2.3 Importing a file from another format
While R is a programming language, RStudio does have some menu options.
NOTE: Best practice is to import using code, not point-and-click. That way you have a record of what you did. However, when you use RStudio’s “File - Import Dataset” menu option, the code will be shown and you can copy the code and save it in an R script file. [If you copy from the console, you may have to delete some > or + characters that appear since the console is showing output.]
Using “Import Dataset” is also a great way to explore the data before importing it so you can see if there are problems (e.g., a column with a mix of character and numeric data). You can try out different delimiters or missing data options and see the result in the preview window before importing.
Using File – Import Dataset, you can import data from a Text file, Excel, SPSS, SAS, or Stata. If the file is from another format, you will need to find a way to convert it into one of these formats before importing into R, or possibly there is another R function out there for that format which you can find online There are two “Text” options, one that uses base R and the other the readr (Wickham, Hester, and Bryan 2024) package, which is part of tidyverse (Wickham et al. 2019; Wickham 2023b; Wickham and Grolemund 2017).
2.3.1 Delimited .txt file
2.3.1.1 From Text (base) (base R)
DO: File – Import Dataset – From Text (base). Browse to where you saved the file RheumArth.txt and select that file. You should then see the following:
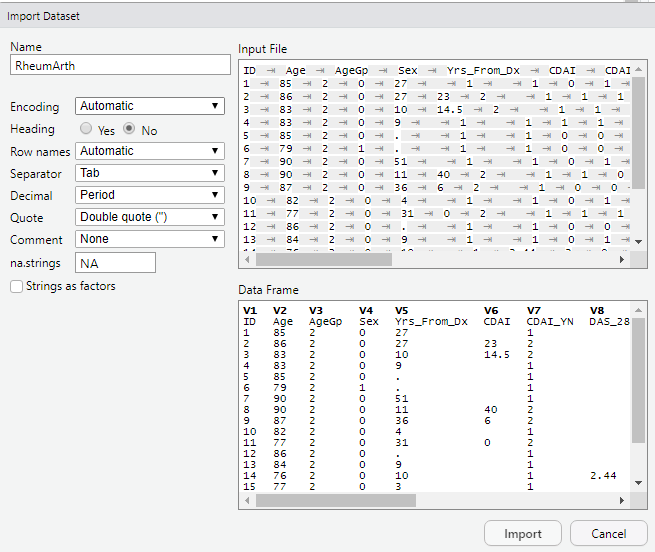
Things you can do in the import dialog box:
- Name: Name your dataset (the default is the name of the file, in this case “RheumArth”)
- Encoding: Change the Encoding (for example, if the file is in a different language)
- Heading: Set Heading = Yes if your variable names are in the first row.
DO: Try Heading = Yes vs. No and see what changes in the Data Frame preview.
- Row names: Specify if there are row names… Typically you will leave this at Automatic.
- Separator: Specify the delimiter, here called a “separator.” In this example, R correctly guessed “Tab.”
DO: Select some other delimiter options and see how the preview changes. For anything other than Tab, there is either no preview or all the data are in one variable (it may look like they are in columns but there is a single variable name in the top row made up of all the column headings squished together). Reset the delimiter to “Tab” before proceeding.
- Decimal: Specify the character used for decimals.
- Quote: Specify if double, single, or no quotes are used.
- Comment: Specify if there is a character that designates a line as being a comment rather than data. Comments are skipped.
- na.strings: Specify what character designates missing data. The default is NA. Sometimes you will want to use a period (.) instead.
- Strings as factors: This affects how R handles a column with characters.
- If left unchecked, a column with ANY characters will be read in as a
charactervariable. - If checked, a column with ANY characters will be read in as a
factorvariable. - I sometimes leave this UNchecked and then convert variables that should be factors later in my code. It is easier to fix misspellings before converting to a factor.
- If left unchecked, a column with ANY characters will be read in as a
DO: Import RheumArth.txt after setting Heading = Yes. The code should appear in your console and a dataset tab should appear.
NOTE: The code automatically displayed in the console may not be portable because of the way the path is specified – it is specific to the computer you are working on. The code below is portable. If you were to move your R Project folder to another location on your computer, or to a different computer and open RStudio from the .Rproj file in the new location, the code below would still work.
