Chapter 19 Plot APA Heatmaps
19.1 Description
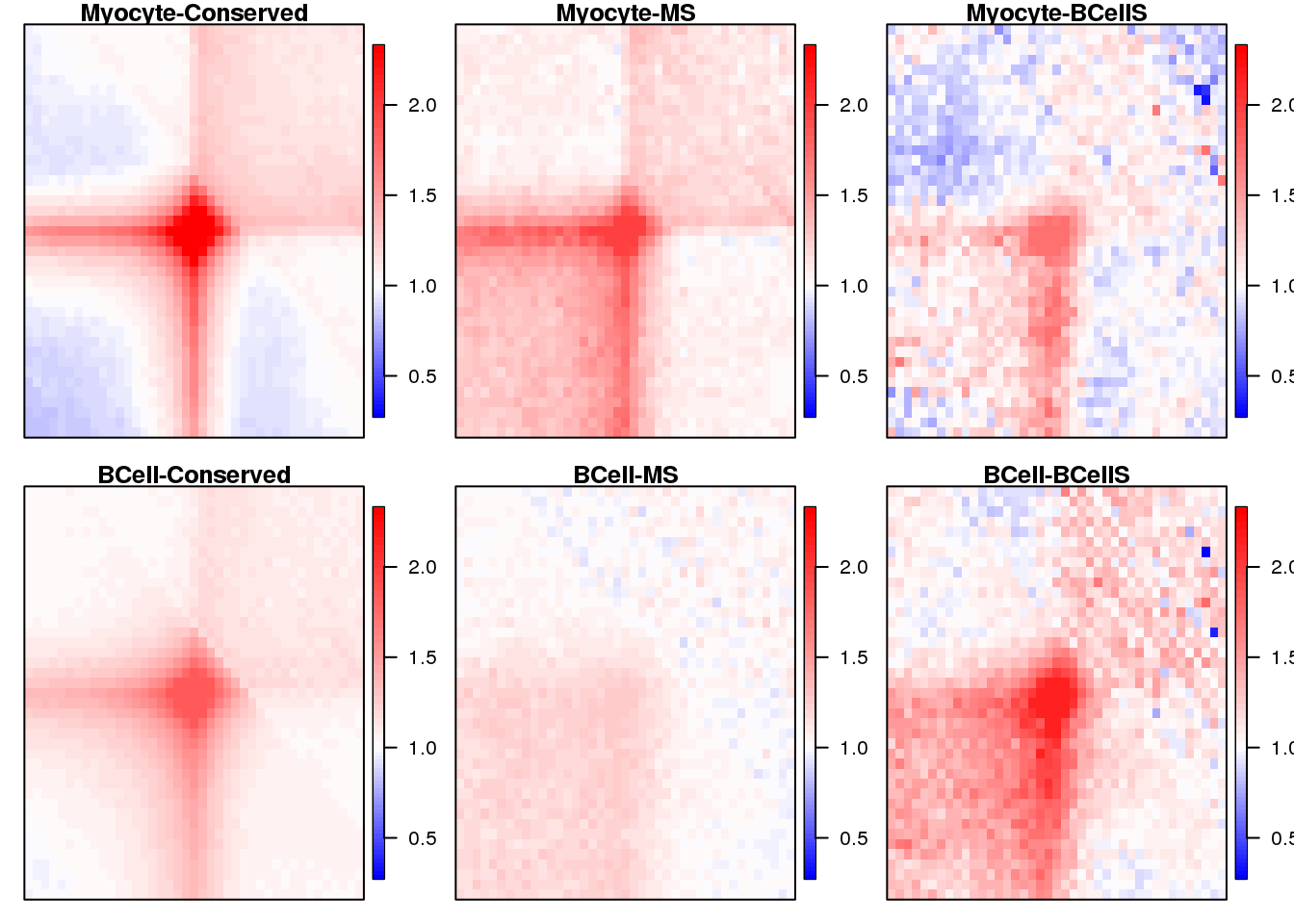
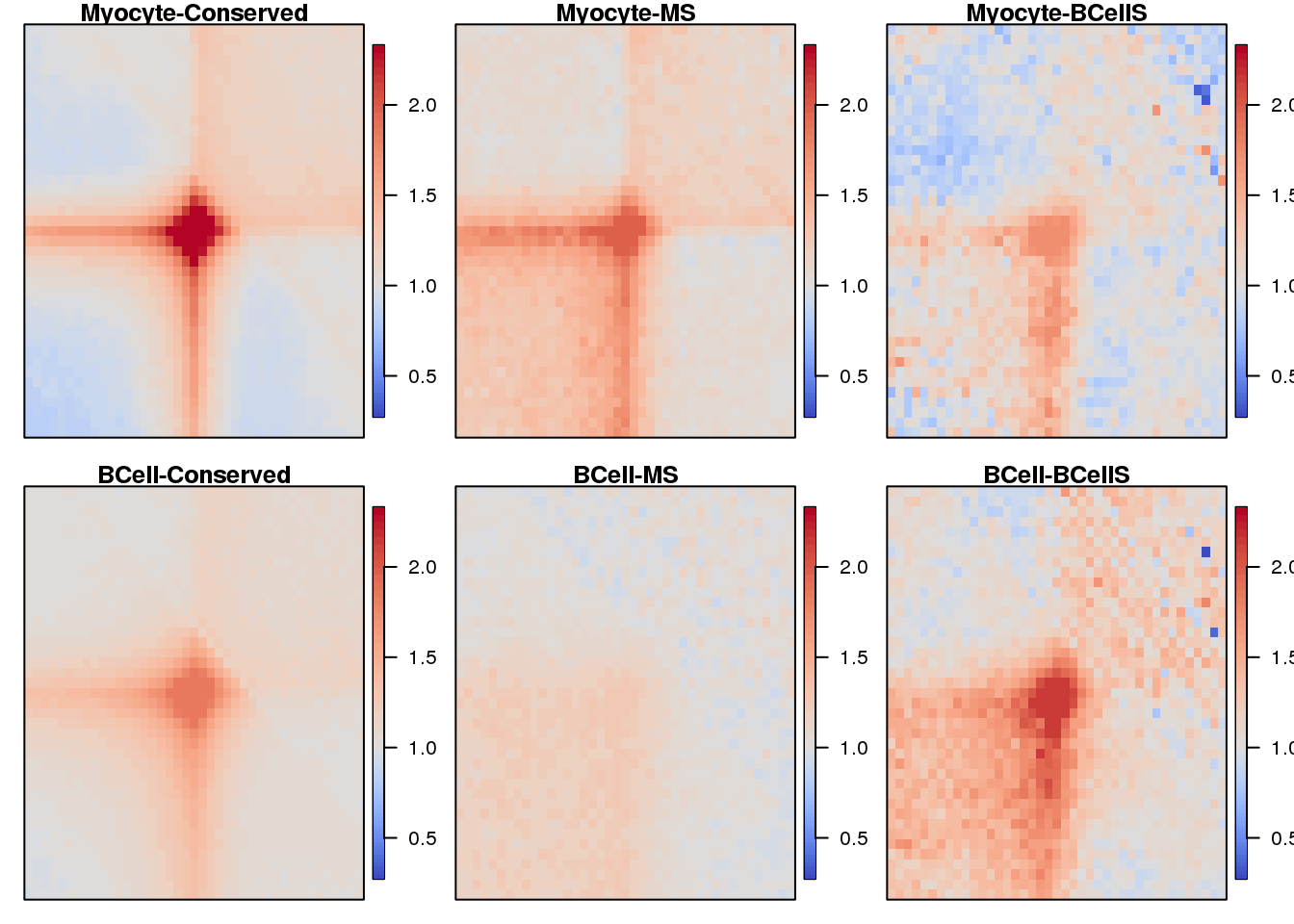
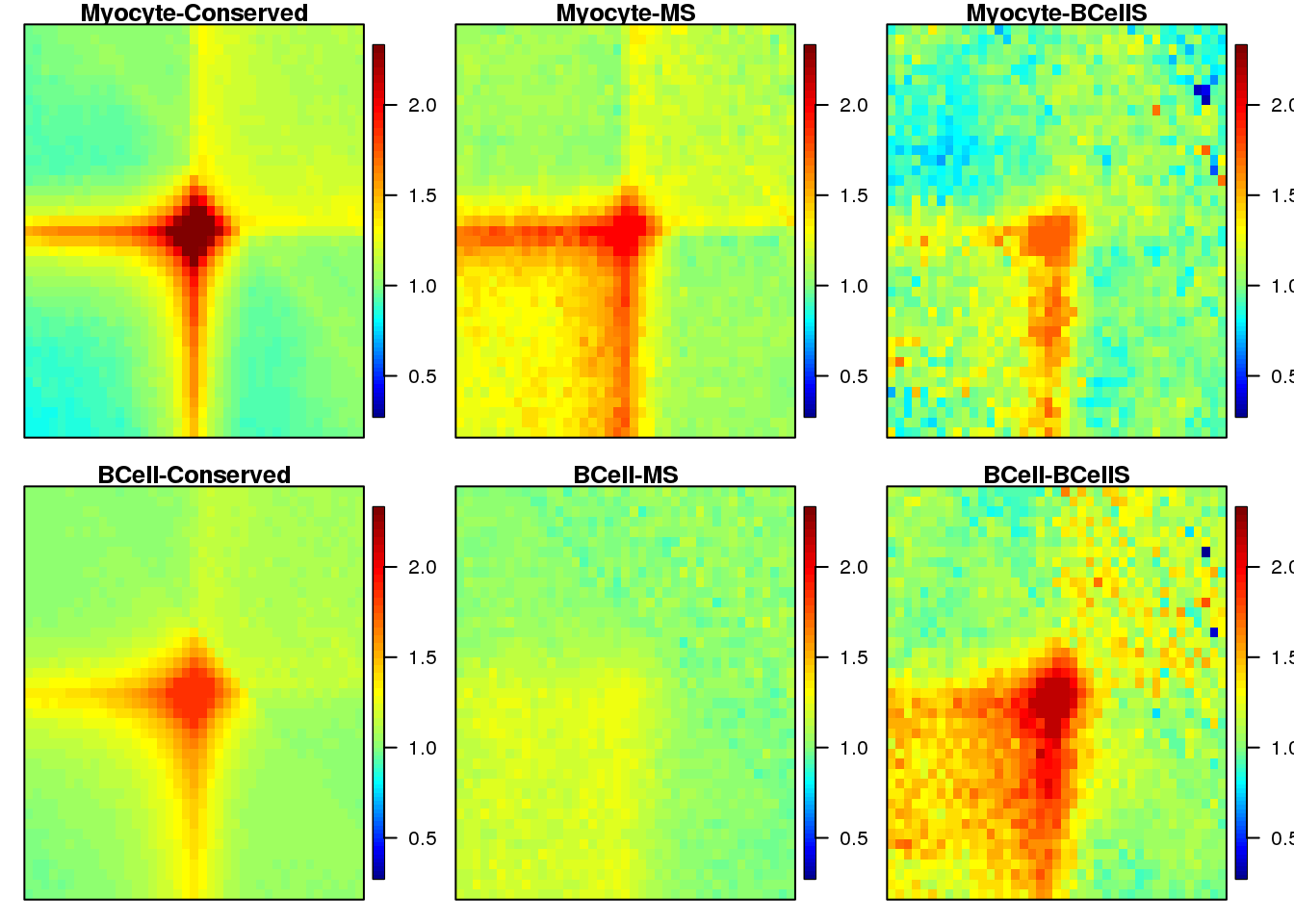
Plot heatmaps for loops APA
19.2 Load data
file.ls <- list.files('data/HiC/Pup-npfiles/', full.names = T)
file.ls <- file.ls[c(8,7,5,4,3,1)]
file.ls## [1] "data/HiC/Pup-npfiles//WT_DM.allValidPairs.res5kb-5.0K_over_WT_DM_3k5k10kb_merged_loops_StSt_CBS_10-shifts_dist_0-inf.np.txt"
## [2] "data/HiC/Pup-npfiles//WT_DM.allValidPairs.res5kb-5.0K_over_WT_DM_3k5k10kb_merged_loops_SpeSpeANDSpeNON_CBS_10-shifts_dist_0-inf.np.txt"
## [3] "data/HiC/Pup-npfiles//WT_DM.allValidPairs.res5kb-5.0K_over_GSE98119_Vian-2018-activated_B_cells_24_hours_WT.hic_5k10kloops_onlyGSM3027985diffBDFDR01_SpeSpeANDSpeNON_CBS_10-shifts_dist_0-inf.np.txt"
## [4] "data/HiC/Pup-npfiles//GSE98119_Vian-2018-activated_B_cells_24_hours_WT.hic.5kb-5.0K_over_WT_DM_3k5k10kb_merged_loops_StSt_CBS_10-shifts_dist_0-inf.np.txt"
