/Testing/Total>
<5771/1443/7214># Extract the two datasets
data_train <- training(data_split)
data_test <- testing(data_split) # Do not touch until the end!
# Further split the training data into analysis (training2) and assessment (validation) dataset
data_folds <- validation_split(data_train, prop = .80)
data_folds# Validation Set Split (0.8/0.2)
# A tibble: 1 x 2
splits id
<list> <chr>
1 <split [4616/1155]> validation data_analysis <- analysis(data_folds$splits[[1]])
data_assessment <- assessment(data_folds$splits[[1]])
# 2.
# Define the model
lr_mod <-
logistic_reg() %>%
set_engine("glm")
# 3.
# Create workflow
ml_wflow <-
workflow() %>%
add_model(lr_mod) %>%
add_formula(is_recid ~ age + priors_count)
ml_wflow== Workflow ====================================================================
Preprocessor: Formula
Model: logistic_reg()
-- Preprocessor ----------------------------------------------------------------
is_recid ~ age + priors_count
-- Model -----------------------------------------------------------------------
Logistic Regression Model Specification (classification)
Computational engine: glm # 4.
# Use workflow to fit/train the model
ml_fit <-
ml_wflow %>%
fit(data = data_analysis)
ml_fit== Workflow [trained] ==========================================================
Preprocessor: Formula
Model: logistic_reg()
-- Preprocessor ----------------------------------------------------------------
is_recid ~ age + priors_count
-- Model -----------------------------------------------------------------------
Call: stats::glm(formula = ..y ~ ., family = stats::binomial, data = data)
Coefficients:
(Intercept) age priors_count
1.02606 -0.04882 0.18054
Degrees of Freedom: 4615 Total (i.e. Null); 4613 Residual
Null Deviance: 6390
Residual Deviance: 5643 AIC: 5649# 5.
# Extract model coefficients
ml_fit %>%
extract_fit_parsnip() %>%
tidy()# A tibble: 3 x 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 1.03 0.102 10.1 6.73e-24
2 age -0.0488 0.00297 -16.5 7.69e-61
3 priors_count 0.181 0.00913 19.8 5.96e-87
Next we use our trained workflow (ml_fit) to predict with the unseen data_assessment (this is not the final test data but rather the validation dataset), which we will do with a single call to predict() or augment():
# Use model to obtain predictions for data_assessment
predict(ml_fit, data_assessment)# A tibble: 1,155 x 1
.pred_class
<fct>
1 0
2 0
3 1
4 1
5 0
6 0
7 0
8 0
9 0
10 0
# ... with 1,145 more rows# Augment test data with predictions and subset variables
data_assessment <- augment(ml_fit, data_test) %>%
select(id, age, priors_count, race, is_recid, .pred_class, .pred_1, .pred_0)
data_assessment# A tibble: 1,443 x 8
id age priors_count race is_recid .pred_cl~1 .pred_1 .pred_0
<dbl> <dbl> <dbl> <chr> <fct> <fct> <dbl> <dbl>
1 6 43 2 Other 0 0 0.331 0.669
2 9 43 3 Other 0 0 0.369 0.631
3 15 23 3 African-American 1 1 0.595 0.405
4 19 47 1 Caucasian 1 0 0.258 0.742
5 33 32 0 Other 0 0 0.369 0.631
6 37 27 8 African-American 1 1 0.741 0.259
7 38 36 3 Caucasian 0 0 0.447 0.553
8 42 32 4 African-American 1 1 0.535 0.465
9 50 49 7 Other 1 0 0.467 0.533
10 66 25 9 Caucasian 1 1 0.788 0.212
# ... with 1,433 more rows, and abbreviated variable name 1: .pred_class- Q: What is
.pred_class,.pred_1and.pred_0?
And calculate different measures of accuracy for the data_assessment.
