Set env, load arrow project and scRNA-seq
## Section: set default para
##################################################
addArchRThreads(threads = 16) # setting default number of parallel threads
## Section: load object
##################################################
proj <- loadArchRProject(path = "data/ArchR/ArrowProject/Merged/")
## Section: imputation
##################################################
proj <- addImputeWeights(proj, reducedDims = "IterativeLSI_peak")
## Section: load scRNA-seq seurat object
##################################################
ct <- readRDS("data/SYM-scRNA-Res-Demo_expression_mat.RDS")
meta <- readRDS("data/SYM-scRNA-Res-Demo_meta.RDS")
rse <- SummarizedExperiment(assays = SimpleList(counts = ct), colData = meta)
table(colData(rse)$cell_type)
##
## Astrocyte Cycling End Ex Ex-SP Ex-U InCGE InMGE
## 10 65 31 1004 152 425 423 533
## InPSB IP Mic OPC oRG Per vRG
## 32 83 13 37 93 12 87
Unconstrained integration
proj <- addGeneIntegrationMatrix(
ArchRProj = proj,
useMatrix = "GeneScoreMatrix",
matrixName = "GeneIntegrationMatrix",
reducedDims = "IterativeLSI_peak",
seRNA = rse,
addToArrow = TRUE,
groupRNA = "cell_type",
nameCell = "predictedCell",
nameGroup = "predictedGroup",
nameScore = "predictedScore",
force = TRUE
)
Visualize the predicted cell types
plotEmbedding(
proj,
embedding = "UMAP_peak",
colorBy = "cellColData",
name = "predictedGroup_Un"
)
## ArchR logging to : ArchRLogs/ArchR-plotEmbedding-25d375ee72bd-Date-2021-11-12_Time-14-59-25.log
## If there is an issue, please report to github with logFile!
## Getting UMAP Embedding
## ColorBy = cellColData
## Plotting Embedding
## 1
## ArchR logging successful to : ArchRLogs/ArchR-plotEmbedding-25d375ee72bd-Date-2021-11-12_Time-14-59-25.log
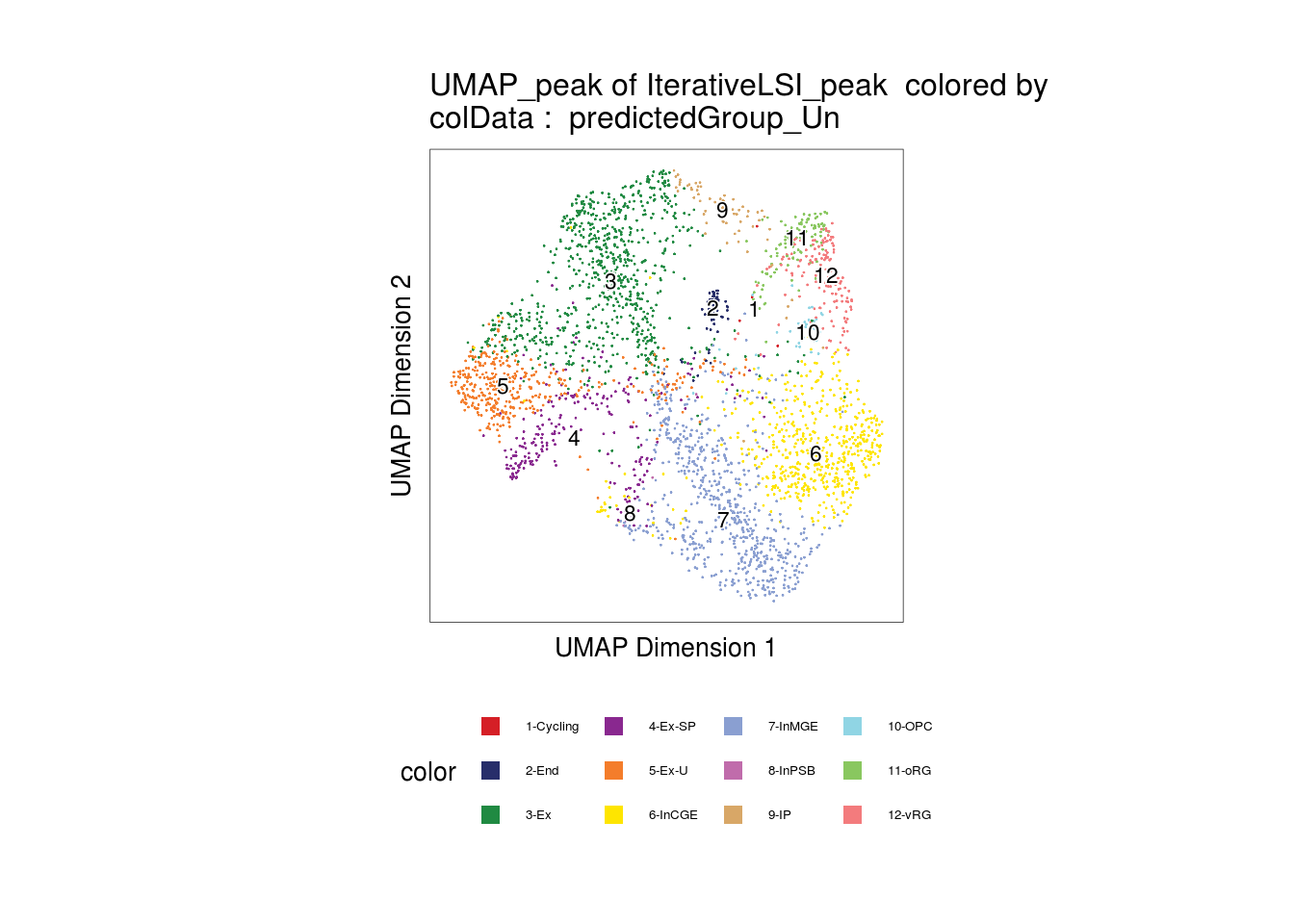
cM <- as.matrix(confusionMatrix(proj$Cluster_peak_res.0.8, proj$predictedGroup_Un))
cM
## End vRG Ex-SP Ex InMGE InCGE Cycling Ex-U OPC IP oRG InPSB
## C1 46 0 0 1 0 0 0 0 0 0 0 0
## C6 0 20 1 17 7 71 0 5 27 3 1 0
## C10 0 0 51 4 107 38 0 2 1 0 0 4
## C8 0 0 1 93 0 0 0 0 0 57 0 0
## C11 0 0 0 0 319 6 0 0 0 0 0 0
## C9 0 0 1 401 0 1 0 0 0 0 0 0
## C2 0 0 62 163 0 3 0 288 0 0 0 0
## C12 0 0 1 1 7 410 0 0 0 1 0 0
## C3 0 0 84 0 0 0 0 1 0 0 0 0
## C7 0 121 0 3 0 0 3 0 3 8 107 0
## C4 3 0 24 69 177 5 0 44 0 0 1 1
## C5 7 2 11 19 23 26 1 30 5 0 1 0
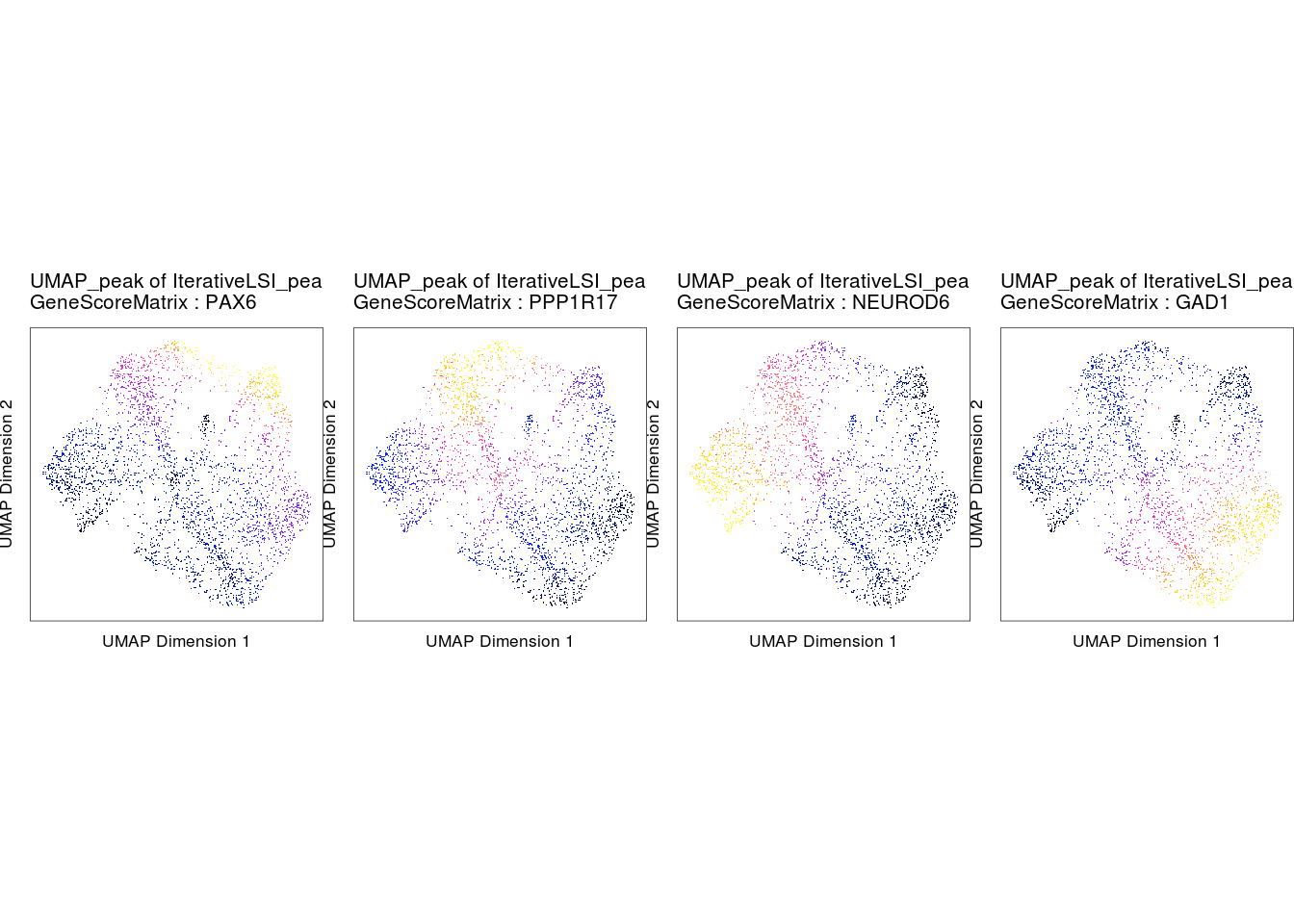
