Chapter 7 diffBD chromatin state across cells
7.1 Description
- explore the chromatin states dynamics arcross cells per CBS sets
- chromHMM files are download from Roadmap project
7.2 Load chromHMM files
## Section: load data
##################################################
file.ls <- list.files('data/chromHMM-Roadmap/', pattern = 'Mid', full = T)
file.ls <- file.ls[c(5,2,3,1)]7.3 Get the chromatin state dynamics
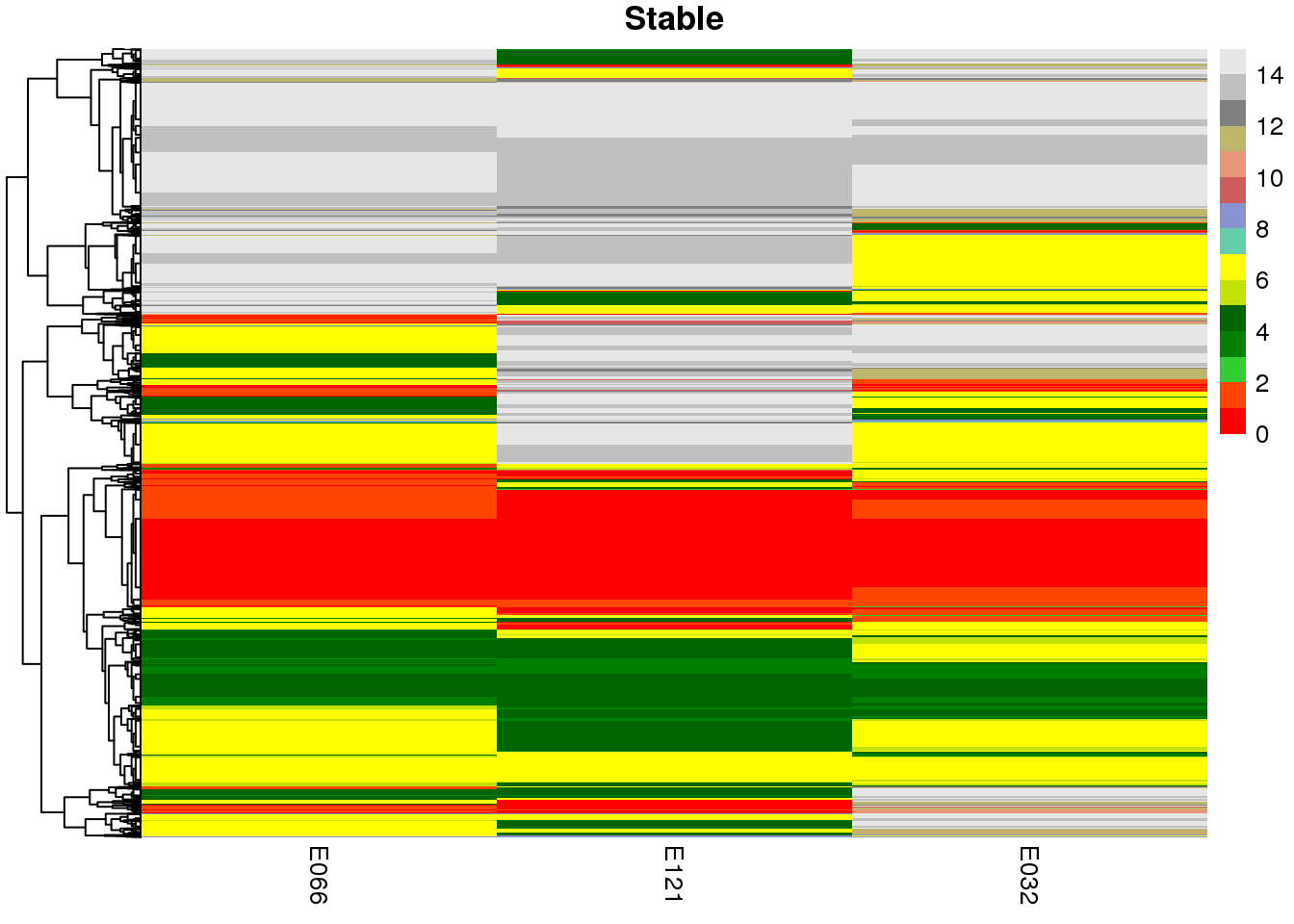
sample.ls <- c('Stable', 'Hepatocyte', 'Myocyte', 'Bcell')
mat.ls <- 1:4 %>% map( ~ {
f <- file.ls[.x]