# Calculate accuracy
data_assessment %>%
accuracy(truth = is_recid, estimate = .pred_class)# A tibble: 1 x 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 accuracy binary 0.688# Calculate F1 score
data_assessment %>%
f_meas(truth = is_recid, estimate = .pred_class)# A tibble: 1 x 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 f_meas binary 0.704# Several metrics
# Multiple regression metrics
classification_metrics <- metric_set(accuracy, mcc, f_meas)
# mcc: Matthews correlation coefficient:
# The returned function has arguments:
data_assessment %>% # training set predictions
classification_metrics(truth = is_recid, estimate = .pred_class)# A tibble: 3 x 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 accuracy binary 0.688
2 mcc binary 0.383
3 f_meas binary 0.704
Figure 4 visualizes both precision and recall as a function of the threshold. The pr_curve() function can be used to calculate the corresponding values.
library(ggplot2)
library(dplyr)
data_assessment %>%
pr_curve(truth = is_recid,
estimate = .pred_1,
event_level = "second") %>%
pivot_longer(cols = c("recall", "precision"),
names_to = "measure",
values_to = "value") %>%
mutate(measure = recode(measure,
"recall" = "Recall (= True pos. rate = TP/P = sensitivity)",
"precision" = "Precision (= Pos. pred. value = TP/P*)")) %>%
ggplot(aes(x = .threshold,
y = value,
color = measure)) +
geom_line() +
xlab("Threshold value") +
ylab("Value of measure") +
theme_bw()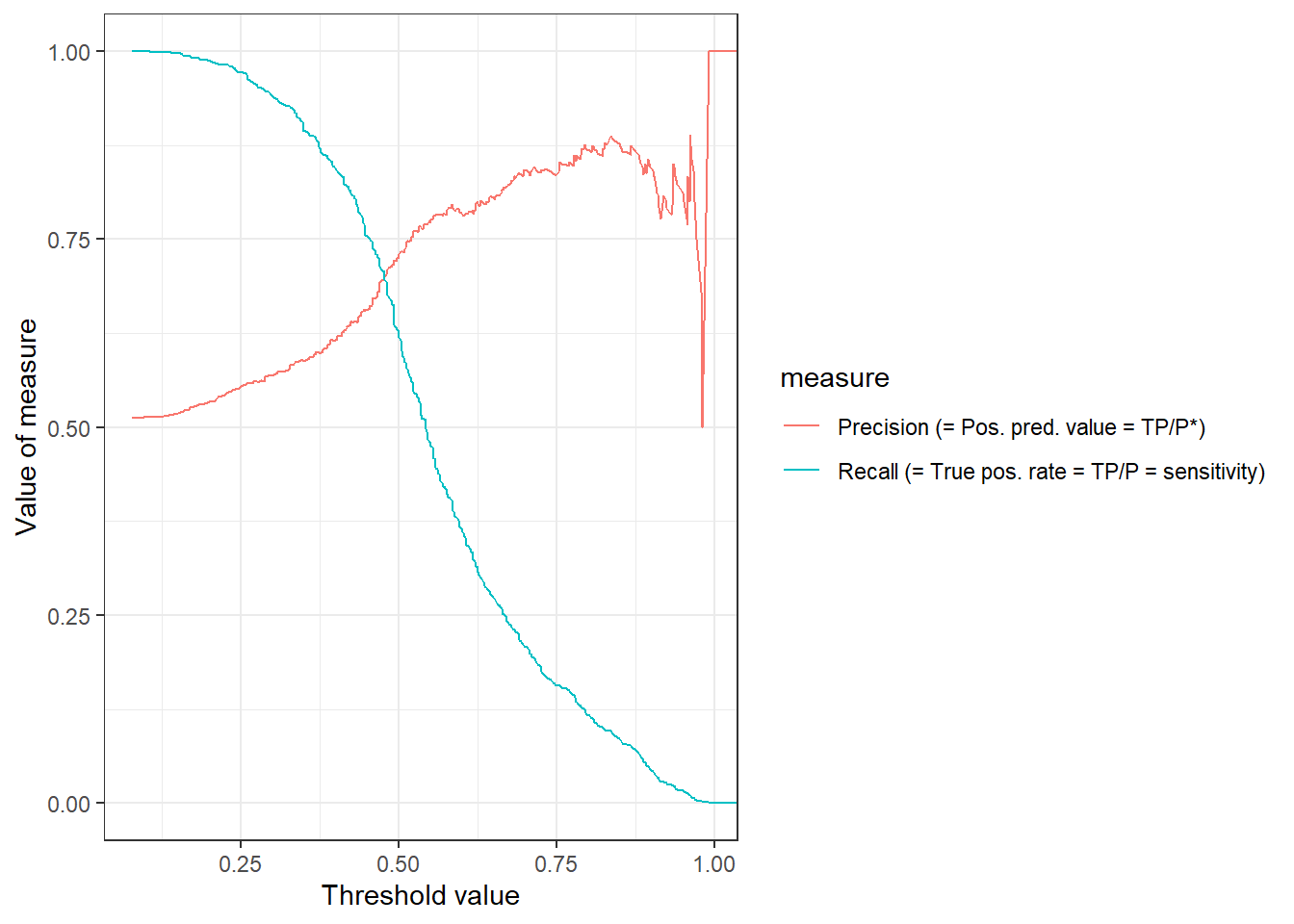
Finally, Figure 5 displays the ROC curve (see the corresponding slide for an overview). The corresponding values can be obtained using the roc_curve() function.
