14 Multi-Modality Matching
14.1 Step 1: Model Selection and Sample Filtering
Initiate by selecting relevant models and apply the “Clinical Filter” to refine the samples within each:
For TCGA, use ‘Cancer_code=STAD’
For CCLE, set ‘Tumor_site=Stomach’
PDX models do not require a filter at this stage.

14.2 Step 2: Hybrid Clustering Method
Proceed by clicking the “Match Me” button, then activate ‘cluster_row=TRUE’. Illustrated below is the hybrid clustering output from multi-modality matching:
TCGA use ‘Cancer_code=STAD’
CCLE set ‘Tumor_site=Stomach’
PDX models do not require a filter at this stage.
Figure. An example of Multi-Modality Matching.
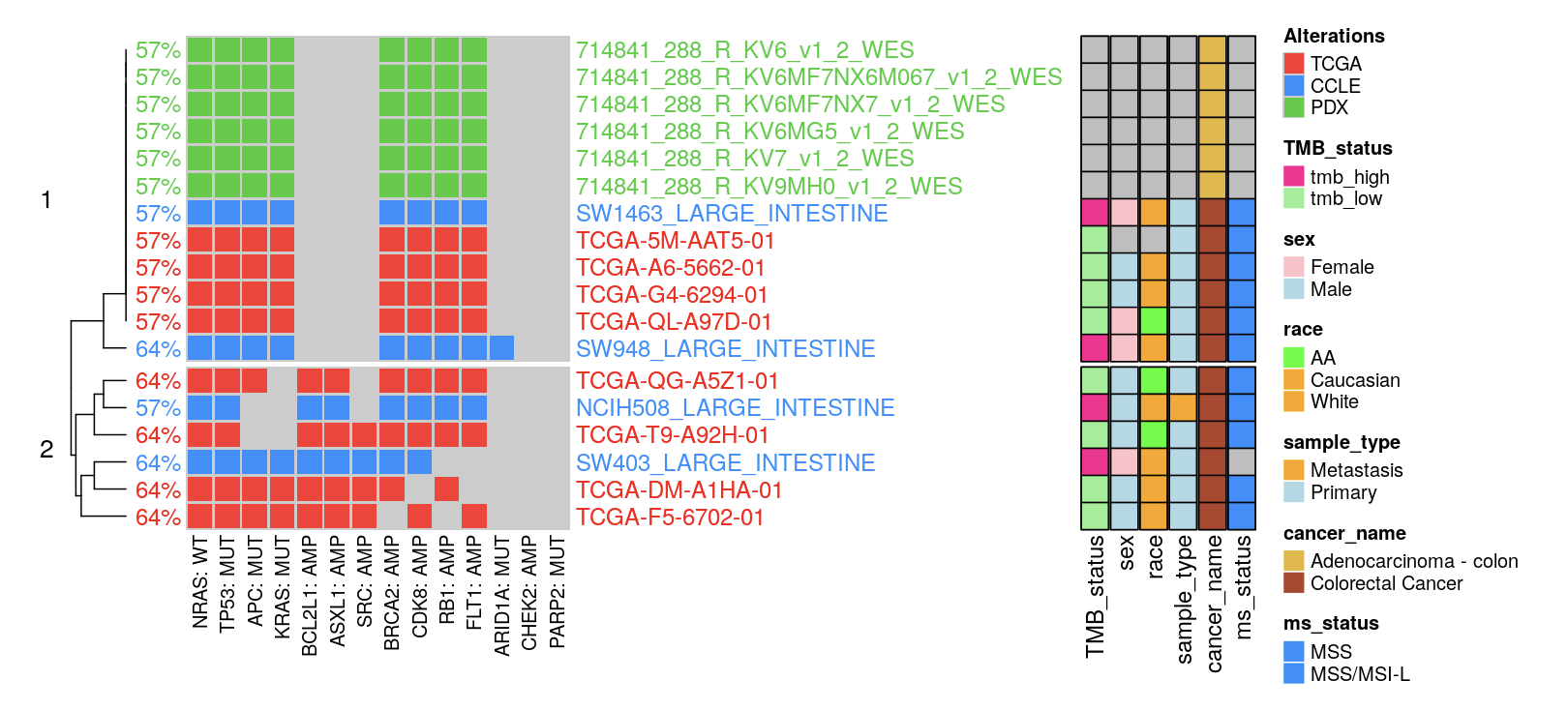
14.3 Step 3: Oncoplot Customization Tools
Post-matching, three tools are offered to further customize oncoplots:
-
Cluster Row: This function clusters matched samples’ rows based on a binary matrix. Users can dictate the number of clusters by entering a value in “N cluster”.