## [5] "data/HiC/Pup-npfiles//GSE98119_Vian-2018-activated_B_cells_24_hours_WT.hic.5kb-5.0K_over_WT_DM_3k5k10kb_merged_loops_SpeSpeANDSpeNON_CBS_10-shifts_dist_0-inf.np.txt"
## [6] "data/HiC/Pup-npfiles//GSE98119_Vian-2018-activated_B_cells_24_hours_WT.hic.5kb-5.0K_over_GSE98119_Vian-2018-activated_B_cells_24_hours_WT.hic_5k10kloops_onlyGSM3027985diffBDFDR01_SpeSpeANDSpeNON_CBS_10-shifts_dist_0-inf.np.txt"mat.ls <- lapply(file.ls, function(f){
mat <- fread(f, skip = 35)
mat <- as.matrix(mat)
mat[is.na(mat)] <- 0
return(mat)
})19.3 Plot Function
plotSqure <- function(xdata.ls, id.ls, quant, colind){
## Section: make diag as na and trim max value
##################################################
xdata.ls <- map2(xdata.ls, quant, ~ {
xdata <- .x
quant <- .y
qmax <- quantile(xdata, quant, na.rm = T)
xdata[xdata>qmax] <- qmax
xdata <- t(xdata)
xdata <- as.matrix(xdata[,ncol(xdata):1])
return(xdata)
})
## Section: colour choice
##################################################
csize <- 1 # dot size
k <- 64
col.ls1 <- colorRampPalette(c('blue','white','red'))(k)
col.ls2 <- coolwarm(k)
col.ls3 <- tim.colors(k)
if(colind == 1){
mycol = col.ls1
}
if(colind == 2){
mycol = col.ls2
}
if(colind == 3){
mycol = col.ls3
}
## Section: breaks
##################################################
minv <- min(unlist(lapply(xdata.ls, function(f){
quantile(unlist(f))
})))
maxv <- max(unlist(lapply(xdata.ls, function(f){
quantile(unlist(f))
})))
myBreaks <- c(seq(minv, 1, length.out=ceiling(k/2) + 1),
seq(maxv/k+1, maxv, length.out=floor(k/2)))
## Section: plot
##################################################
par(mfrow = c(2,3))
seq_along(xdata.ls) %>% map(~ {
xdata <- xdata.ls[[.x]]
id <- id.ls[[.x]]
