2.3 Optimizing Program Performance
In the this digital age, datasets are getting larger, and extracting insights from them in the shortest possible time is of growing demand
Optimizing program performance enables the handling of larger datasets and more computationally intensive tasks.
The following introduces some techniques to optimize your program performance.
2.3.1 Organizing Code
There are two traps that are easy to fall into when trying to make your code faster:
- Writing faster but incorrect code.
- Writing code that you think is faster but is actually no better.
Writing well-structured code improves readability, maintainability, and performance.
Key principles include:
Modularization: Break down code into reusable functions.
Avoiding redundant calculations: Store intermediate results instead of recomputing them.
Profiling: Use
profvis()orRprof()to identify bottlenecks.
2.3.2 Preallocating Memory
Avoid growing objects dynamically inside loops, as it leads to memory reallocation overhead.
A good rule of thumb is to compare your objects before and after a loop; have they increased in length?
In R, you don’t have to explicitly allocate memory before you use it.
You can pre-allocate memory by setting the length,
nrow,ncol, ordimattributes for an object.You should also consider pre-allocating memory for data frames and lists.
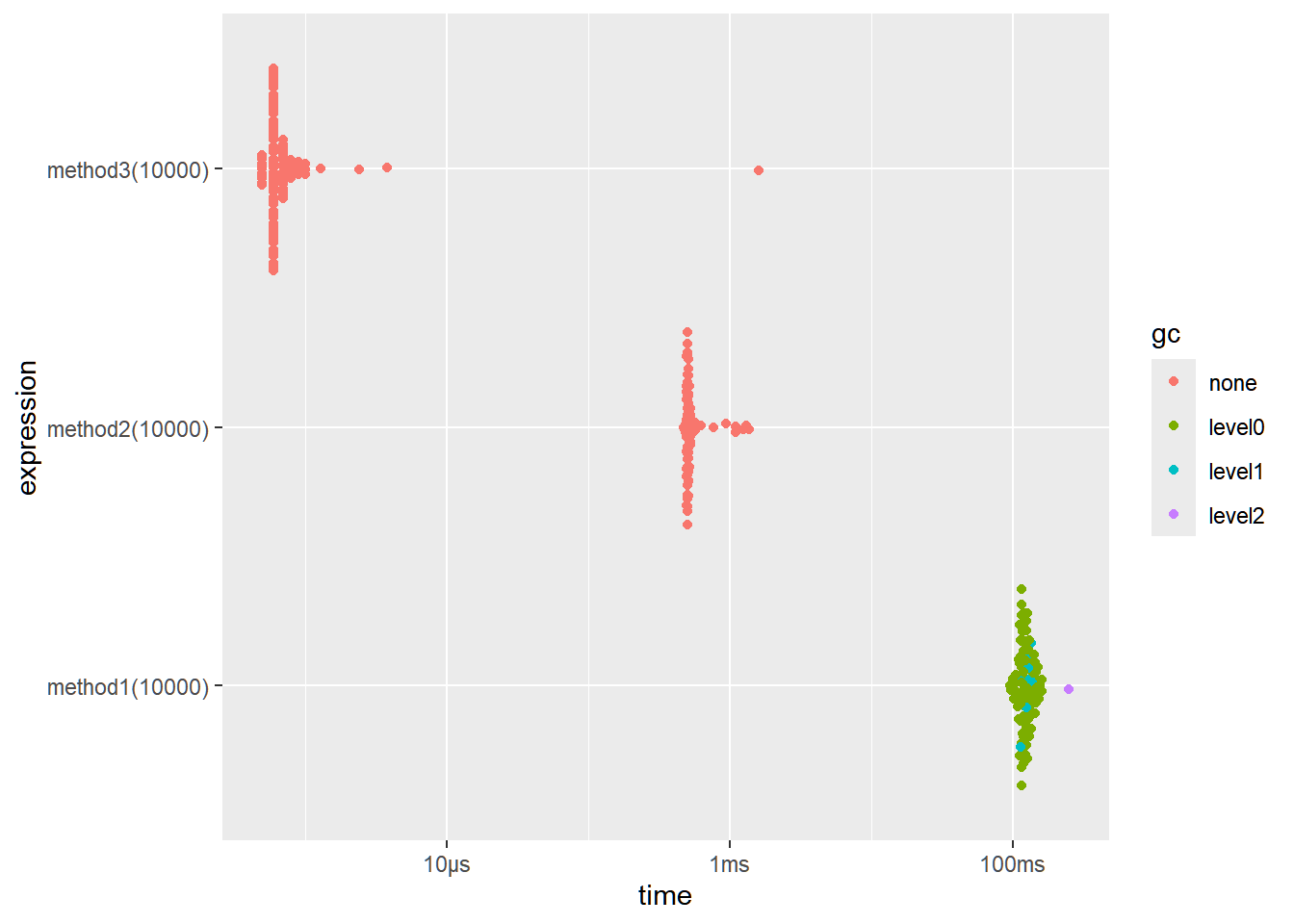
Example
We will create a function that generates a vector of integers from 1 up to n.