- Important (
?roc_curve): “There is no common convention on which factor level should automatically be considered the”event” or “positive” result when computing binary classification metrics. In yardstick, the default is to use the first level. To alter this, change the argument event_level to “second” to consider the last level of the factor the level of interest.”- Here we pick the 1s, i.e., recidivating as the “event” or “positive” result.
data_assessment %>%
roc_curve(truth = is_recid, estimate = .pred_1,
event_level = "second") %>%
autoplot() +
xlab("1 - specificity (FP/N, false positives rate)") +
ylab("sensitivity (TP/P, true positives rate)")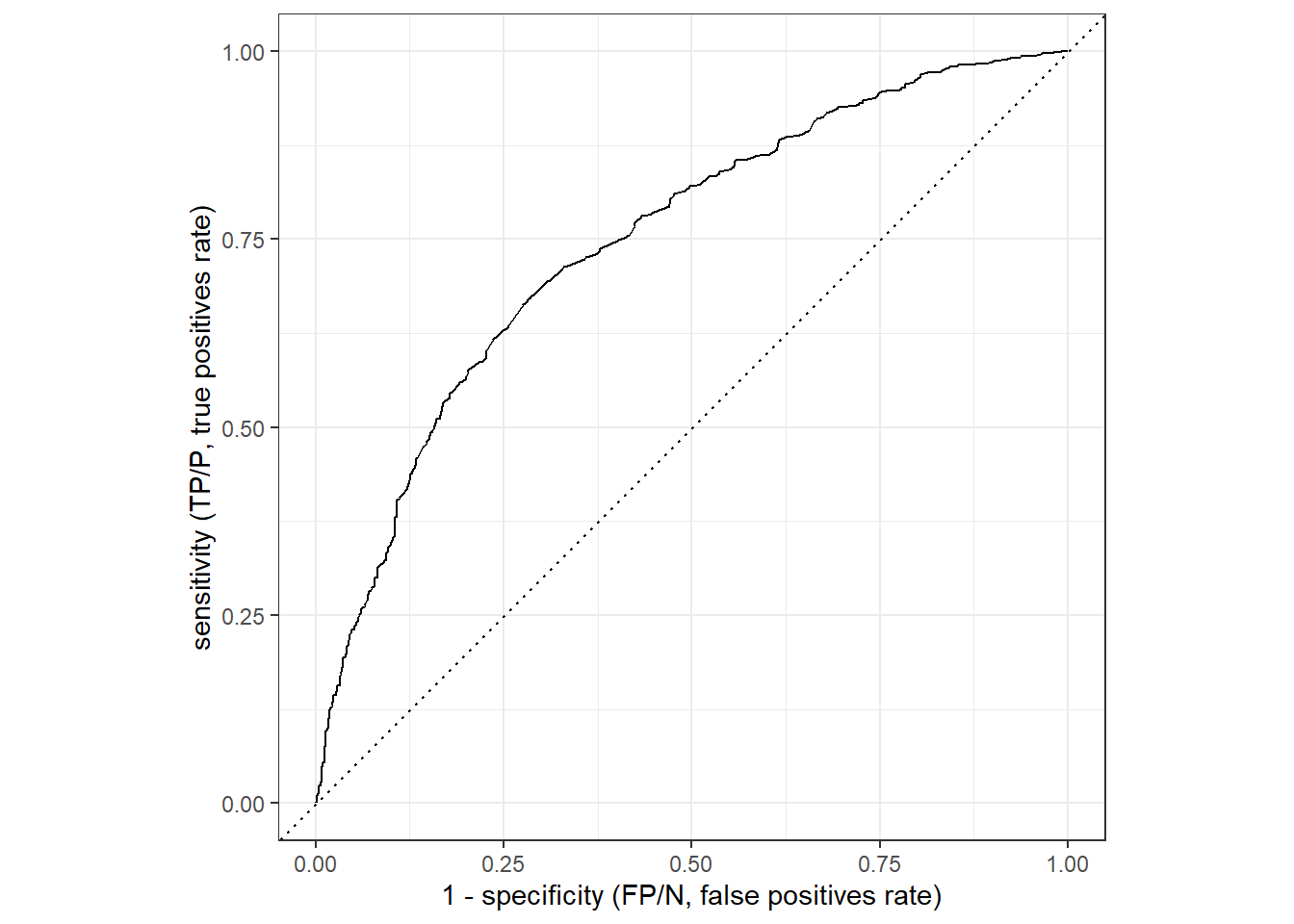
And we can also calculate the area under the ROC curve (the higher the better with 1 being the maximum as outlined here):
# Compute area under ROC curve
data_assessment %>%
roc_auc(truth = is_recid, estimate = .pred_1,
event_level = "second")# A tibble: 1 x 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 roc_auc binary 0.74810.2 Resampling and cross-validation
Below we estimate the same model from above, however, now we employ resampling to be better able to evaluate its accuracy.
# Build and evaluate model WITH resampling
data <- read_csv(sprintf("https://docs.google.com/uc?id=%s&export=download",
"1plUxvMIoieEcCZXkBpj4Bxw1rDwa27tj"))Below we:
- Split the data
- Create resampled partitions with