sample <- sample.ls[.x]
mat <- fread(f)
mat <- mat[, c(4, 5, 9)]
mat <- dcast(mat, V4 ~ V5)
colnames(mat)[1] <- 'id'
mat <-
mgsub::mgsub(
mat,
c(
"1_TssA",
"2_TssAFlnk",
"3_TxFlnk",
"4_Tx",
"5_TxWk",
"6_EnhG",
"7_Enh",
"8_ZNF/Rpts",
"9_Het",
"10_TssBiv",
"11_BivFlnk",
"12_EnhBiv",
"13_ReprPC",
"14_ReprPCWk",
"15_Quies"
),
seq(1, 15)
)
mat$E007 <- as.numeric(mat$E007)
mat$E032 <- as.numeric(mat$E032)
mat$E066 <- as.numeric(mat$E066)
mat$E121 <- as.numeric(mat$E121)
return(mat)
})## Using 'V9' as value column. Use 'value.var' to override
## Using 'V9' as value column. Use 'value.var' to override
## Using 'V9' as value column. Use 'value.var' to override
## Using 'V9' as value column. Use 'value.var' to override7.4 Heatmap showing the dynamics
myColor <-
c(
rgb(255, 0, 0, maxColorValue = 255),
rgb(255, 69, 0, maxColorValue = 255),
rgb(50, 205, 50, maxColorValue = 255),
rgb(0, 128, 0, maxColorValue = 255),
rgb(0, 100, 0, maxColorValue = 255),
rgb(194, 225, 5, maxColorValue = 255),
rgb(255, 255, 0, maxColorValue = 255),
rgb(102, 205, 170, maxColorValue = 255),
rgb(138, 145, 208, maxColorValue = 255),
rgb(205, 92, 92, maxColorValue = 255),
