17 Clinical trials
This module helps visualize clinical trials listed in the clinical sequence reports.
It connects to the clinicaltrials.gov database to extract the most updated information on clinical trials and summarize them into one single visual, in both technical and non-technical versions.
17.0.1 Input format

There are three ways to provide clinical trials.
For Upload function:
It accepts input in in .csv format:
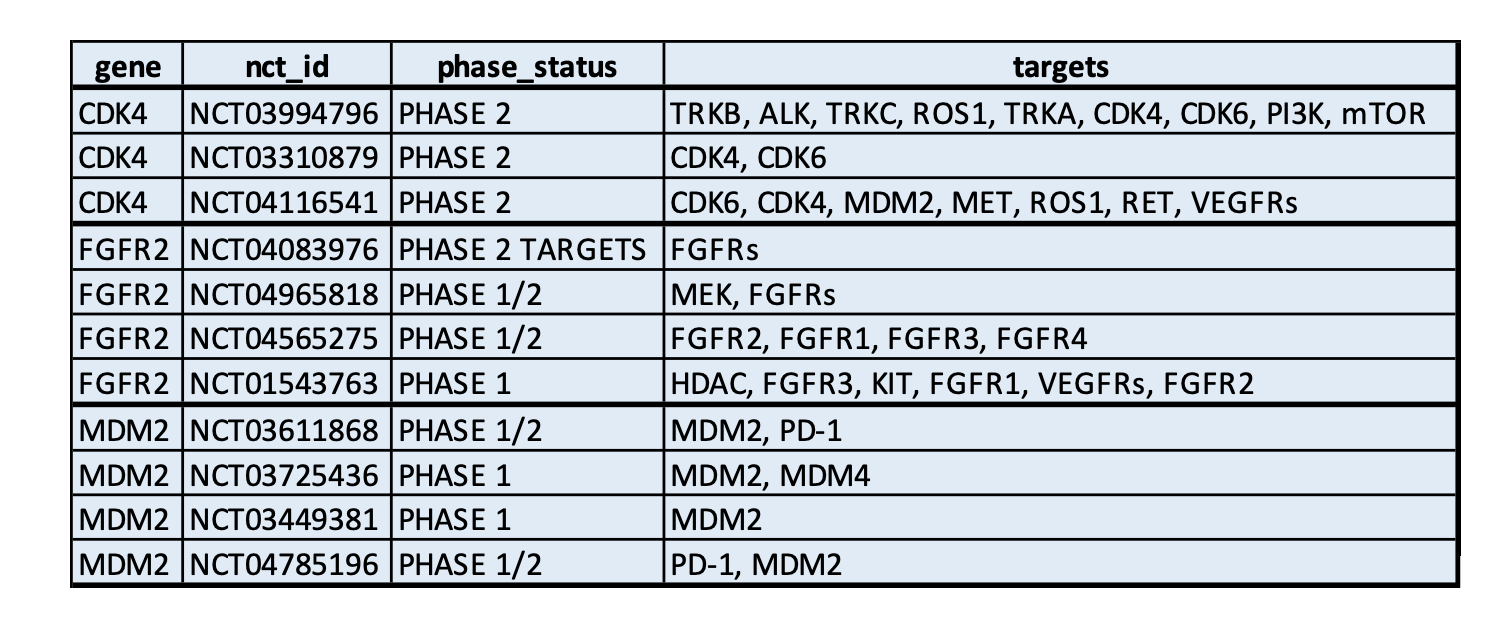
- CSV: Users can upload a table with clinical trials and targets.
Example:
The input table file requires at least three columns, including:
gene: genes in the genome profile that are associated with the clinical trialsnct_id: clinical trial id starting with “NCT”.targets: the gene targets are shown in trials, either the same the genes or associated pathways
Note: If the user uploads the pdf version of de-identified PDF reports that include the pages of clinical trials. The app will extract these information and no manual input will be needed.
For NCT.gov function:
The app will automatically take the input genomic profile and search trials using clinicaltrials.gov API for the most updated trials.
Users can add more information such as cancer type or other keyword to search trials.