vfold_cv() - Define the model (
lr_mod= “logistic regression model”) - Bundle the model/formula into workflow (
ml_wflow= “machine learning workflow)
# Convert outcome to factor
data <- data %>%
mutate(is_recid = factor(is_recid))
# 1.
# Split the data into training and test data
data_split <- initial_split(data, prop = 0.80)
data_split # Inspect<Training/Testing/Total>
<5771/1443/7214># Extract the two datasets
data_train <- training(data_split)
data_test <- testing(data_split) # Do not touch until the end!
# 2.
# Create resampled partitions
set.seed(345)
data_folds <- vfold_cv(data_train, v = 10) # V-fold/k-fold cross-validation
data_folds # data_folds now contains several resamples of our training data # 10-fold cross-validation
# A tibble: 10 x 2
splits id
<list> <chr>
1 <split [5193/578]> Fold01
2 <split [5194/577]> Fold02
3 <split [5194/577]> Fold03
4 <split [5194/577]> Fold04
5 <split [5194/577]> Fold05
6 <split [5194/577]> Fold06
7 <split [5194/577]> Fold07
8 <split [5194/577]> Fold08
9 <split [5194/577]> Fold09
10 <split [5194/577]> Fold10 # You can also try loo_cv(data_train) instead
# 3.
# Define the model
lr_mod <-
logistic_reg() %>%
set_engine("glm")
# 4.
# Create workflow
ml_wflow <-
workflow() %>%
add_model(lr_mod) %>%
add_formula(is_recid ~ age + priors_count)Then we use this workflow to fit/train our model using the fit_resamples() function.