# Import RheumArth.txt from the working directory
# using read.delim with the default options
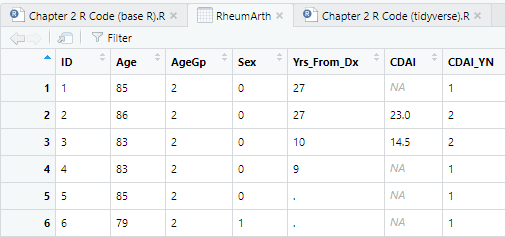
RheumArth <- read.delim("Data/RheumArth.txt")You can see in the dataset tab that blanks were imported as NA (missing data), but periods were left as periods. These should also be missing data.
DO: Re-import RheumArth.txt, but this time type a period (.) in na.strings.
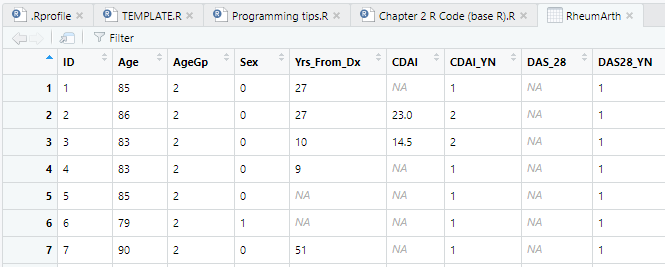
Looking in the new dataset tab, both periods and blanks are now set to NA, signifying a missing value. In this dataset, that method worked. But if the dataset contained two types of missing value codes, not including blanks and NAs, you would not be able to handle it using the Import Dataset dialog because you cannot specify more than one missing data code. However, you can do this in the code using a vector created by c() for na.strings. For example:
# Import RheumArth.txt using read.delim with the na.strings option
# In the Import Dataset dialog, you cannot specify more than one
# missing data code, but you can using code.
RheumArth <- read.delim("Data/RheumArth.txt", na.strings=c("NA", "."))NOTE: It is possible that none of the options will lead to a readable dataset. In that case, you will have to do some detective work to figure out what is causing the problem. It could be there are some extra rows or columns in the file that should not be there, or some unreadable characters. Sometimes the “simple” task of reading in a raw dataset can be very time consuming!
2.3.1.2 From Text (redr) (tidyverse)
The base R function read.delim() has a tidyverse counterpart called read_delim() which is in the readr package (loaded automatically along with tidyverse so you do not need to load it if you have already run library(tidyverse)).
DO: File – Import Dataset – From Text (readr).
DO: Click on “Browse” at the upper right and navigate to where you saved the file RheumArth.txt and select that file. You should then see the following:
