Chapter 13 HiC CBS Density around Boundary CurvePlot
13.1 Description
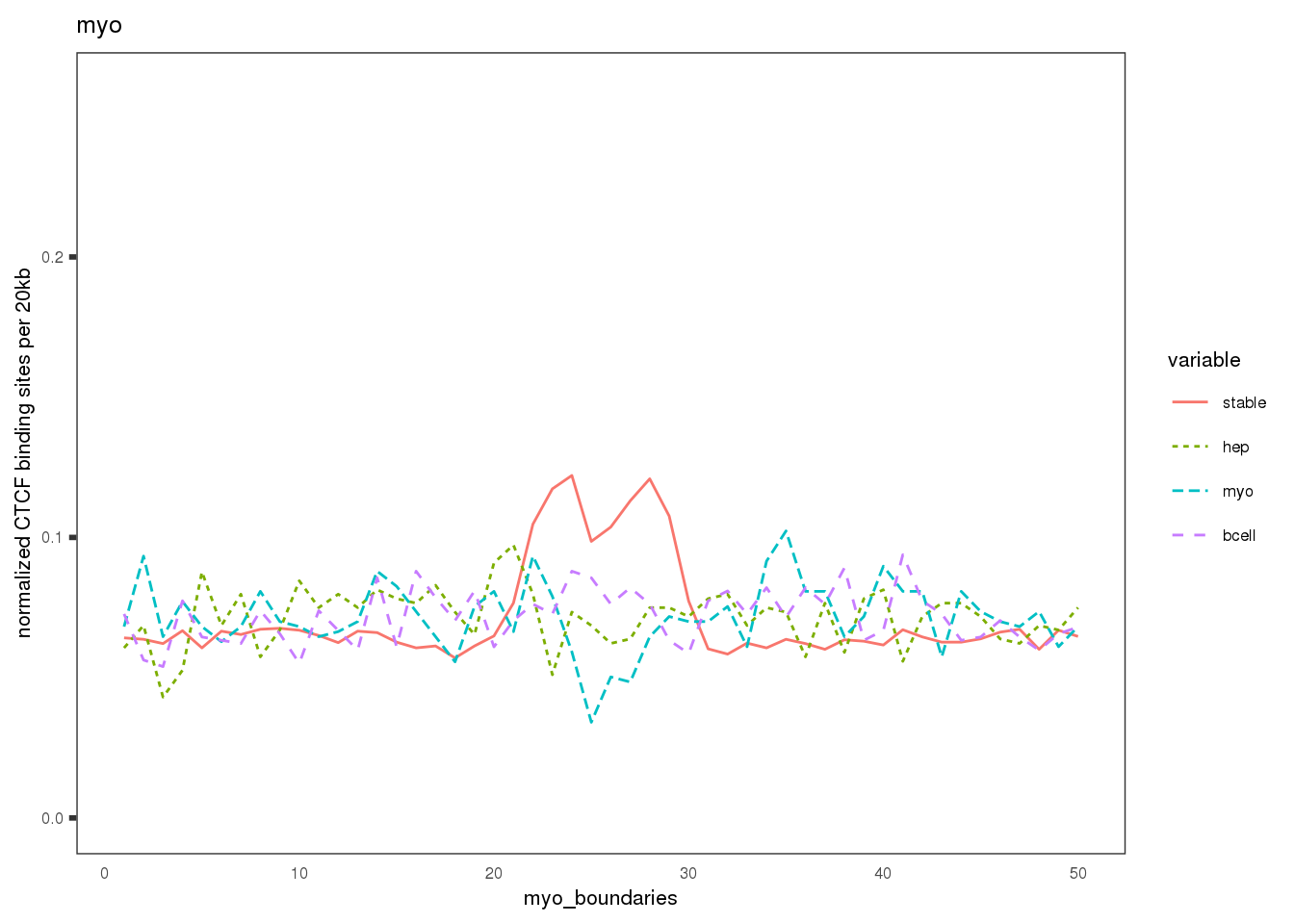
Plot the CBS density distribution around boundaries in three lineages; lineage-conserved and lineage-specific CBS sets.
