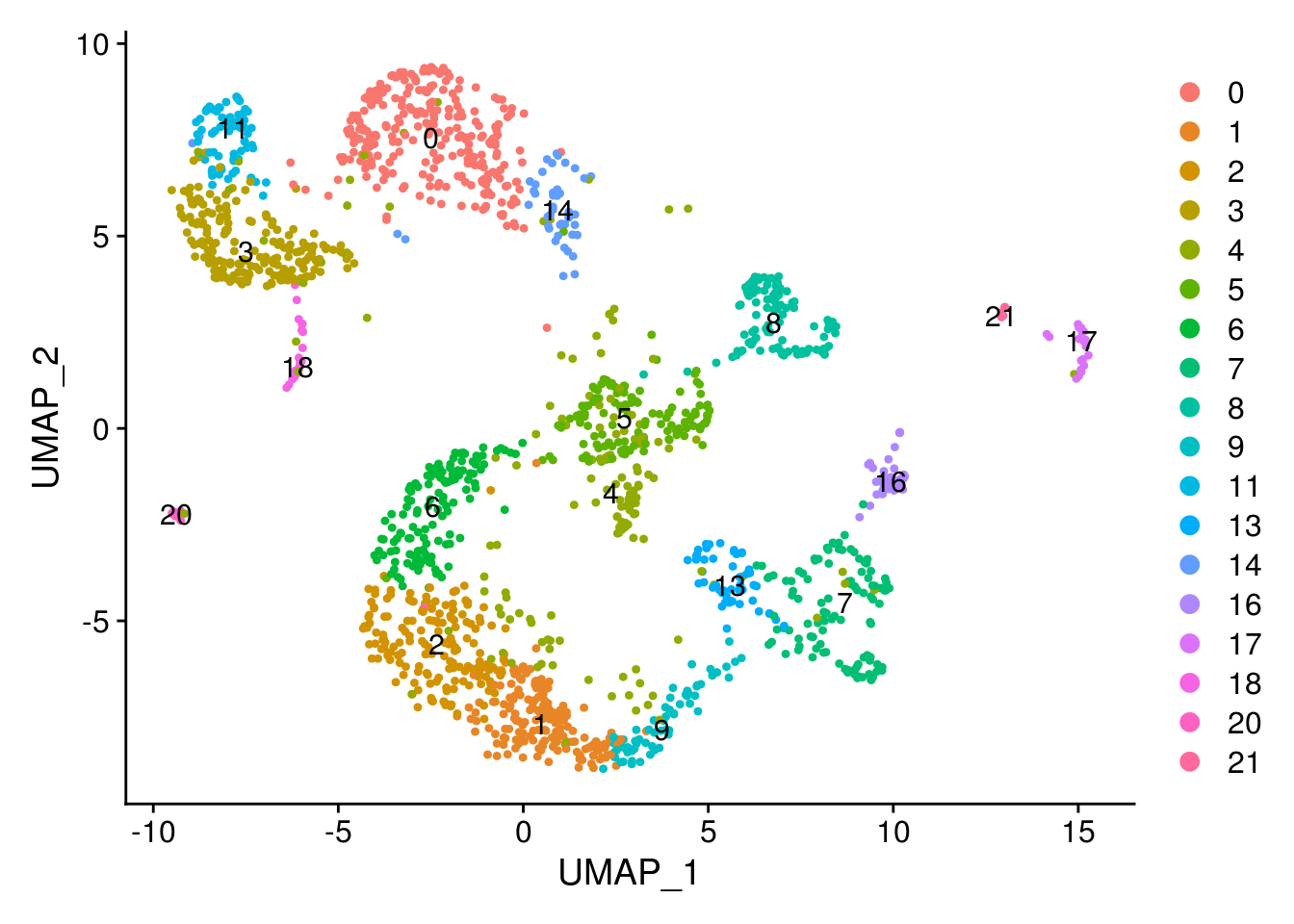
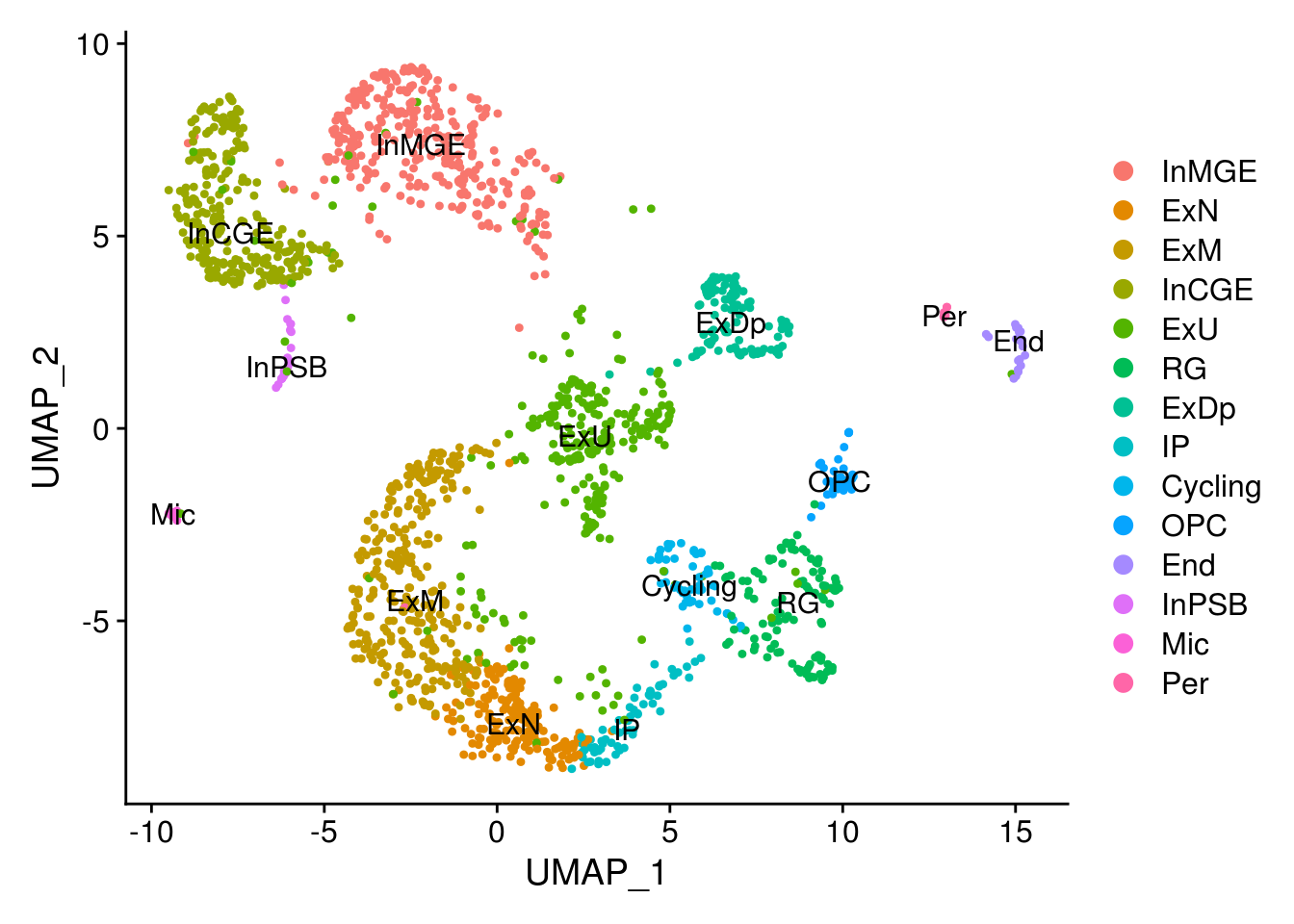
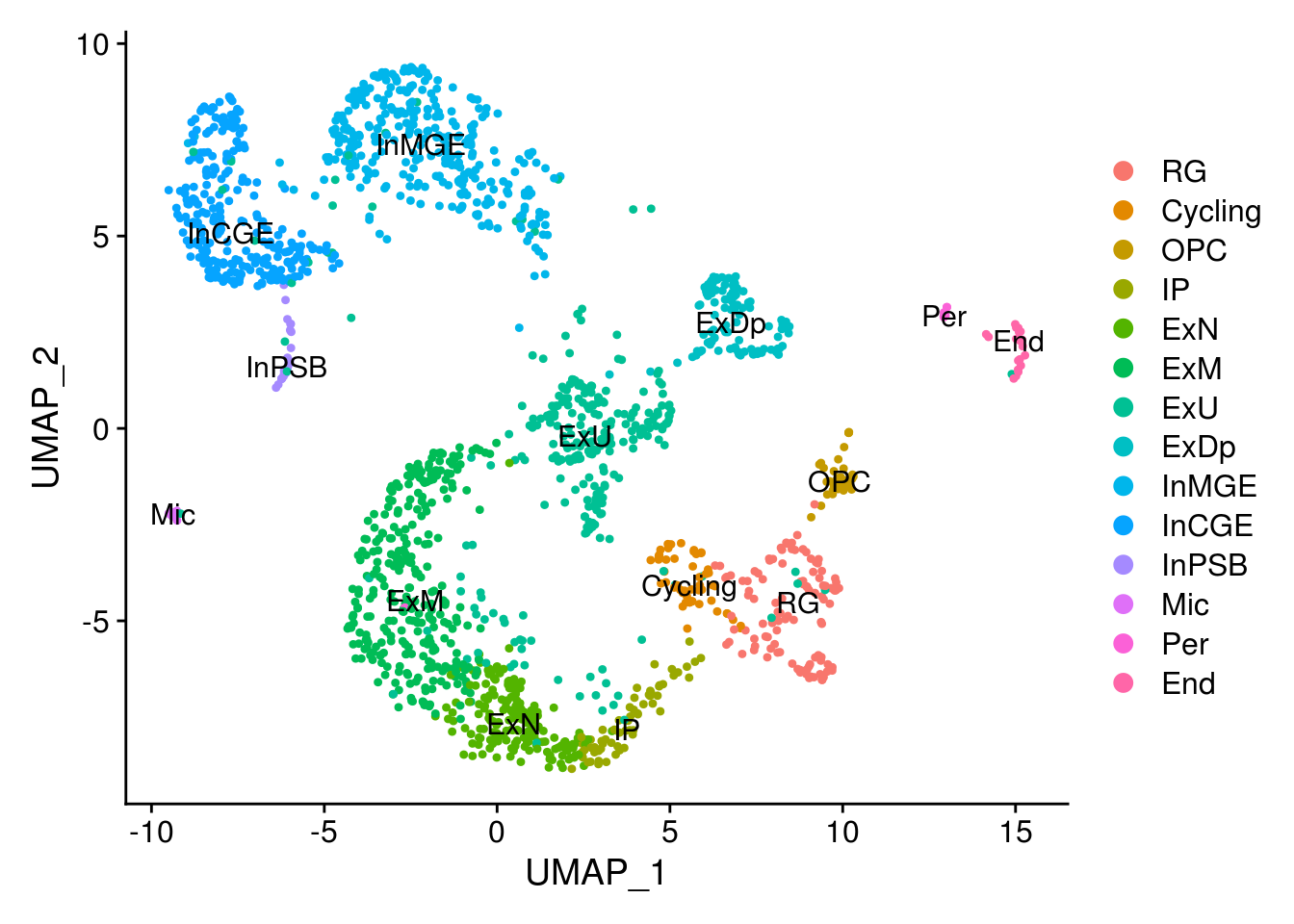
new.cl <- c('InMGE', 'ExN', 'ExM', 'InCGE', 'ExU', 'ExU', 'ExM', 'RG', 'ExDp', 'IP',
'InCGE', 'Cycling', 'InMGE', 'OPC', 'End', 'InPSB', 'Mic', 'Per')
names(new.cl) <- levels(combined)
combined <- RenameIdents(combined, new.cl)
DimPlot(combined, label = T)
RG = c(7)
OPC = c(16)
Cycling = c(13)
IP = c(9)
ExN = c(1)
ExM = c(2,6)
ExU = c(5,4)
ExDp = c(8)