# Use workflow to fit the to train the model
# There is no need in specifying data_analysis/data_assessment as
# the functions understand the corresponding parts
ml_fit_rs <-
ml_wflow %>%
fit_resamples(resamples = data_folds,
control = control_resamples(save_pred = TRUE,
extract = function (x) extract_fit_parsnip(x)))
ml_fit_rs# Resampling results
# 10-fold cross-validation
# A tibble: 10 x 6
splits id .metrics .notes .extra~1 .predi~2
<list> <chr> <list> <list> <list> <list>
1 <split [5193/578]> Fold01 <tibble [2 x 4]> <tibble [0 x 3]> <tibble> <tibble>
2 <split [5194/577]> Fold02 <tibble [2 x 4]> <tibble [0 x 3]> <tibble> <tibble>
3 <split [5194/577]> Fold03 <tibble [2 x 4]> <tibble [0 x 3]> <tibble> <tibble>
4 <split [5194/577]> Fold04 <tibble [2 x 4]> <tibble [0 x 3]> <tibble> <tibble>
5 <split [5194/577]> Fold05 <tibble [2 x 4]> <tibble [0 x 3]> <tibble> <tibble>
6 <split [5194/577]> Fold06 <tibble [2 x 4]> <tibble [0 x 3]> <tibble> <tibble>
7 <split [5194/577]> Fold07 <tibble [2 x 4]> <tibble [0 x 3]> <tibble> <tibble>
8 <split [5194/577]> Fold08 <tibble [2 x 4]> <tibble [0 x 3]> <tibble> <tibble>
9 <split [5194/577]> Fold09 <tibble [2 x 4]> <tibble [0 x 3]> <tibble> <tibble>
10 <split [5194/577]> Fold10 <tibble [2 x 4]> <tibble [0 x 3]> <tibble> <tibble>
# ... with abbreviated variable names 1: .extracts, 2: .predictions# Extract components
ml_fit_rs$.metrics[1:2] # get first two rows of .metrics[[1]]
# A tibble: 2 x 4
.metric .estimator .estimate .config
<chr> <chr> <dbl> <chr>
1 accuracy binary 0.678 Preprocessor1_Model1
2 roc_auc binary 0.705 Preprocessor1_Model1
[[2]]
# A tibble: 2 x 4
.metric .estimator .estimate .config
<chr> <chr> <dbl> <chr>
1 accuracy binary 0.664 Preprocessor1_Model1
2 roc_auc binary 0.710 Preprocessor1_Model1 ml_fit_rs$.extracts [1:2] # get first two rows of .extracts[[1]]
# A tibble: 1 x 2
.extracts .config
<list> <chr>
1 <fit[+]> Preprocessor1_Model1
[[2]]
# A tibble: 1 x 2
.extracts .config
<list> <chr>
1 <fit[+]> Preprocessor1_Model1 ml_fit_rs$.extracts[[1]]$.extracts # Extract models[[1]]
parsnip model object
Call: stats::glm(formula = ..y ~ ., family = stats::binomial, data = data)
Coefficients:
(Intercept) age priors_count
1.0170 -0.0481 0.1724
Degrees of Freedom: 5192 Total (i.e. Null); 5190 Residual
Null Deviance: 7192
Residual Deviance: 6387 AIC: 6393Then we can evaluate our models averaging across our resamples, accounting for the natural variability across different analysis datasets that we could possibly use to built the model.
# Collect metrics across resamples
collect_metrics(ml_fit_rs)# A tibble: 2 x 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.681 10 0.00669 Preprocessor1_Model1
2 roc_auc binary 0.728 10 0.00629 Preprocessor1_Model1# Compare to single fold approach
data_assessment %>% # training set predictions
classification_metrics(truth = is_recid, estimate = .pred_class)# A tibble: 3 x 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 accuracy binary 0.682
2 mcc binary 0.363
3 f_meas binary 0.697There does not seem to be much of a difference.
Importantly, we can also continue work with the single folds using the map() function, extracting datasets, fitting models, extracting coefficients, unnesting etc.