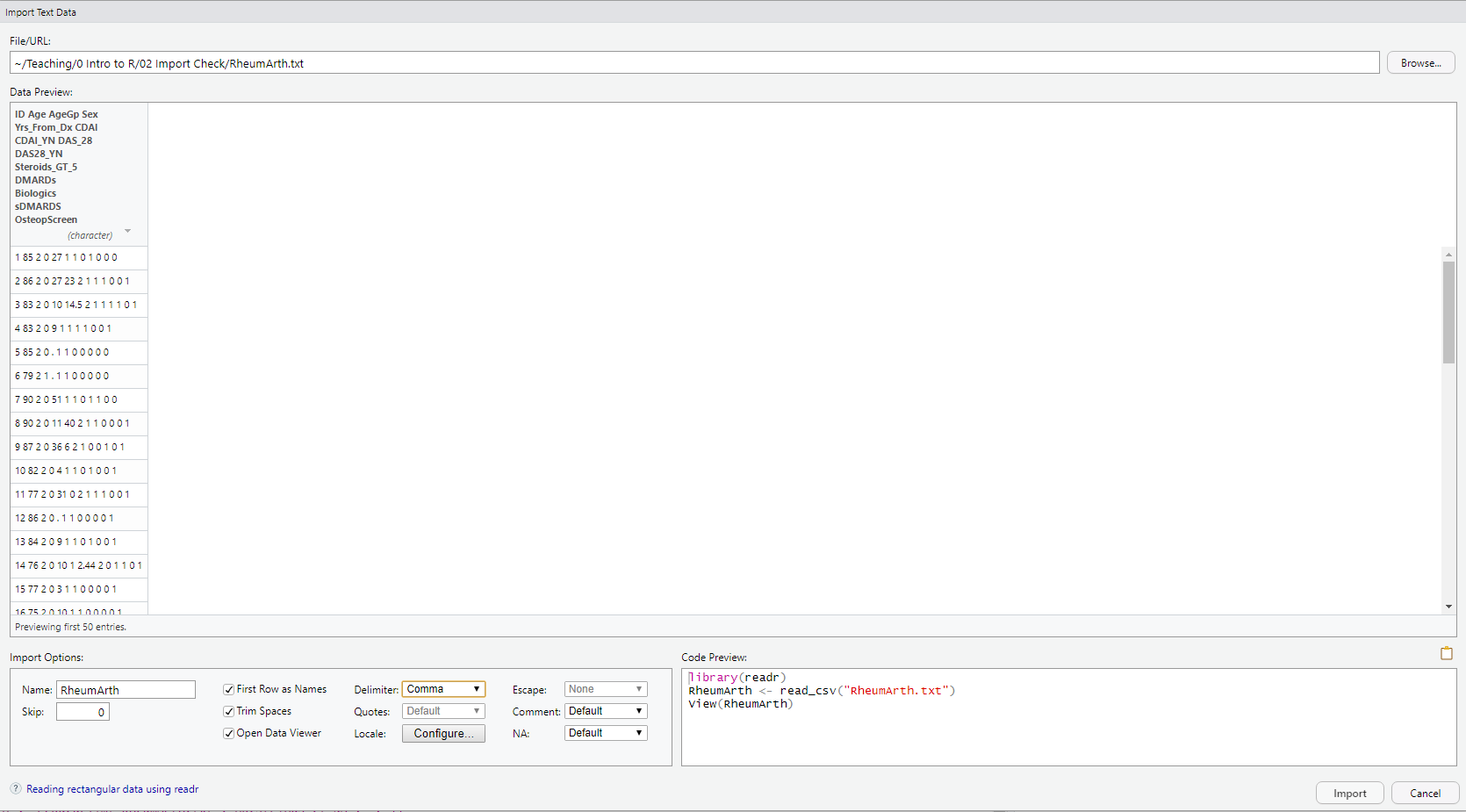
The Data Preview shows that the data have been read into a single variable. The “Import Options” part of the window has a number of options, and the default settings do not seem to be working in this case.
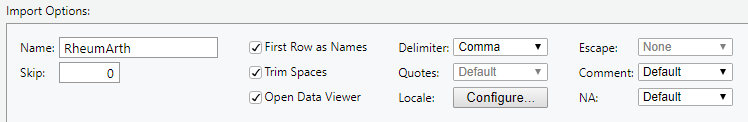
- Name: Name your dataset (the default is the name of the file, in this case “RheumArth”)
DO: Change the name to “RheumArth_tibble”.
- Skip: How many rows to skip at the top of the file. Usually 0.
- First Row as Names: Check this if your variable names are in the first row.
- Trim Spaces: Check this to trim off white space at the beginning and end of values.
- Open Data Viewer: Check this to run
View()after importing so you can view the data in a data tab. - Delimiter: Specify the delimiter to use.
DO: Select some other delimiter options and see how the preview changes. Select Tab before proceeding.
- Locale: Opens a dialog where you can specify various formats.
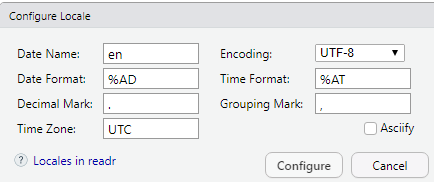
- Escape: Certain characters, like quotes, can have special meanings (like designating what is text). An “escape” character tells R to read in the next character literally, not as a special character (e.g., \” would tell R that the text to read is the quote, rather than the quote being a special character).
- Comment: Specify if there is a character that designates a line as being a comment rather than data. Comments are skipped.
- NA: Specify what character designates missing data. The default is NA and there are only a few options, but you can use code to specify other missing value codes (shown below).
This procedure does NOT convert character data to factors (recall, with base R, we had to check or uncheck a box called “Strings as factors” – that is not an option here).
DO: Import RheumArth.txt with the name “RheumArth_tibble”, use Tab as the delimiter, and leave the other options at their default values. The following should appear in your console and a dataset tab should appear.