rgb(233, 150, 122, maxColorValue = 255),
rgb(189, 183, 107, maxColorValue = 255),
rgb(128, 128, 128, maxColorValue = 255),
rgb(192, 192, 192, maxColorValue = 255),
rgb(230, 230, 230, maxColorValue = 255)
)
myBreaks <- seq(0, 15, 1)
map2(mat.ls, sample.ls, ~ {
pp <-
pheatmap(
.x[, c(4, 5, 3)],
color = myColor,
breaks = myBreaks,
show_rownames = F,
cluster_cols = F,
main = .y
)
pp
})
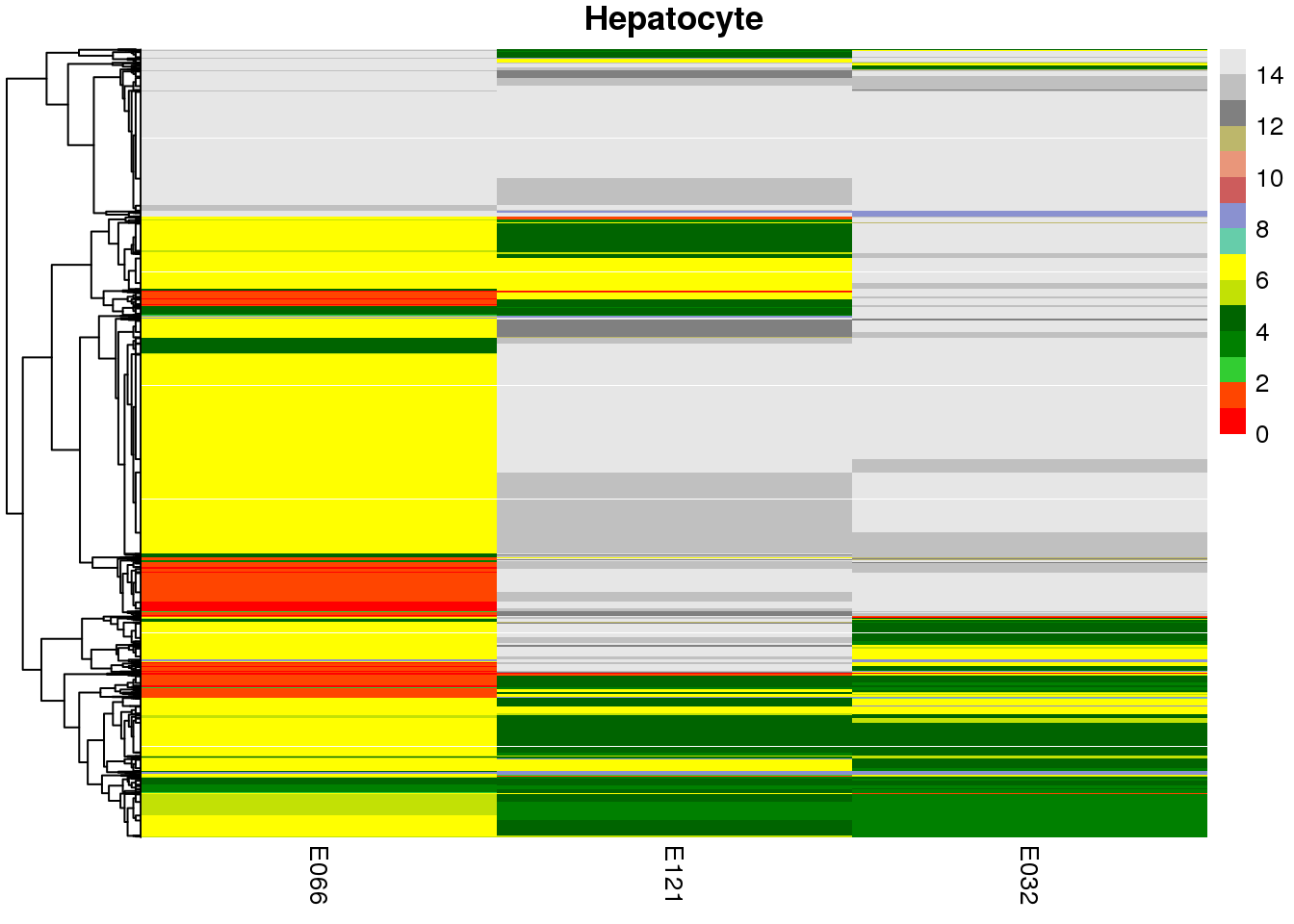
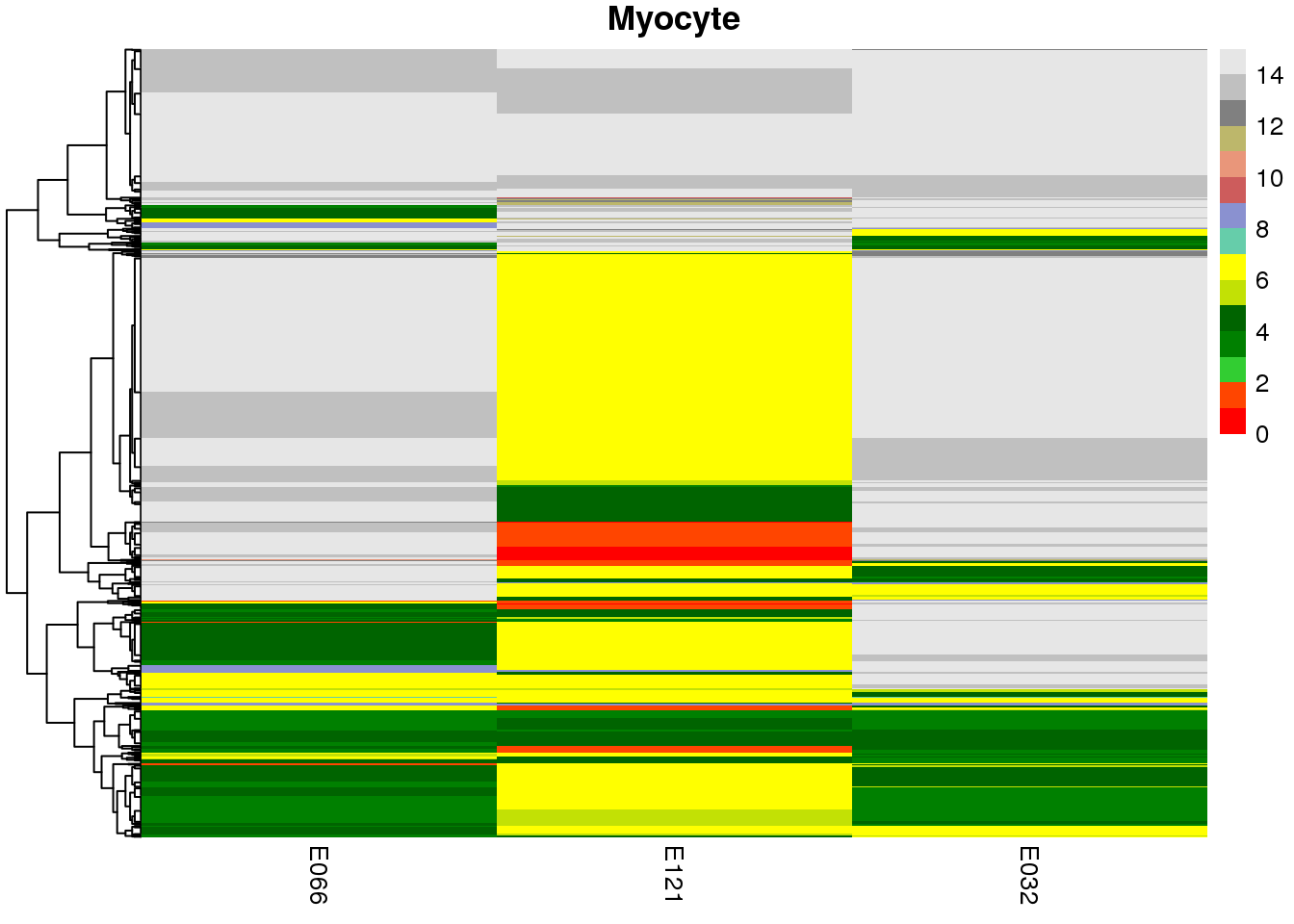
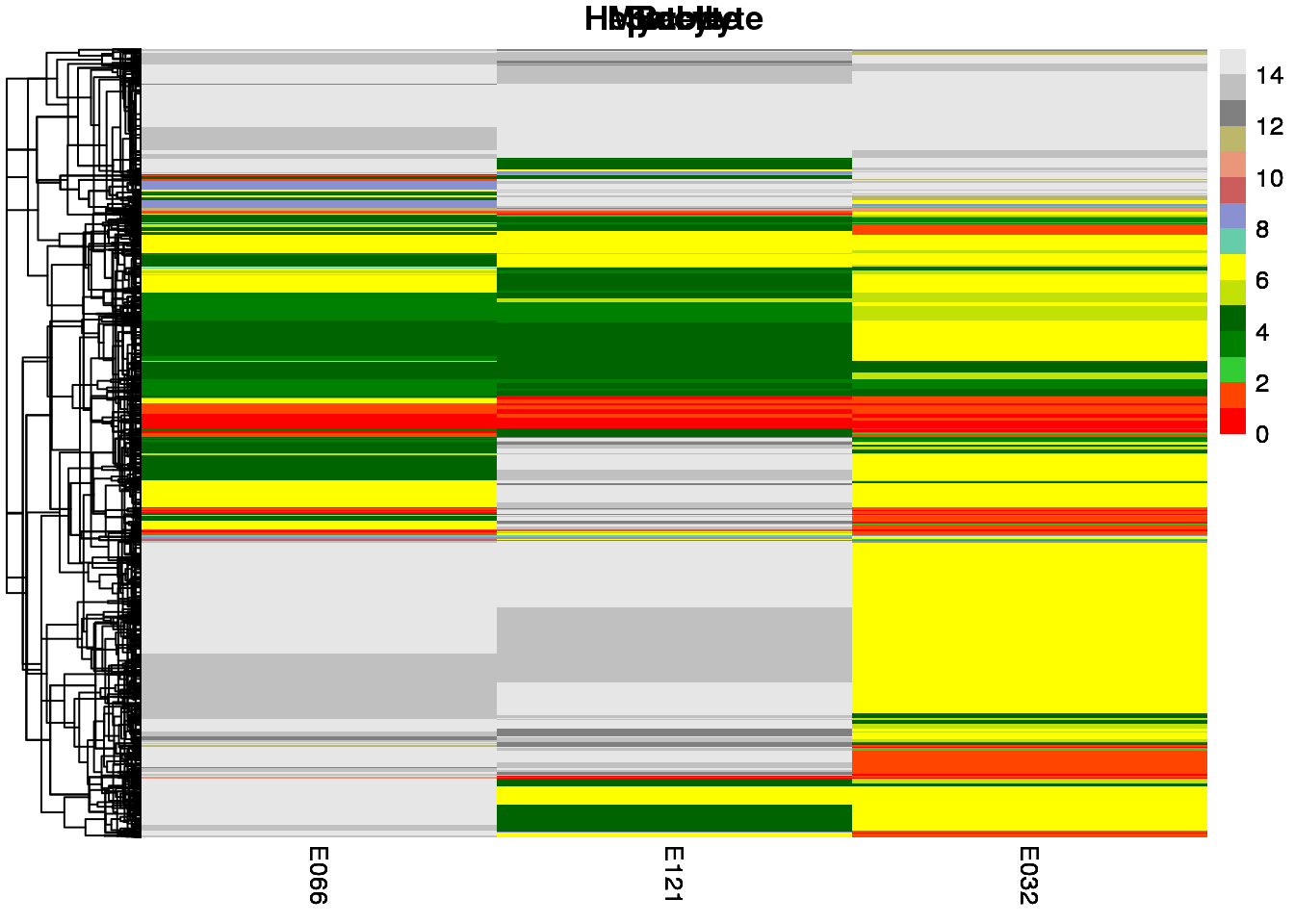
## [[1]]
##
## [[2]]
##
## [[3]]
##
## [[4]]