13.2 Load CBS density data
## Load data
##################################################
file.ls <- list.files('data/countmat/StableSpecificCBS_peakdensity_on_diamondBoundaries/', full.names = T)
file.ls## [1] "data/countmat/StableSpecificCBS_peakdensity_on_diamondBoundaries//Mouse.stable.specific.CBS.peakDensity_ON_800kbAround_GSE93431_UNTR.50kb.cool.HDF5_diamond-insulation.800kb.1000kb.txt_filter-0.1.matrix"
## [2] "data/countmat/StableSpecificCBS_peakdensity_on_diamondBoundaries//Mouse.stable.specific.CBS.peakDensity_ON_800kbAround_GSE98119_Vian-2018-activated_B_cells_24_hours_WT.hic.50kb_diamond-insulation.800kb.1000kb.txt_filter-0.1.matrix"
## [3] "data/countmat/StableSpecificCBS_peakdensity_on_diamondBoundaries//Mouse.stable.specific.CBS.peakDensity_ON_800kbAround_WT_DM.allValidPairs.res50kb_diamond-insulation.800kb.1000kb.txt_filter-0.1.matrix"13.3 Calculate density
## Section: read the CBS density distribution
##################################################
# absolute density
mat.ls <- lapply(file.ls, function(f){
df <- fread(f, skip = 1)
df <- df[, 7:ncol(df)]
df[is.na(df)] <- 0
den <- colMeans(df)
n <- ncol(df)
n.ls <- seq(0,n, length = 5)[-1]
mat <- data.frame(dist = seq(1,n/4), stable = den[1:n.ls[1]], hep = den[(n.ls[1]+1):n.ls[2]],
myo = den[(n.ls[2]+1):n.ls[3]], bcell = den[(n.ls[3]+1):n.ls[4]])
mat <- melt(mat, id.vars = 'dist')
mat$variable <- factor(mat$variable, levels = c('stable','hep','myo','bcell'))
mat$value <- as.numeric(as.character(mat$value))
return(mat)
})
# normalized density
mat.ls <- lapply(file.ls, function(f){
df <- fread(f, skip = 1)
df <- df[, 7:ncol(df)]
df[is.na(df)] <- 0
n <- ncol(df)
n.ls <- seq(0,n, length = 5)[-1]
stable <- df[,1:n.ls[1]]
hep <- df[, (n.ls[1]+1):n.ls[2]]
myo <- df[, (n.ls[2]+1):n.ls[3]]
bcell <- df[, (n.ls[3]+1):n.ls[4]]
stable <- colMeans(stable)/sum(stable)*1e4
hep <- colMeans(hep)/sum(hep)*1e04
myo <- colMeans(myo)/sum(myo)*1e04
bcell <- colMeans(bcell)/sum(bcell)*1e04
mat <- data.frame(dist = seq(1,n/4), stable = stable, hep = hep, myo = myo, bcell = bcell)