Without pre-allocation (slow)
method1 <- function(n) { vec <- c() for (i in 1:n) { vec <- c(vec, i) # Reallocates memory every iteration } return(vec) }This creates an empty vector and gradually increases (or grows) the length of the vector.
With pre-allocation (fast)
This creates an object of with pre-defined length and then changes the values in the object by indexing.
directly creating the final object
All these methods yield the same outcome yet differ in terms of efficiency.
If we try to compare using microbenchmarking:
result <- bench::mark( method1(1e4), method2(1e4), method3(1e4), iterations = 100, time_unit = "ms" ) result[c("expression", "min", "median", "itr/sec", "n_gc")]
<bnch_xpr> 94.9564 121.24500 8.047288 <bnch_xpr> 0.4704 0.50785 1810.957743 <bnch_xpr> 0.0005 0.00060 60067.275347
2.3.3 Vectorizing
Vectorization in R leverages optimized C-based operations instead of loops, significantly improving speed.
Many R functions are vectorized, that is the function’s inputs and/or outputs naturally work with vectors, reducing the number of function calls required. For example, the code
x <- runif(n) + 1performs two vectorized operations. First runif() returns n random numbers. Second, we add 1 to each element of the vector. In general, it is a good idea to exploit vectorized functions.
Example
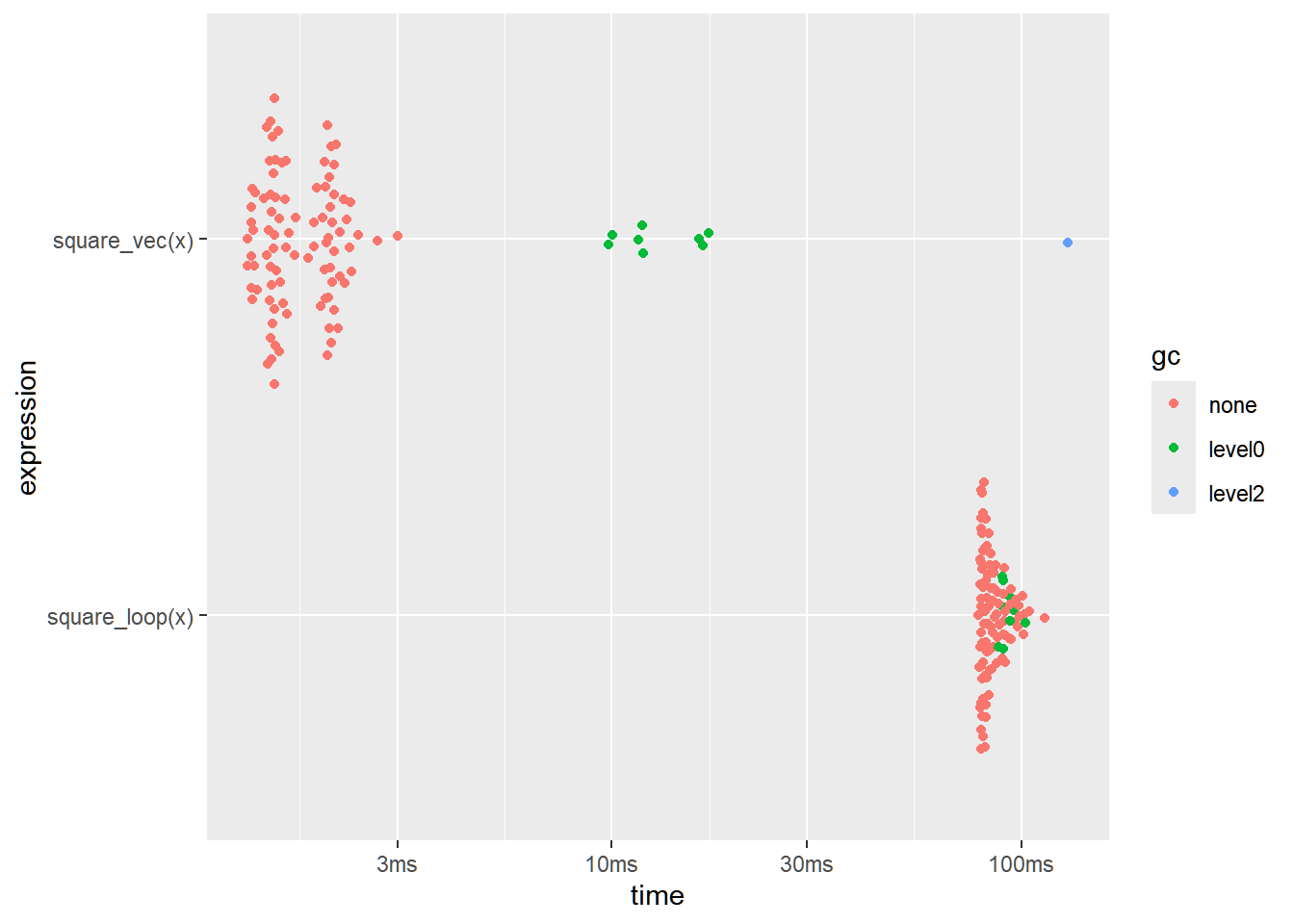
Consider this piece of R code that calculates the square of each element of x:
# Using a loop (slow)
square_loop <- function(x) {
res <- numeric(length(x))
for (i in seq_along(x)) {
res[i] <- x[i]^2
}
return(res)
}How can we vectorize this?