InMGE = c(0,14)
InCGE = c(3,11)
InPSB = c(18)
Mic = c(20)
Per = c(21)
End = c(17)
cl.mat <- cbind(c(rep("RG", length(RG)), rep("OPC", length(OPC)), rep("Cycling", length(Cycling)), rep("IP", length(IP)),
rep("ExN", length(ExN)), rep("ExM", length(ExM)), rep("ExU", length(ExU)), rep("ExDp", length(ExDp)),
rep("InMGE", length(InMGE)), rep("InCGE", length(InCGE)), rep("InPSB", length(InPSB)),
rep("Mic", length(Mic)), rep("Per", length(Per)), rep("End", length(End))
),
c(RG, OPC, Cycling, IP, ExN, ExM, ExU, ExDp, InMGE, InCGE, InPSB, Mic, Per, End))
cl.vec <- combined$SCT_snn_res.0.5
ct.vec <- rep(NA, length(cl.vec))
for(x in unique(cl.mat[,1])){
cl.x <- cl.mat[cl.mat[,1]==x,2]
ct.vec[which(cl.vec%in%cl.x)] <- x
}
combined$cluster <- ct.vec
combined$cluster <- factor(combined$cluster, levels = c('RG', 'Cycling','OPC', 'IP',
'ExN', 'ExM', 'ExU', 'ExDp',
'InMGE', 'InCGE', 'InPSB',
'Mic', 'Per', 'End'))
DimPlot(combined, group.by = 'cluster', label = T)