13 Enhanced Sample Pool
13.1 Enhanced Sample Pool Selection in myCMIE 2.0 (New Feature)
Prior to the matching analysis, the “Sample Pool” feature enables users to choose from a range of public patient profiles within our comprehensive database. Whereas myCMIE 1.0 allowed users to refine their selection by filtering through clinical data such as cancer type, tumor types, and TMB status, myCMIE 2.0 introduces a dual filtering system: the “Clinical Filter” and the “Molecular Filter,” expanding the customization of searches.
13.2 Clinical Filter
Accessible via the “Connect -> Start” menu, the Clinical Filter is enhanced to accommodate multi-model matching. Users are required to refine the sample pool for their desired model by navigating through the dropdown menu at “Connecting -> Dataset.”
Figure 1 illustrates the array of models available for matching.
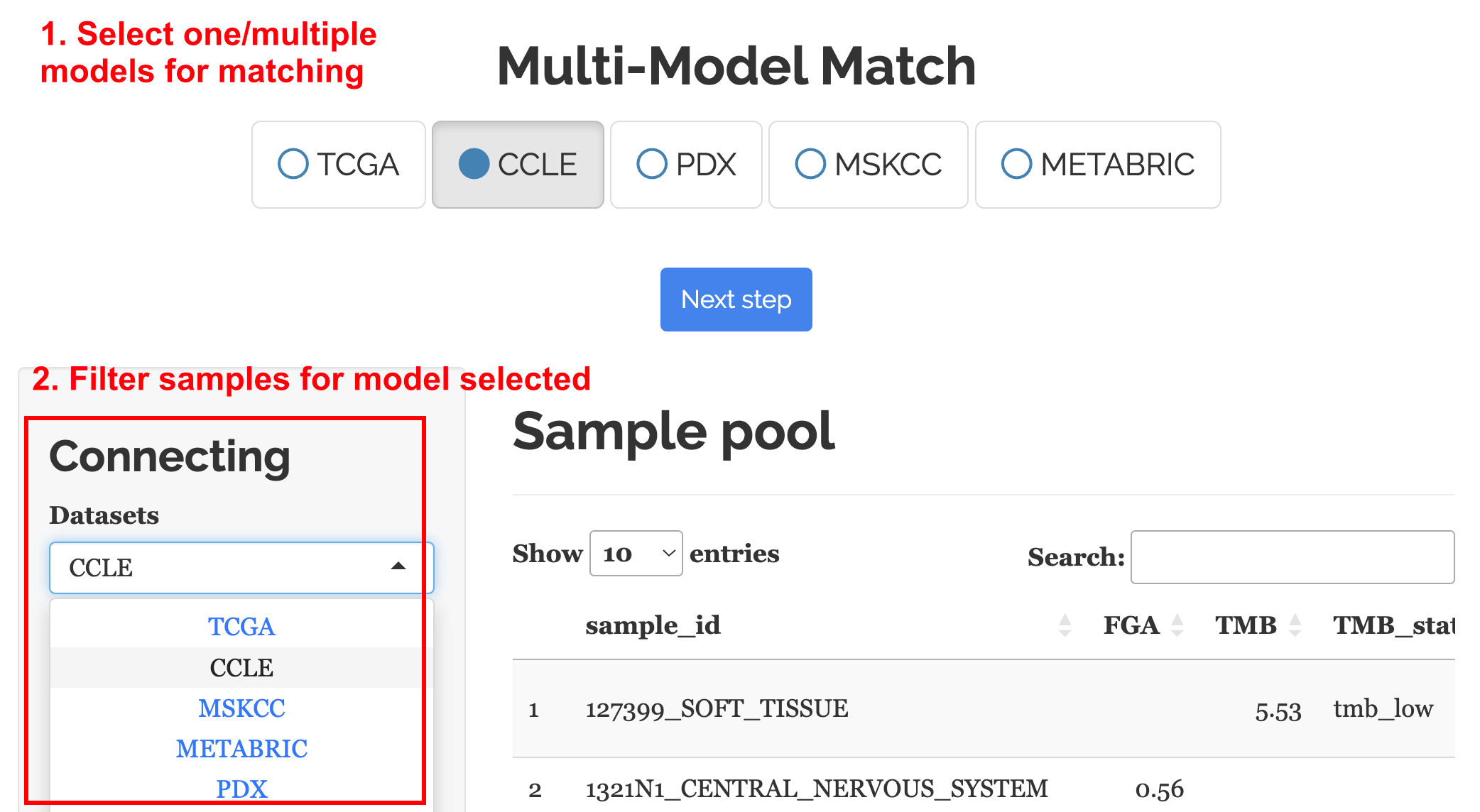
Recommended Primary Filters for Cancer Type Queries:
CCLE: Preferred filtering by ‘Tumor_site’
TCGA: Utilize ‘Cancer_code’ as the main filter
PDX: Filter through ‘Cancer_name’
13.3 Molecular Filter
Molecular filter is under “Connect -> Digital twin community”. Within the “Connect -> Digital twin community” section, an upgraded Molecular Filter is now available. This filter revolutionizes sample selection by allowing users to pinpoint samples with precise genetic alterations, such as ‘ERBB2: AMP’.