# Output
> library(readr)
> RheumArth_tibble <- read_delim("Data/RheumArth.txt",
+ "\t", escape_double = FALSE, trim_ws = TRUE)
Parsed with column specification:
cols(
ID = col_double(),
Age = col_double(),
AgeGp = col_double(),
Sex = col_double(),
Yrs_From_Dx = col_character(),
CDAI = col_double(),
CDAI_YN = col_double(),
DAS_28 = col_double(),
DAS28_YN = col_double(),
Steroids_GT_5 = col_double(),
DMARDs = col_double(),
Biologics = col_double(),
sDMARDS = col_double(),
OsteopScreen = col_double()
)
> View(RheumArth_tibble)Some things to notice about this output…
- The Import dialog automatically added
library(readr)to load the readr package (Wickham, Hester, and Bryan 2024), the only part oftidyverseneeded forread_delim(). If you already ranlibrary(tidyverse)thenreadris actually already loaded. - Unlike the base R Import dialog, this one produces reproducible code (since the file was in the working directory, there is no path specified).
- Tab delimiters are specified using “\t”.
NOTE: If the name of an R script in the RStudio tab appears in red with an asterisk, that means you have unsaved changes. Use File – Save to save your file.
Blanks were imported as NA (missing data), but periods were left as periods. These should also be missing data.
DO: Re-import RheumArth.txt but this time tell R to set periods (.) to missing. You cannot do this in the Import Text Data dialog, but you can with the following code.
Now all the periods are NA (missing value).
read_delim() automatically sets the resulting data.frame to be of class tbl_df (which, if you read the first part out loud, sounds like “tibble”).
