Chapter 6 Enhancer conservation across tissues grouped by SE/Ehn-stableCBS/Enh-specificCBS
6.1 Description
Compare the tissue-specificity for enhancers: SEs, enhancer with stable CBS, enhancer with specific CBS
6.2 Get overlap for three enhancer sets
## Section: get the overlap: three tissues
##################################################
f.ls <- list.files('data/peakfiles/Enhancer-groupby-CBS/', full.names = T, pattern = "specific")
f.ls <- f.ls[c(2,3,1)]
f.ls## [1] "data/peakfiles/Enhancer-groupby-CBS//Hep.with.specificCBS.bed"
## [2] "data/peakfiles/Enhancer-groupby-CBS//Myocyte.with.specificCBS.bed"
## [3] "data/peakfiles/Enhancer-groupby-CBS//Bcell.with.specificCBS.bed"gr.ls <- lapply(f.ls, function(f){
mat <- fread(f)
mat <- mat[,1:3]
names(mat) <- c('chr', 'start', 'end')
gr <- makeGRangesFromDataFrame(mat)
return(gr)
})
getOvlp <- function(gr.ls){
a <- gr.ls[[1]]
b <- gr.ls[[2]]
c <- gr.ls[[3]]
comm.ab <- unique(queryHits(findOverlaps(a,b)))
comm.ac <- unique(queryHits(findOverlaps(a,c)))
comm.bc <- unique(queryHits(findOverlaps(b,c)))
abc <- intersect(comm.ab, comm.ac)
return(c(length(a), length(b), length(c), length(comm.ab),
length(comm.ac), length(comm.bc), length(abc)))
}
n.ls <- getOvlp(gr.ls)6.3 Venn diagram
## Section: triple venn plot
##################################################
myCol <- brewer.pal(3, "Pastel2")
getVen <- function(n.ls){
draw.triple.venn(n.ls[1], n.ls[2], n.ls[3], n.ls[4], n.ls[5],n.ls[6], n.ls[7],
# Circles
lwd = 2,
lty = 'blank',
fill = myCol,
# Numbers
cex = .6,
fontface = "bold",
fontfamily = "sans",
# Set names
cat.cex = 0.6,
cat.fontface = "bold",
cat.default.pos = "outer",
cat.pos = c(-27, 27, 135),
cat.dist = c(0.055, 0.055, 0.085),
cat.fontfamily = "sans",
rotation = 1,
category = c('Bcell','hep', 'myocyte'))
}
getVen(n.ls)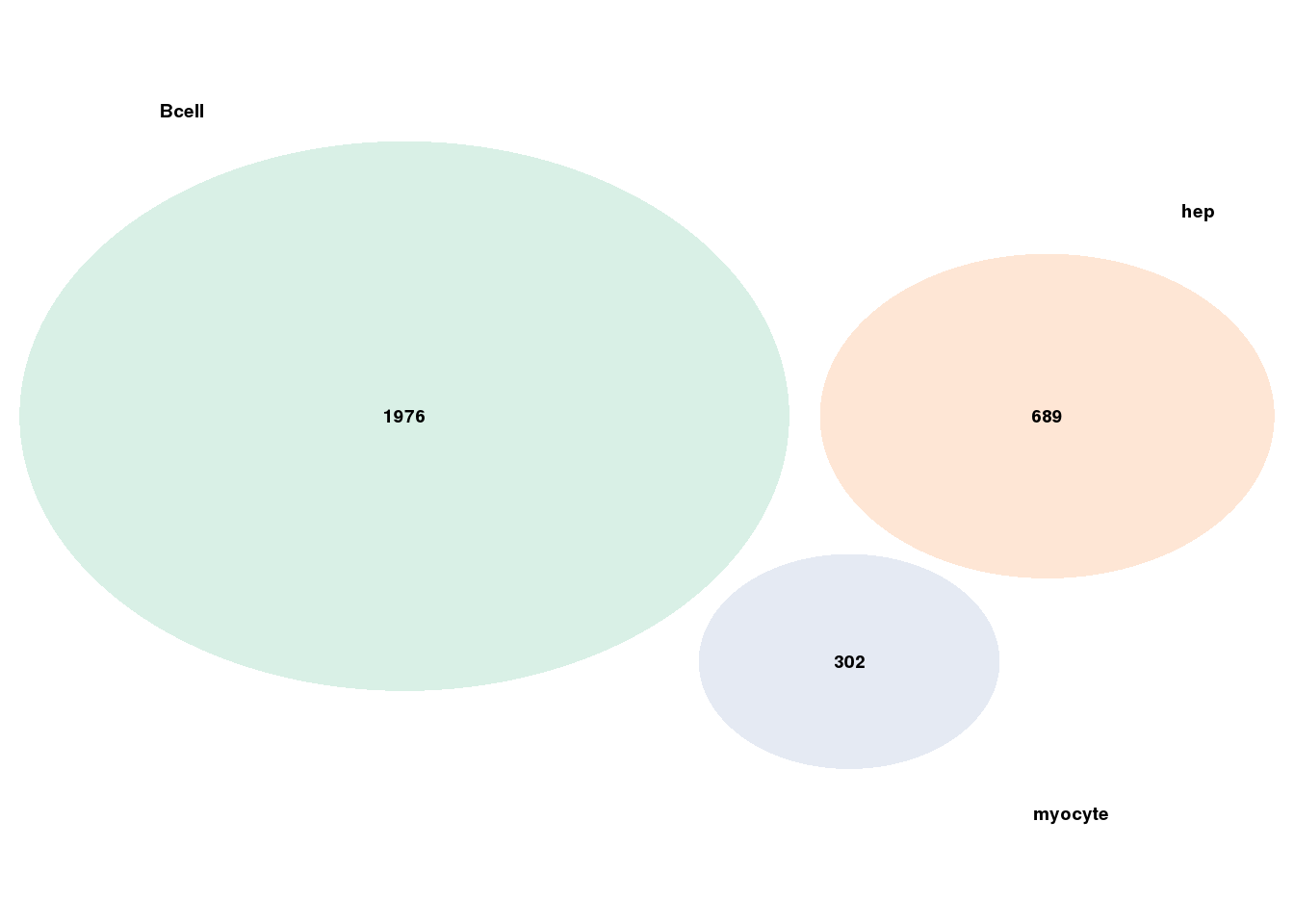
## (polygon[GRID.polygon.1], polygon[GRID.polygon.2], polygon[GRID.polygon.3], polygon[GRID.polygon.4], polygon[GRID.polygon.5], polygon[GRID.polygon.6], text[GRID.text.7], text[GRID.text.8], text[GRID.text.9], text[GRID.text.10], text[GRID.text.11], text[GRID.text.12])## Section: triple venn plot
##################################################
pdf('results/Features/N6-Human-enhancer-conservation-AllEnh-withspecificCBS-vennplot.pdf', width = 2, height = 2)
getVen(n.ls)
dev.off()