Chapter 3 DiffBD-GenomeAnnotation
3.1 Description
Using ChIPseeker to annotate the lineage-conserved/specific CBSs based on genomic regions
3.2 Load data
file.ls <- list.files('data/DiffBD/', full.names = T, pattern = "bed")
peaks <- list(
stable = file.ls[5],
liver = file.ls[2],
muscle = file.ls[3],
bcell = file.ls[1])3.3 Annotate
txdb = TxDb.Hsapiens.UCSC.hg19.knownGene
promoter <- getPromoters(TxDb = txdb, upstream = 2000, downstream = 2000)
peakAnnoList <- lapply(peaks, annotatePeak, TxDb=txdb,
tssRegion=c(-3000, 3000), verbose=FALSE)3.4 Plot
plotAnnoBar(peakAnnoList)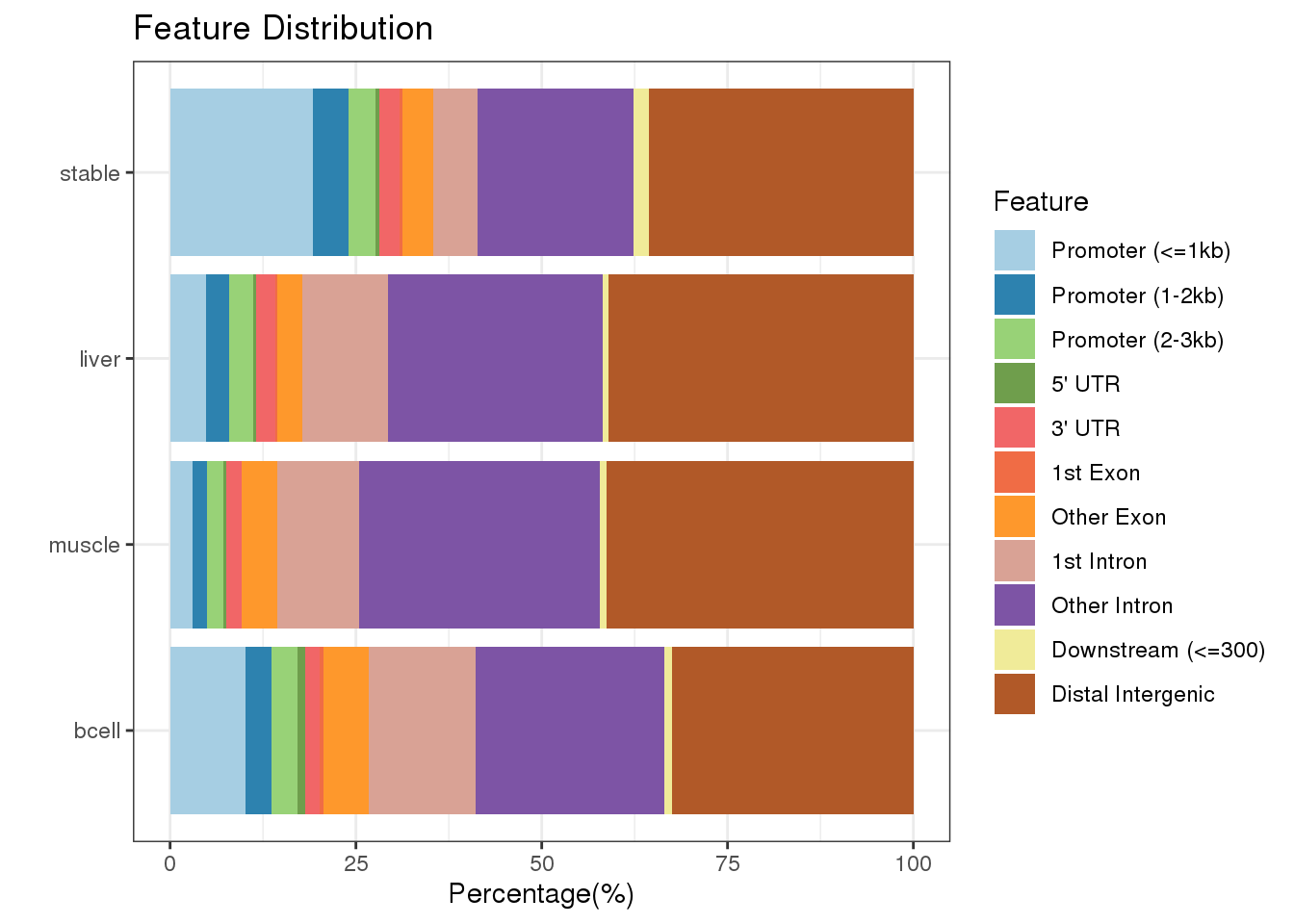
plotDistToTSS(peakAnnoList)
pdf('results/Features/Hep.Myo.Bcell-specificANDstable-CTCF-GenomciAnnotation-barplot.pdf', width = 5, height = 3)
plotAnnoBar(peakAnnoList)
plotDistToTSS(peakAnnoList)
dev.off()