## [1] "spec_tbl_df" "tbl_df" "tbl" "data.frame"Also, the output window explicitly tells you the type of each column that was read in.
cols(
ID = col_double(),
Age = col_double(),
AgeGp = col_double(),
Sex = col_double(),
Yrs_From_Dx = col_double(),
CDAI = col_double(),
CDAI_YN = col_double(),
DAS_28 = col_double(),
DAS28_YN = col_double(),
Steroids_GT_5 = col_double(),
DMARDs = col_double(),
Biologics = col_double(),
sDMARDS = col_double(),
OsteopScreen = col_double()
)2.3.2 Comma delimited .csv file
A .csv file is a special kind of text file. It is comma delimited and if you have Excel then .csv files by default open in Excel. If you use the methods described in Section 2.3.1 to import a .csv file, R will use functions specifically designed for .csv files. The code for these options is described here.
2.3.3 Excel (.xls, .xlsx)
The R function readxl() is from the readxl package (Wickham and Bryan 2025). tidyverse does not load it automatically, but it is part of the tidyverse in the sense that it creates a tibble, not a data frame.
DO: File – Import Dataset – From Excel
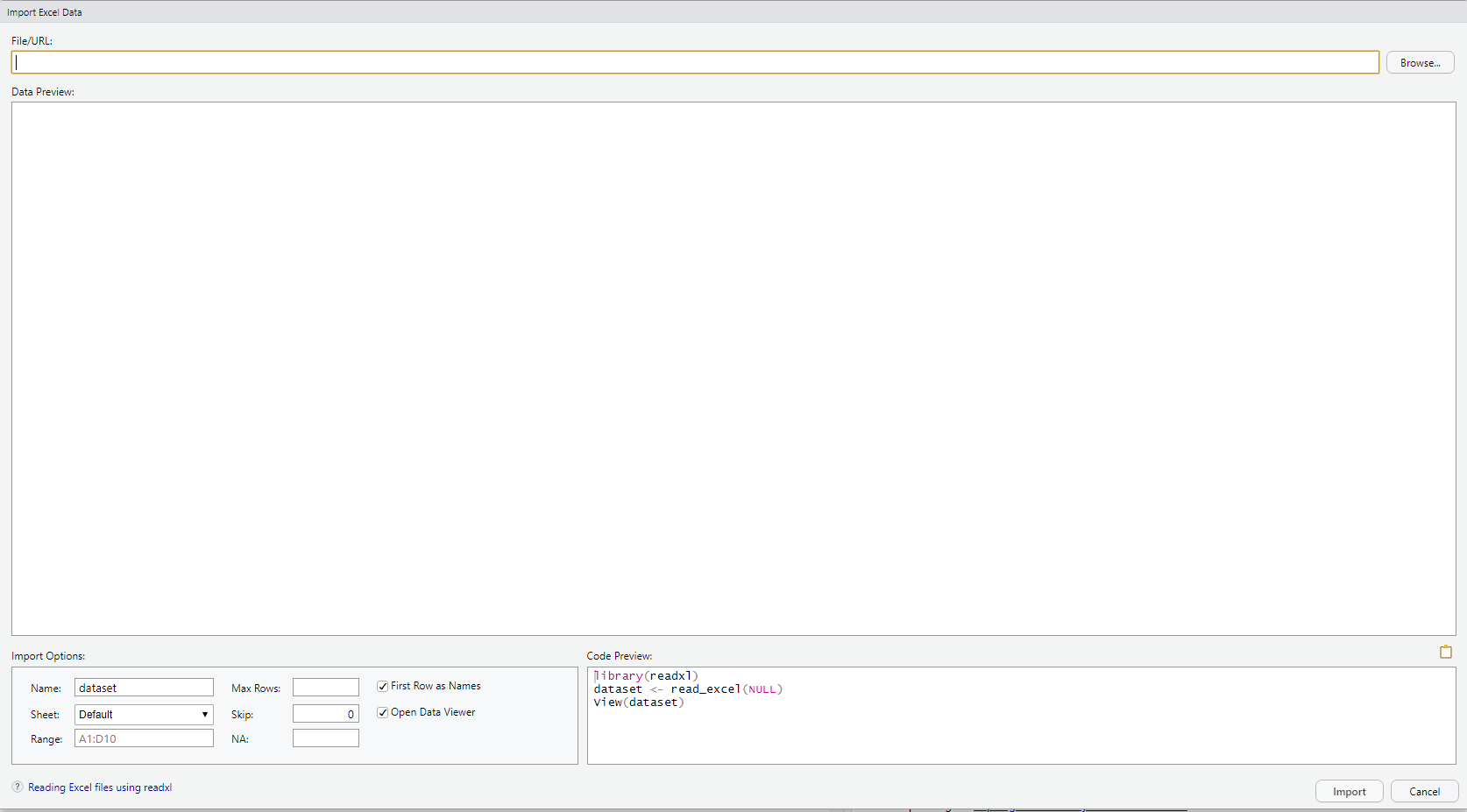
DO: Click on “Browse” at the upper right and navigate to where you saved the file RheumArth.xlsx and select that file. You should then see the following:
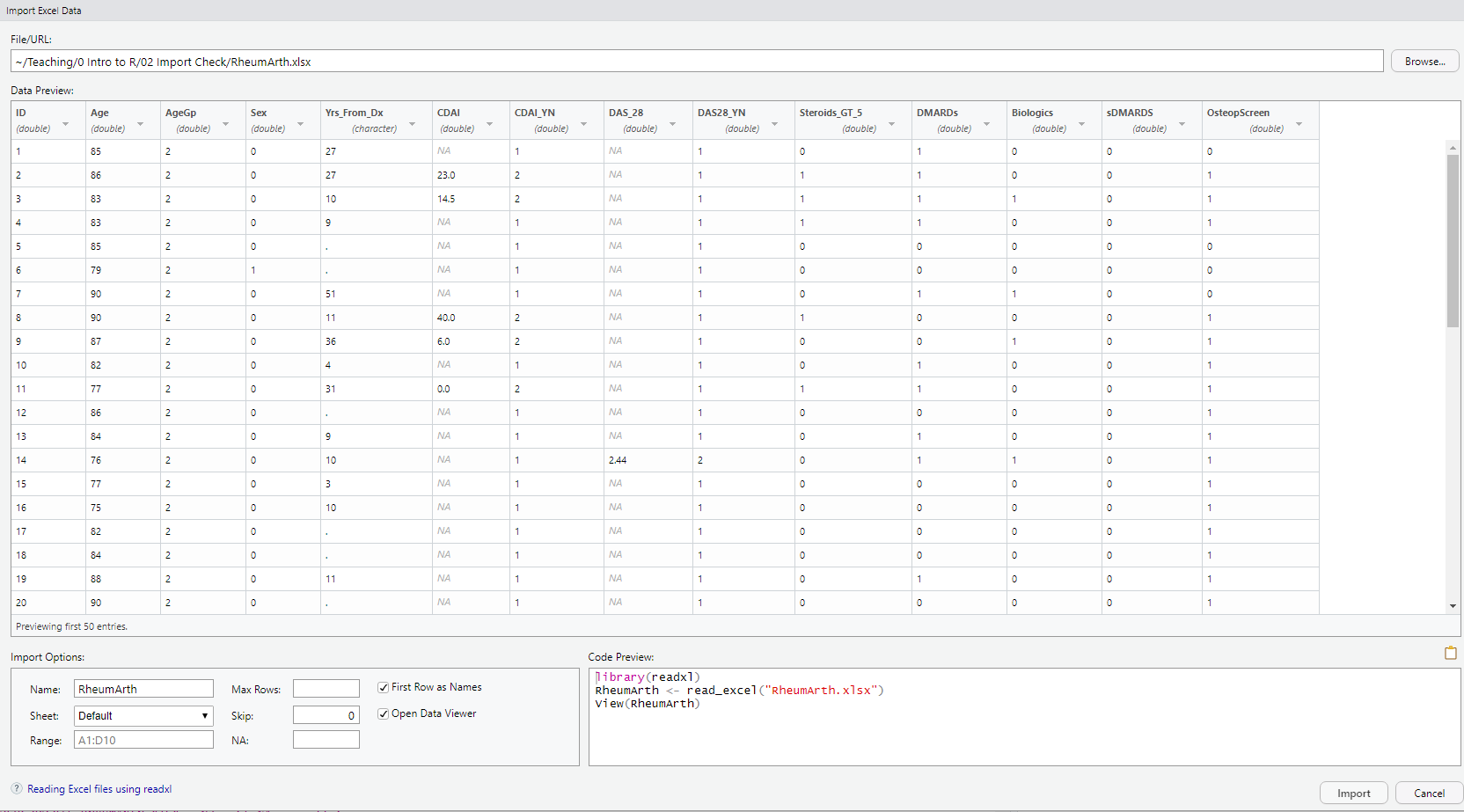
Import Options has the following:
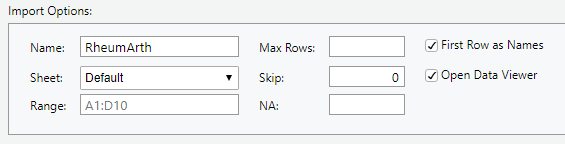
- Name: Name your dataset (the default is the name of the file, in this case “RheumArth”)
- Sheet: R can only read in one worksheet at a time, so if there is more than one worksheet in your Excel file, choose one.