mat <- melt(mat, id.vars = 'dist')
mat$variable <- factor(mat$variable, levels = c('stable','hep','myo','bcell'))
mat$value <- as.numeric(as.character(mat$value))
return(mat)
})13.4 Plot
## Section: plot density
##################################################
names(mat.ls) <- c('hep', 'Bcell', 'myo')
mat.ls %>% imap( ~ {
fontsize = 8
linesize = 1
df <- .x
sample <- .y
ggplot(df, aes(x = dist, y = value)) +
geom_line(aes(color = variable, linetype = variable))+
theme_bw() +
theme(panel.grid.major = element_blank(),panel.grid.minor = element_blank(),
strip.background = element_blank())+
theme(text = element_text(size = fontsize), line = element_line(size = linesize),
axis.ticks.length = unit(.1, "cm"), axis.ticks.x = element_blank()) +
ylab('normalized CTCF binding sites per 20kb')+
xlab(paste0(sample, '_boundaries'))+
ggtitle(label = sample)+ # labs
coord_cartesian(ylim = c(0,0.26))
})## $hep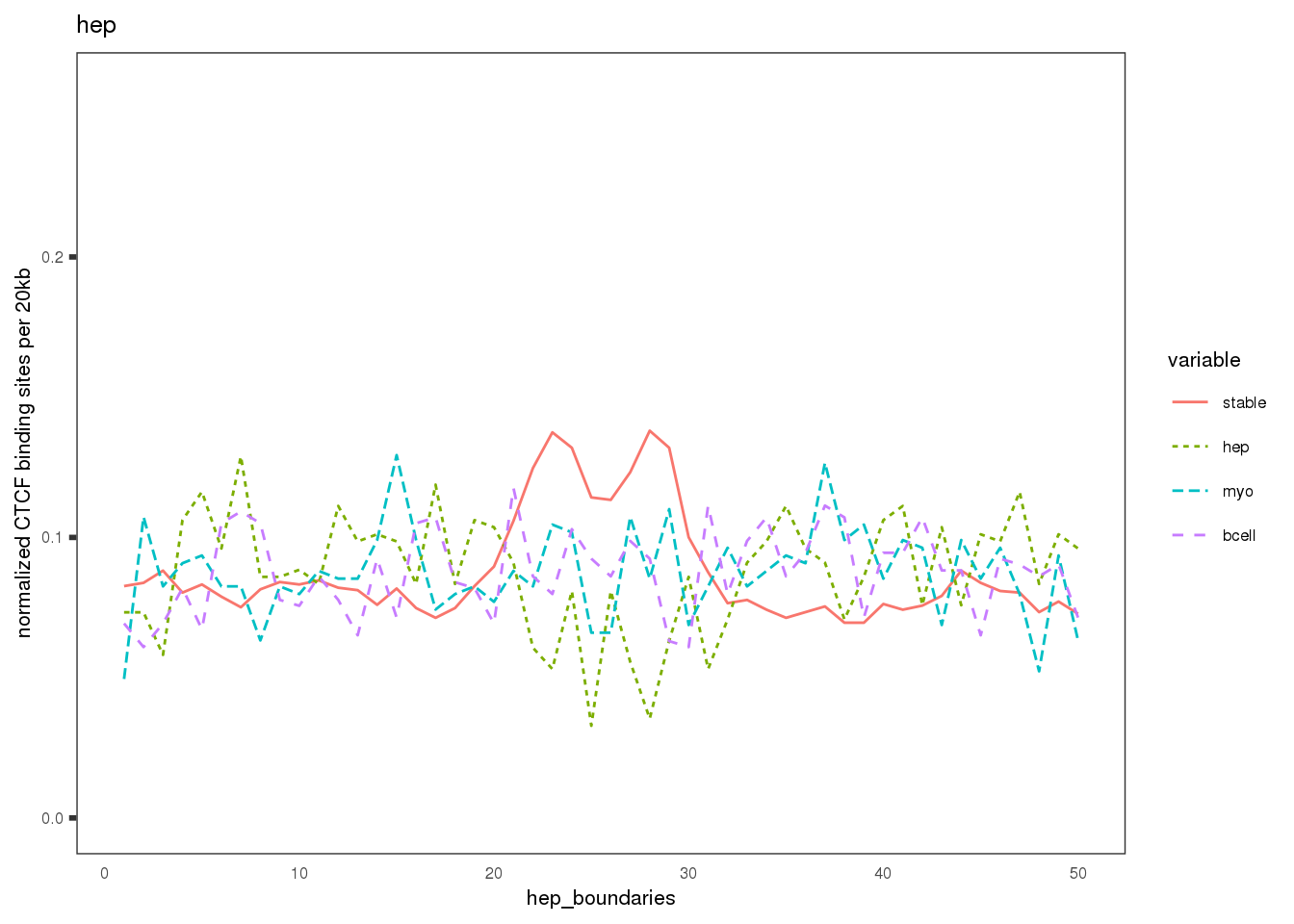
##
## $Bcell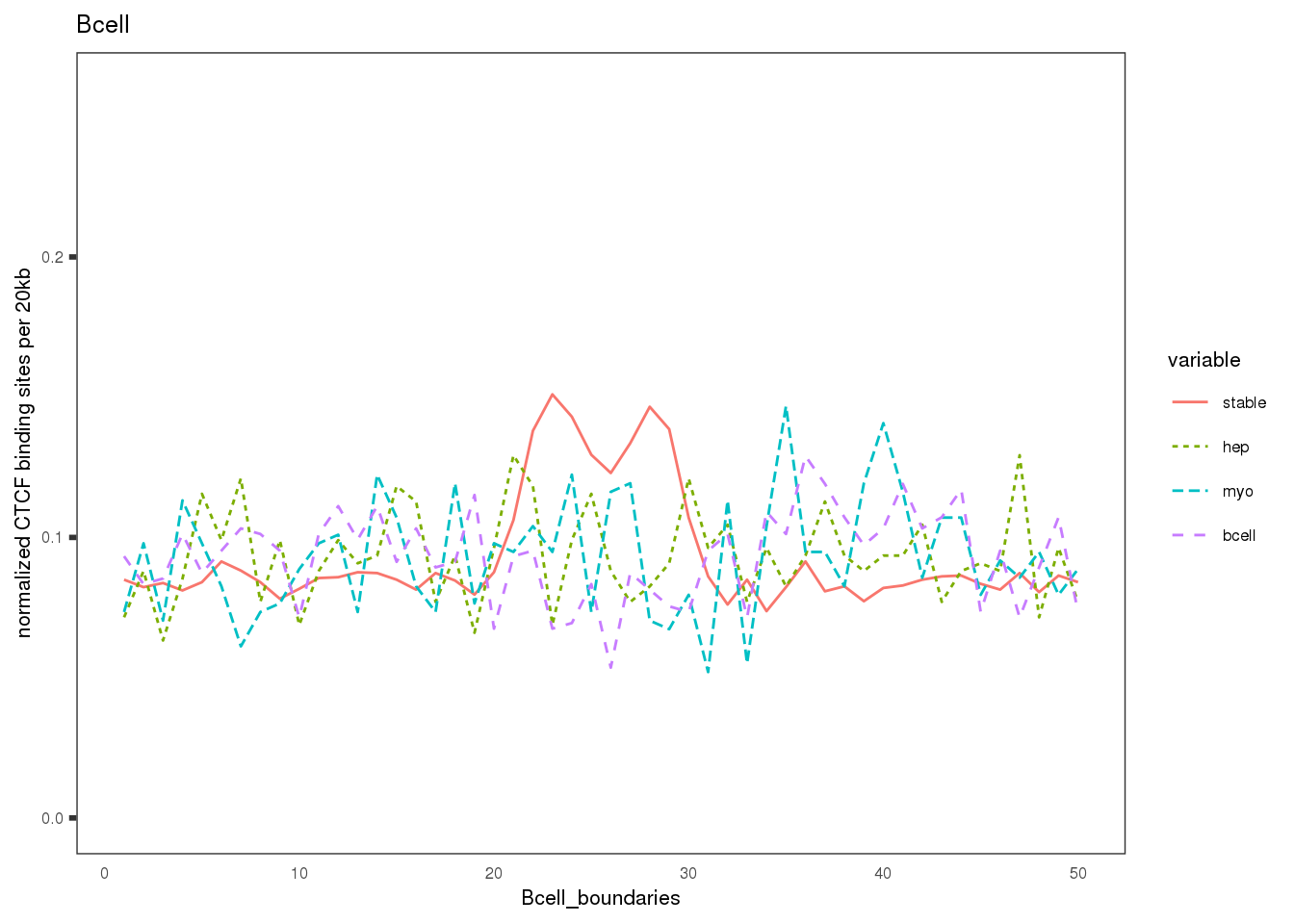
##
## $myo