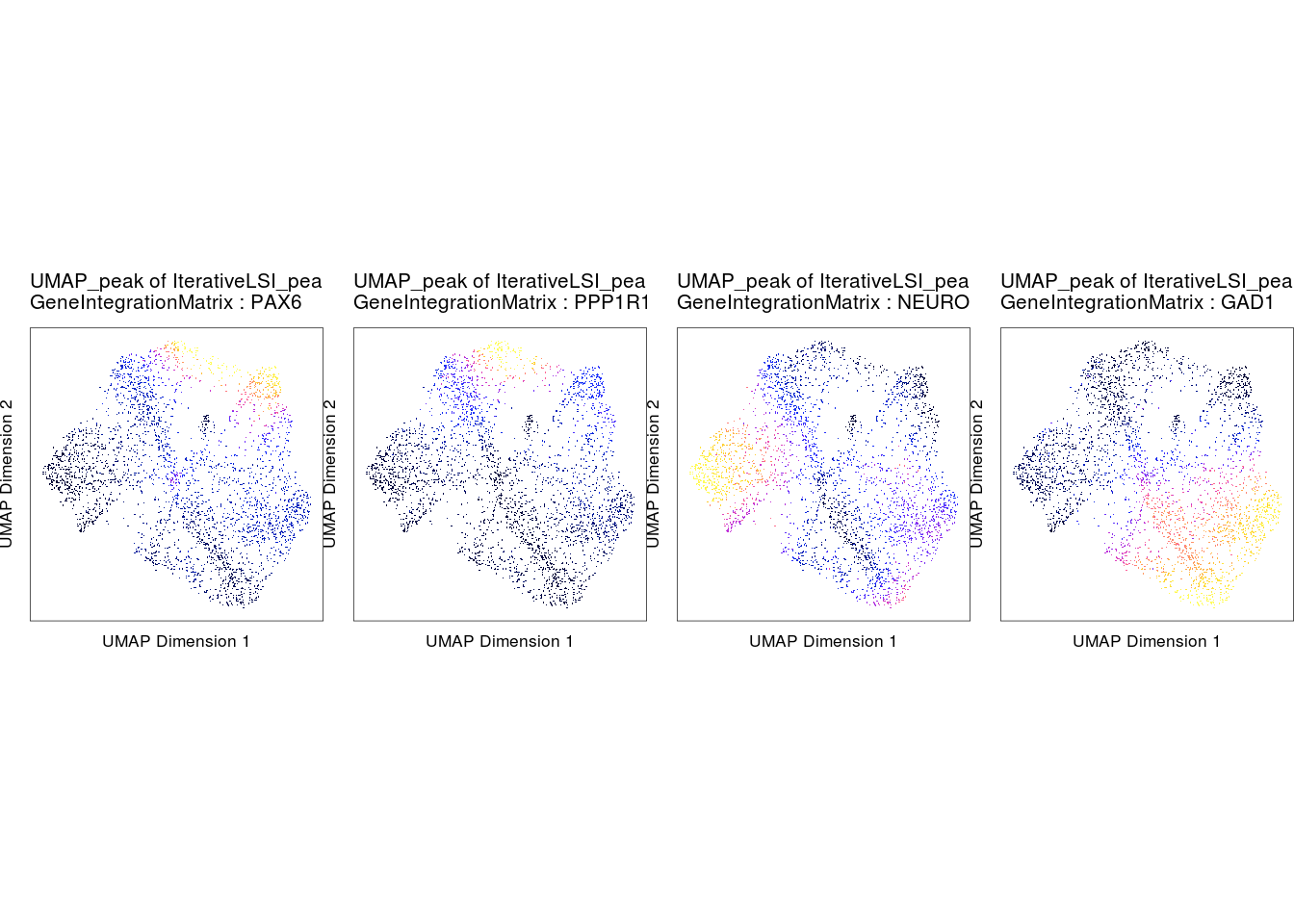
Compare gene scores and gene expression
markerGenes <- c("PAX6", "PPP1R17", "NEUROD6", "GAD1")
p1 <- plotEmbedding(
ArchRProj = proj,
colorBy = "GeneIntegrationMatrix",
name = markerGenes,
continuousSet = "horizonExtra",
embedding = "UMAP_peak",
imputeWeights = getImputeWeights(proj)
)
## Getting ImputeWeights
## ArchR logging to : ArchRLogs/ArchR-plotEmbedding-25d34b4c695a-Date-2021-11-12_Time-14-59-31.log
## If there is an issue, please report to github with logFile!
## Getting UMAP Embedding
## ColorBy = GeneIntegrationMatrix
## Getting Matrix Values...
## 2021-11-12 14:59:32 :
##
## Imputing Matrix
## Using weights on disk
## 1 of 1
## Plotting Embedding
## 1 2 3 4
## ArchR logging successful to : ArchRLogs/ArchR-plotEmbedding-25d34b4c695a-Date-2021-11-12_Time-14-59-31.log
p2 <- plotEmbedding(
ArchRProj = proj,
colorBy = "GeneScoreMatrix",
continuousSet = "horizonExtra",
name = markerGenes,
embedding = "UMAP_peak",
imputeWeights = getImputeWeights(proj)
)