par(mar = c(1,1,1,1))
image.plot(x = 1:nrow(xdata), y = 1:nrow(xdata), z = xdata[nrow(xdata):1, nrow(xdata):1],
breaks = myBreaks, axis = F, axes = F, legend.cex = .2, main = id, col = mycol)
box()
})
} 19.4 Plot
sample <- c("Myocyte-Conserved", "Myocyte-MS", "Myocyte-BCellS",
"BCell-Conserved", "BCell-MS", "BCell-BCellS")
seq(1,3) %>% map(~ {
plotSqure(mat.ls, id.ls = sample, quant = 0.99, colind = .x)
})
## [[1]]
## [[1]][[1]]
## NULL
##
## [[1]][[2]]
## NULL
##
## [[1]][[3]]
## NULL
##
## [[1]][[4]]
## NULL
##
## [[1]][[5]]
## NULL
##
## [[1]][[6]]
## NULL
##
##
## [[2]]
## [[2]][[1]]
## NULL
##
## [[2]][[2]]
## NULL
##
## [[2]][[3]]
## NULL
##
## [[2]][[4]]
## NULL
##
## [[2]][[5]]
## NULL
##
## [[2]][[6]]
## NULL
##
##
## [[3]]
## [[3]][[1]]
## NULL
##
## [[3]][[2]]
## NULL
##
## [[3]][[3]]
## NULL
##
## [[3]][[4]]
## NULL
##
## [[3]][[5]]
## NULL
##
## [[3]][[6]]
## NULL