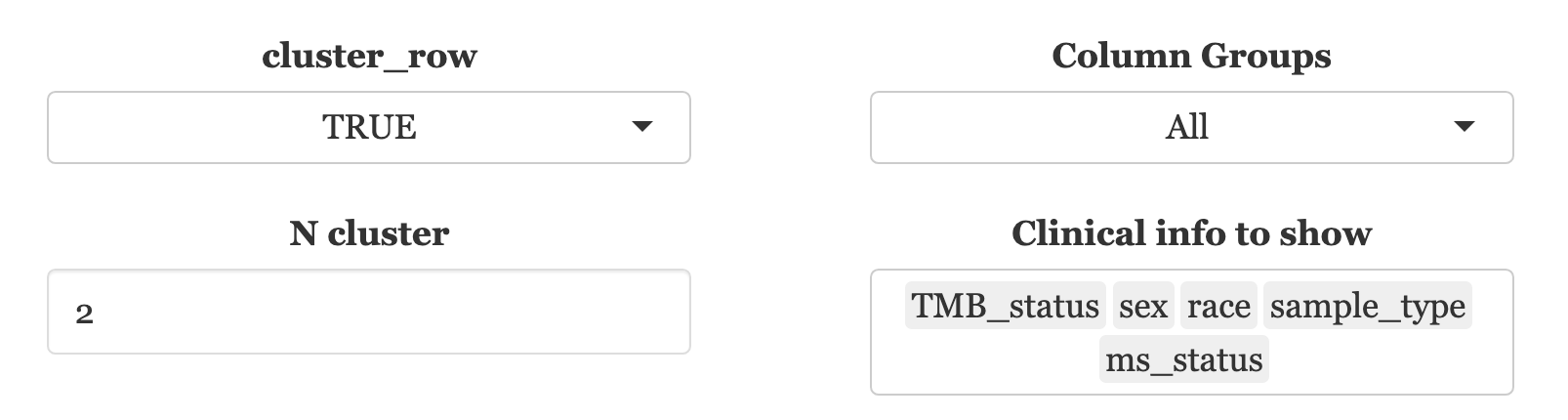
-
Column Groups: This allows for the clustering of columns, which represent gene alterations from patient profiles, again based on a binary matrix.
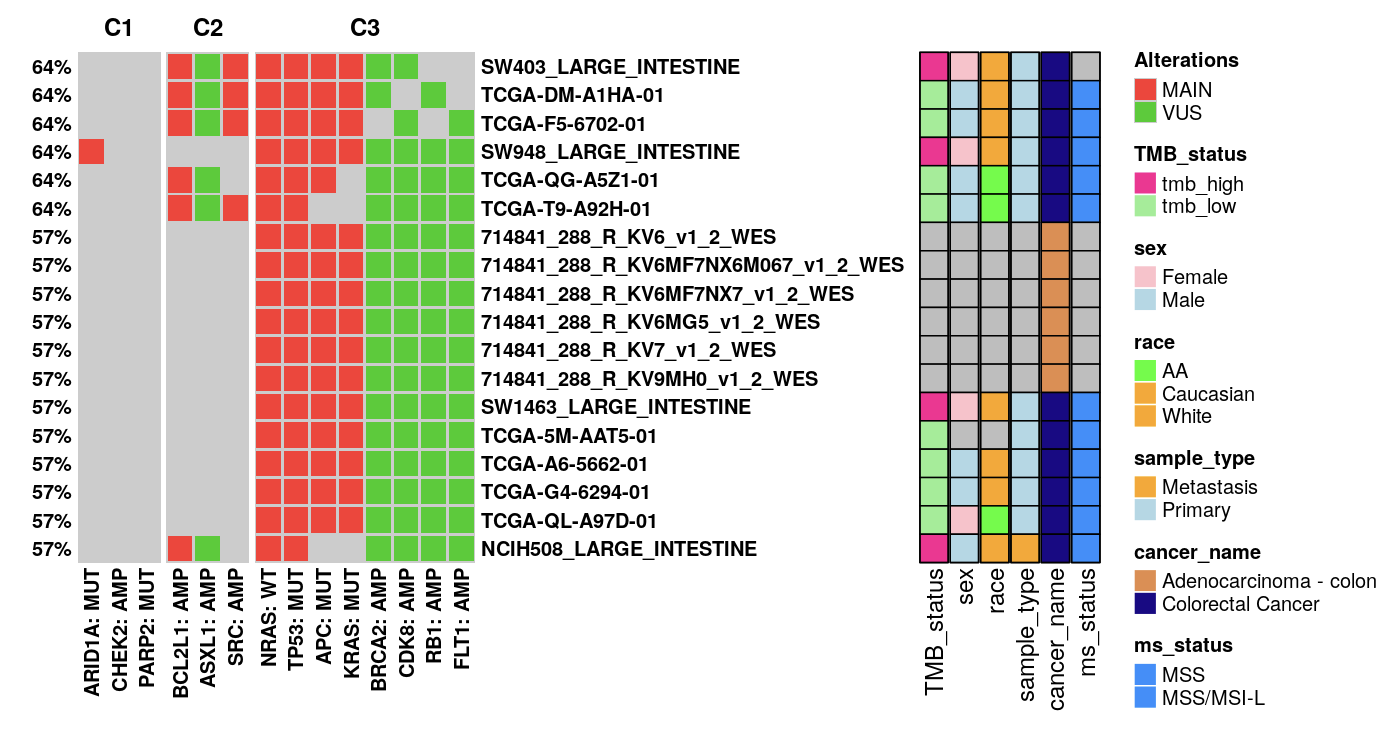
Clinical Info Display: Choose clinical data to augment the oncoplot row annotations. Unavailable data for any model will be marked as ‘NA’ in grey.