data_folds %>%
mutate(
# Extract the train data frame for each split
data_analysis = map(.x = splits, ~training(.x)),
# Extract the validate data frame for each split
data_assessment = map(.x = splits, ~testing(.x))
) %>%
# Fit/train the model on data_analysis
mutate(fit = map(.x = data_analysis,
~lr_mod %>%
fit(is_recid ~ age + priors_count,
data = .x))) %>%
# Extract the coefficients
mutate(coefficients = map(.x = fit, ~tidy(.x))) %>%
# Unnest the dataframe to get the coefficients for the different folds
unnest(coefficients)# A tibble: 30 x 10
splits id data_anal~1 data_a~2 fit term estim~3 std.e~4
<list> <chr> <list> <list> <list> <chr> <dbl> <dbl>
1 <split [5193/578]> Fold01 <tibble> <tibble> <fit[+]> (Int~ 1.02 0.0959
2 <split [5193/578]> Fold01 <tibble> <tibble> <fit[+]> age -0.0481 0.00280
3 <split [5193/578]> Fold01 <tibble> <tibble> <fit[+]> prio~ 0.172 0.00831
4 <split [5194/577]> Fold02 <tibble> <tibble> <fit[+]> (Int~ 1.06 0.0956
5 <split [5194/577]> Fold02 <tibble> <tibble> <fit[+]> age -0.0490 0.00281
6 <split [5194/577]> Fold02 <tibble> <tibble> <fit[+]> prio~ 0.164 0.00808
7 <split [5194/577]> Fold03 <tibble> <tibble> <fit[+]> (Int~ 0.971 0.0954
8 <split [5194/577]> Fold03 <tibble> <tibble> <fit[+]> age -0.0465 0.00279
9 <split [5194/577]> Fold03 <tibble> <tibble> <fit[+]> prio~ 0.164 0.00806
10 <split [5194/577]> Fold04 <tibble> <tibble> <fit[+]> (Int~ 0.993 0.0952
# ... with 20 more rows, 2 more variables: statistic <dbl>, p.value <dbl>, and
# abbreviated variable names 1: data_analysis, 2: data_assessment,
# 3: estimate, 4: std.error11 Homework
- Start by re-reading the code presented above (you can find all the code below).
- Above we re-evaluated the accuracy of our model using 10-fold cross validation. Please re-evaluate the model but now use Leave-one-out cross-validation (LOOCV) instead. Do you find differences in terms of accuracy?
- Subsequently, compare the setting where you use k-Fold Cross-Validation using 5 folds (k = 5) and 20 folds (k = 20). What differences (if any) do you find?
- Finally, shorty outline the advantages and disadvantages of the validation set approach (one analysis data, one assessment data), Leave-one-out cross-validation (LOOCV) and k-Fold Cross-Validation.
12 Exercise: What’s predicted?
- Q: What do we predict using the techniques below? What is the input/what is the output? How could we use those ML models for our own research? What phenomena may they create that we need to research? (discuss 2!)
Insights
- Image recognition: Predict whether an image shows a sunsetSome examples
- Deepart: “Predict” what an image would look like if it was painted by…
- Speech recognition: Predict which (written) words someone just used
- Translation: Predict which English word/sentence corresponds to a German word/sentence (predict language!)
- Pose estimation (2018!): Predict body pose from image
- Deep fakes (2019): ?
- Text analysis/Natural language processing (NLP): Predict entities, sentiment, syntax, categories in text
13 All the code
knitr::include_graphics("06_300px-LOOCV.gif")#
knitr::include_graphics("06_350px-KfoldCV.gif")#
# install.packages(pacman)
pacman::p_load(tidyverse,
tidymodels,
naniar,
knitr,
kableExtra)
# Build and evaluate model without resampling
data <- read_csv(sprintf("https://docs.google.com/uc?id=%s&export=download",
"1plUxvMIoieEcCZXkBpj4Bxw1rDwa27tj"))
# Convert outcome to factor
data <- data %>%
mutate(is_recid = factor(is_recid))
# 1.
# Split the data into training and test data
data_split <- initial_split(data, prop = 0.80)
data_split # Inspect
# Extract the two datasets
data_train <- training(data_split)
data_test <- testing(data_split) # Do not touch until the end!
# Further split the training data into analysis (training2) and assessment (validation) dataset
data_folds <- validation_split(data_train, prop = .80)
data_folds
data_analysis <- analysis(data_folds$splits[[1]])
data_assessment <- assessment(data_folds$splits[[1]])
# 2.
# Define the model
lr_mod <-
logistic_reg() %>%
set_engine("glm")
# 3.
# Create workflow
ml_wflow <-
workflow() %>%
add_model(lr_mod) %>%
add_formula(is_recid ~ age + priors_count)
ml_wflow
# 4.
# Use workflow to fit/train the model
ml_fit <-
ml_wflow %>%
fit(data = data_analysis)