- Range: You can tell R to only import data from a subset of the cells. It defaults to the smallest rectangle of cells with anything that could be data. It is common to see an Excel file with cells filled with metadata like where the data came from, etc. and summaries like row or column averages. Using Range, you can limit what you read in to the rectangle containing the data.
- Max Rows: Tells R to stop after a certain number of rows (e.g., if there is extra information at the end of the file you do not want to read in). Usually leave blank.
- Skip: How many rows to skip at the top of the file. Usually 0.
- NA: Specify what character designates missing data.
DO: Type . (a period) in the NA box and see how the Data Preview changes.
- First Row as Names: Check this if your variable names are in the first row.
- Open Data Viewer: Check this to run View() after importing so you can view the data in a data tab.
DO: Import RheumArth.xlsx, naming it “RheumArth_tibble” and setting NA to period (.).
# readxl is part of tidyverse so will create a tibble,
# but you can use as.data.frame to convert to base R
# (some functions do not work with tibbles)
library(readxl)
RheumArth_tibble <- read_excel("Data/RheumArth.xlsx", na = ".")
class(RheumArth_tibble)## [1] "tbl_df" "tbl" "data.frame"## [1] "data.frame"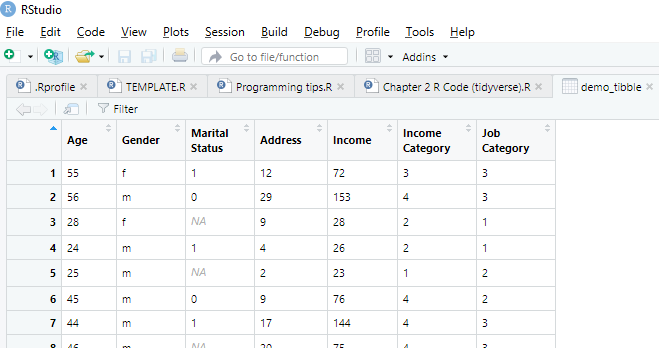
2.3.4 SPSS file (.sav)
You can use File – Import Dataset – From SPSS or the following code:
The haven package (Wickham, Miller, and Smith 2023) results in a tibble that has both variable values and labels.