2 Genomic profiles
It supports either a single case or a case series.
2.1 Example
The app has two built-in examples, one for “coloreactal cancer” case and the other one for a rare cancer case, “Cholangiocarcinoma”.

2.2 Upload
The app accepts patient genomic profiles in .csv format.
The app also takes table format as an input as introduced below.
2.2.1 A single case
A basic query of a single case study will take a table listing genomic alteration reported. It should include at least four columns:
gene: gene symbols reported
type: The alteration groups, such as
MainandVUS. The app currently supports profiles with 2 groups.alteration: The specific alterations such as mutation sites, amplification, or deletion (allow NA).
alt_type: Needs to be one of below:
MUT,AMP,DEL
(The column names are required to be identical)

2.2.2 A case series
If there is a case series, the app will take one .csv table file as the input.
Users can concatenate all the profiles in one table and add an Id column specifying the profile names.
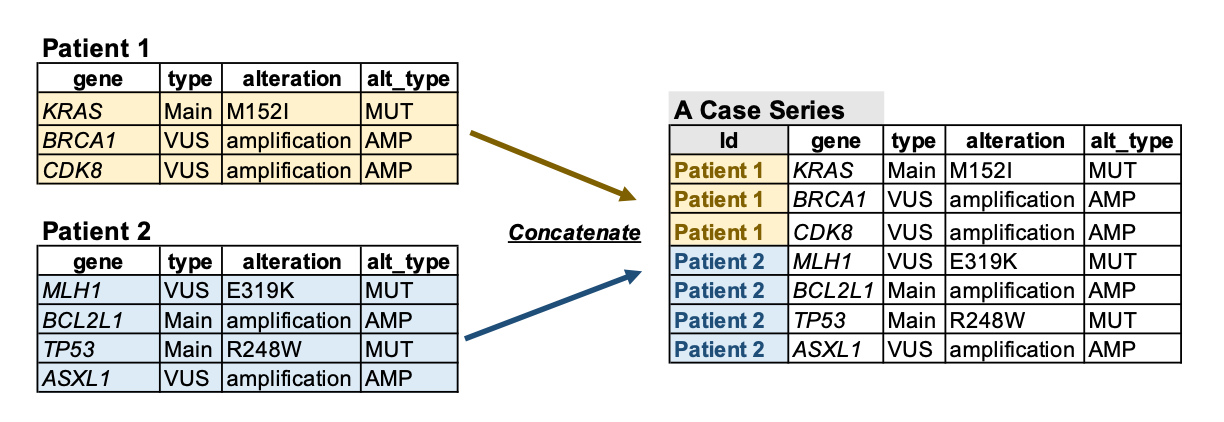
Note: The profile names (Id column) can be from patients, cell lines, etc. myCMIE takes genomic alteration profiles from multiple sources as long as they stay in the format we specify above.
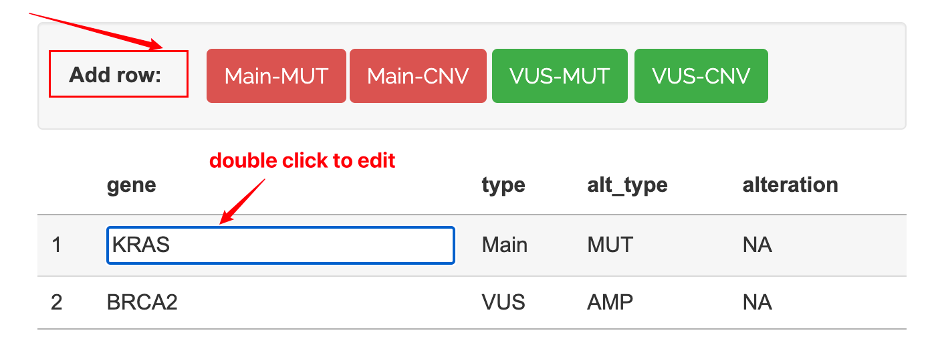
2.3 Online Input
For quick query, the app has a editable table for users to input the genomic profile.
You can directly double click cells edit the table. There are pre-set buttons to add rows for mutation/copy number change options.
2.4 Example
Supplement table for easy copy.
Example in table format:
| gene | type | alteration | alt_type |
|---|---|---|---|
| MLH1 | VUS | E319K | MUT |
| BCL2L1 | Main | amplification | AMP |
| TP53 | Main | R248W | MUT |
| ASXL1 | VUS | amplification | AMP |
| BRCA2 | VUS | amplification | AMP |
| CDK8 | VUS | amplification | AMP |
| NBN | VUS | M152I | MUT |