## Getting ImputeWeights
## ArchR logging to : ArchRLogs/ArchR-plotEmbedding-25d3613c9d45-Date-2021-11-12_Time-14-59-36.log
## If there is an issue, please report to github with logFile!
## Getting UMAP Embedding
## ColorBy = GeneScoreMatrix
## Getting Matrix Values...
## 2021-11-12 14:59:37 :
##
## Imputing Matrix
## Using weights on disk
## 1 of 1
## Plotting Embedding
## 1 2 3 4
## ArchR logging successful to : ArchRLogs/ArchR-plotEmbedding-25d3613c9d45-Date-2021-11-12_Time-14-59-36.log
p1c <- lapply(p1, function(x){
x + guides(color = FALSE, fill = FALSE) +
theme_ArchR(baseSize = 6.5) +
theme(plot.margin = unit(c(0, 0, 0, 0), "cm")) +
theme(
axis.text.x=element_blank(),
axis.ticks.x=element_blank(),
axis.text.y=element_blank(),
axis.ticks.y=element_blank()
)
})
p2c <- lapply(p2, function(x){
x + guides(color = FALSE, fill = FALSE) +
theme_ArchR(baseSize = 6.5) +
theme(plot.margin = unit(c(0, 0, 0, 0), "cm")) +
theme(
axis.text.x=element_blank(),
axis.ticks.x=element_blank(),
axis.text.y=element_blank(),
axis.ticks.y=element_blank()
)
})
do.call(cowplot::plot_grid, c(list(ncol = 4), p1c))
do.call(cowplot::plot_grid, c(list(ncol = 4), p2c))