ml_fit
# 5.
# Extract model coefficients
ml_fit %>%
extract_fit_parsnip() %>%
tidy()
# Use model to obtain predictions for data_assessment
predict(ml_fit, data_assessment)
# Augment test data with predictions and subset variables
data_assessment <- augment(ml_fit, data_test) %>%
select(id, age, priors_count, race, is_recid, .pred_class, .pred_1, .pred_0)
data_assessment
# Calculate accuracy
data_assessment %>%
accuracy(truth = is_recid, estimate = .pred_class)
# Calculate F1 score
data_assessment %>%
f_meas(truth = is_recid, estimate = .pred_class)
# Several metrics
# Multiple regression metrics
classification_metrics <- metric_set(accuracy, mcc, f_meas)
# mcc: Matthews correlation coefficient:
# The returned function has arguments:
data_assessment %>% # training set predictions
classification_metrics(truth = is_recid, estimate = .pred_class)
library(ggplot2)
library(dplyr)
data_assessment %>%
pr_curve(truth = is_recid,
estimate = .pred_1,
event_level = "second") %>%
pivot_longer(cols = c("recall", "precision"),
names_to = "measure",
values_to = "value") %>%
mutate(measure = recode(measure,
"recall" = "Recall (= True pos. rate = TP/P = sensitivity)",
"precision" = "Precision (= Pos. pred. value = TP/P*)")) %>%
ggplot(aes(x = .threshold,
y = value,
color = measure)) +
geom_line() +
xlab("Threshold value") +
ylab("Value of measure") +
theme_bw()
data_assessment %>%
roc_curve(truth = is_recid, estimate = .pred_1,
event_level = "second") %>%
autoplot() +
xlab("1 - specificity (FP/N, false positives rate)") +
ylab("sensitivity (TP/P, true positives rate)")
# Compute area under ROC curve
data_assessment %>%
roc_auc(truth = is_recid, estimate = .pred_1,
event_level = "second")
# Build and evaluate model WITH resampling
data <- read_csv(sprintf("https://docs.google.com/uc?id=%s&export=download",
"1plUxvMIoieEcCZXkBpj4Bxw1rDwa27tj"))
# Convert outcome to factor
data <- data %>%
mutate(is_recid = factor(is_recid))
# 1.
# Split the data into training and test data
data_split <- initial_split(data, prop = 0.80)
data_split # Inspect
# Extract the two datasets
data_train <- training(data_split)
data_test <- testing(data_split) # Do not touch until the end!
# 2.
# Create resampled partitions
set.seed(345)
data_folds <- vfold_cv(data_train, v = 10) # V-fold/k-fold cross-validation
data_folds # data_folds now contains several resamples of our training data
# You can also try loo_cv(data_train) instead
# 3.
# Define the model
lr_mod <-
logistic_reg() %>%
set_engine("glm")
# 4.
# Create workflow
ml_wflow <-
workflow() %>%
add_model(lr_mod) %>%
add_formula(is_recid ~ age + priors_count)
# Use workflow to fit the to train the model
# There is no need in specifying data_analysis/data_assessment as
# the functions understand the corresponding parts
ml_fit_rs <-
ml_wflow %>%
fit_resamples(resamples = data_folds,
control = control_resamples(save_pred = TRUE,
extract = function (x) extract_fit_parsnip(x)))
ml_fit_rs
# Extract components
ml_fit_rs$.metrics[1:2] # get first two rows of .metrics
ml_fit_rs$.extracts [1:2] # get first two rows of .extracts
ml_fit_rs$.extracts[[1]]$.extracts # Extract models
# Collect metrics across resamples
collect_metrics(ml_fit_rs)
# Compare to single fold approach
data_assessment %>% # training set predictions
classification_metrics(truth = is_recid, estimate = .pred_class)
data_folds %>%
mutate(
# Extract the train data frame for each split
data_analysis = map(.x = splits, ~training(.x)),
# Extract the validate data frame for each split
data_assessment = map(.x = splits, ~testing(.x))
) %>%
# Fit/train the model on data_analysis
mutate(fit = map(.x = data_analysis,
~lr_mod %>%
fit(is_recid ~ age + priors_count,
data = .x))) %>%
# Extract the coefficients
mutate(coefficients = map(.x = fit, ~tidy(.x))) %>%
# Unnest the dataframe to get the coefficients for the different folds
unnest(coefficients)