We designed three search modes with different searching key word:
Simple: gene symbols
Medium: gene+alteration type. (e.g. KRAS-Mutation)
Strict: gene + specific alteration (e.g. KRAS + G12C)

17.0.2 Informative Clinical trials visualization
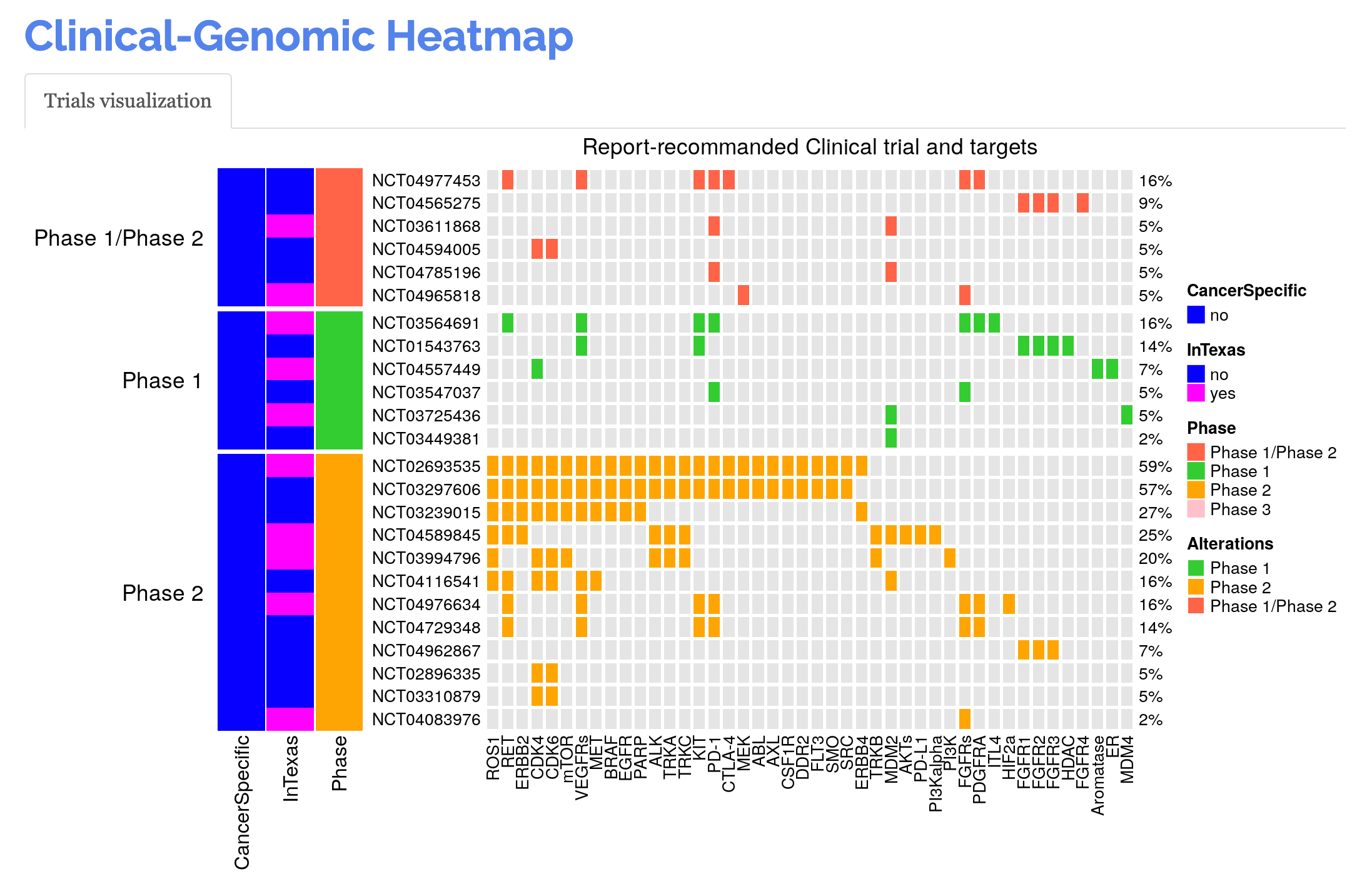
The clinical trials will be visualized in a heat map with altered genes as columns and trials as rows. Cancer type, location, and phase information are annotated for each trial with available data extracted from ClinicalTrials.gov.