The newly implemented ‘Filter Mode’ adds an advanced layer of selection, empowering users with the option to apply either an inclusive (OR) or exclusive (AND) search logic when multiple genetic criteria are involved.
The Molecular Filter operates on a principle that only includes samples possessing the genetic alterations specified by the user, such as gene mutations or copy number alterations, in the matching process. Conversely, samples lacking any of these specified genetic changes are excluded from the search for digital twins.
Figure. Molecular filter with different Filter Modes.
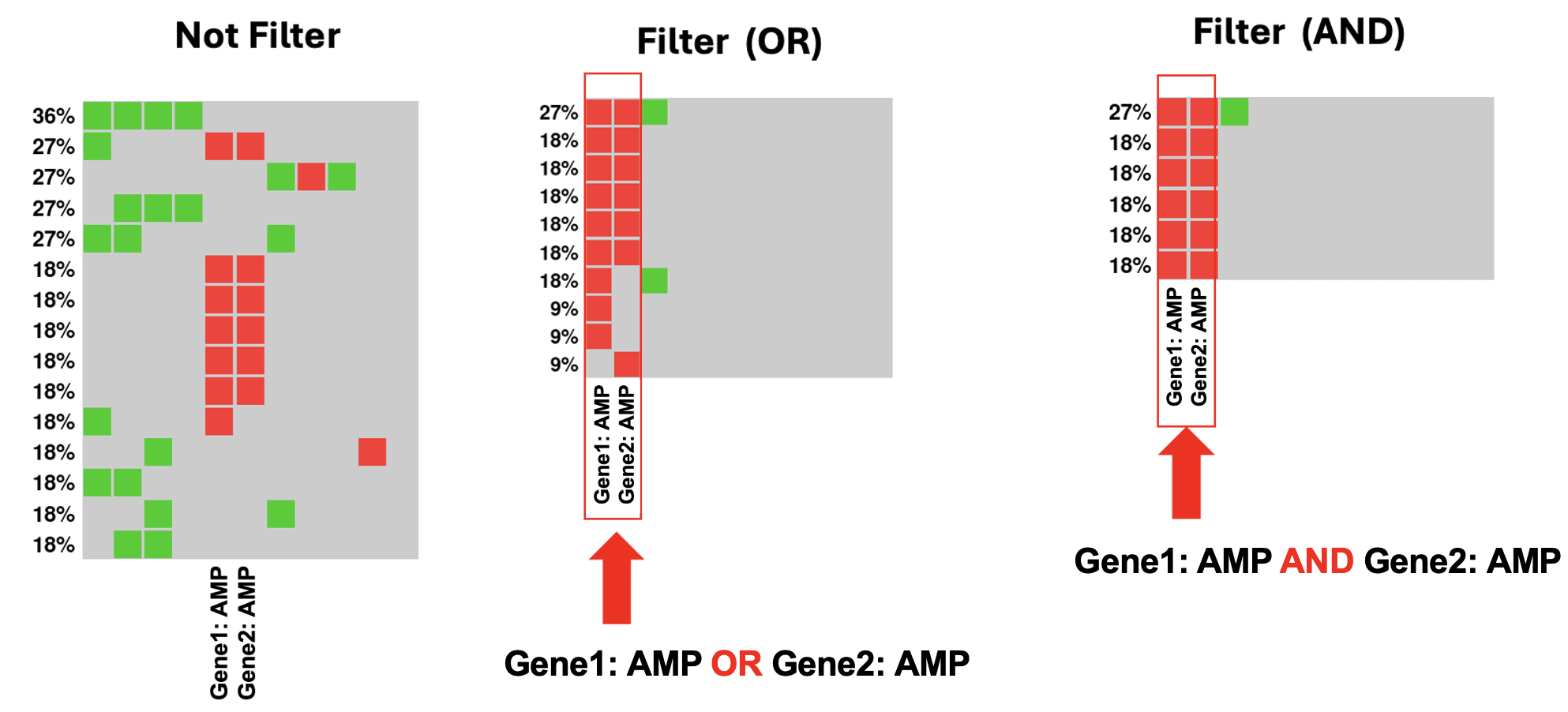
About OQL input: Users have the capability to input numerous “alterations” within the molecular filter. The expected format is “gene: alteration”, with a space following the colon. For instance, ‘ERBB2: AMP’ or ‘MYC: MUT’.
About Filter Mode: When multiple OQL alterations are entered, users have the flexibility to retrieve samples that either contain “all” listed alterations or have “at least one” of the alterations, catering to diverse research needs.