Answer
Straightforward, using the operation x^2
For a quick test, we have the following benchmarking:
x <- 1:1000000
result <- bench::mark(square_loop(x),
square_vec(x),
iterations = 100,
time_unit = "ms"
)
result[c("expression", "min", "median", "itr/sec")]expression <bnch_xpr> | min <dbl> | median <dbl> | itr/sec <dbl> | |
|---|---|---|---|---|
| <bnch_xpr> | 78.3229 | 83.1027 | 11.68007 | |
| <bnch_xpr> | 1.2912 | 1.5997 | 569.75633 |
Note: There are three levels of collections.
Level 0 collects only the youngest generation, level 1 collects the two youngest generations and level 2 collects all generations.
After 20 level-0 collections the next collection is at level 1,
and after 5 level-1 collections at level 2.
Further, if a level-n collection fails to provide 20% free space (for each of nodes and the vector heap), the next collection will be at level n+1.
The R-level function
gc()performs a level-2 collection.
Writing code this way has a number of benefits.
- It’s faster. When n=106, the R way is about four times faster.
- It’s neater.
- It doesn’t contain a bug when
xis of length 0.
Important note:
The slowdown isn’t due to the for loop. Instead, it’s because there are many more function calls.
So instead of going through a loop and performing many function calls, it’s better if we can exploit vectorized operations in R.
2.3.4 Using Matrix Operations
Another reason why matrix operations is taught is that they are sometimes more efficient when applied in programs.
This is an extension of vectorizing.
For example, using nested loops for matrix multiplication
A <- matrix(rnorm(10000), 100, 100)
B <- matrix(rnorm(10000), 100, 100)
mat_mul <- function(X,Y){
Z <- matrix(0, 100, 100)
for (i in 1:100) {
for (j in 1:100) {
for (k in 1:100) {
Z[i, j] <- Z[i, j] + X[i, k] * Y[k, j] # Very slow
}
}
}
return(Z)
}expression <bnch_xpr> | min <dbl> | median <dbl> | itr/sec <dbl> | mem_alloc <bnch_byt> | gc/sec <dbl> |
|---|---|---|---|---|---|
| <bnch_xpr> | 168705.1 | 175928.8 | 5.681797 | 124.96094KB | 0 |
| <bnch_xpr> | 491.4 | 510.6 | 1815.551922 | 78.17188KB | 0 |
WARNING: Problems in Precision
However, be careful for possible rounding off errors due to floating-point arithmetic.
This is expected when performing matrix operations like
Jbar %*% mat, since matrix multiplication involves multiple floating-point operations.Suppose we have the following dataset
2 7 1 3 9 3 4 11 6 5 13 9 6 15 11 You want to subtract each element from the mean of each column. In other words, you are “centering” this dataset.
You may premultiply the centering matrix I−ˉJ to the matrix.
## [,1] [,2] [,3] [,4] [,5] ## [1,] 1 0 0 0 0 ## [2,] 0 1 0 0 0 ## [3,] 0 0 1 0 0 ## [4,] 0 0 0 1 0 ## [5,] 0 0 0 0 1 ## [,1] [,2] [,3] [,4] [,5] ## [1,] 0.2 0.2 0.2 0.2 0.2 ## [2,] 0.2 0.2 0.2 0.2 0.2 ## [3,] 0.2 0.2 0.2 0.2 0.2 ## [4,] 0.2 0.2 0.2 0.2 0.2 ## [5,] 0.2 0.2 0.2 0.2 0.2Now obtaining (I−ˉJ)X
## x1 x2 x3 ## [1,] -2 -4.000000e+00 -5.000000e+00 ## [2,] -1 -2.000000e+00 -3.000000e+00 ## [3,] 0 4.440892e-16 8.881784e-16 ## [4,] 1 2.000000e+00 3.000000e+00 ## [5,] 2 4.000000e+00 5.000000e+00This happened because the centering matrix (I−ˉJ) contains decimal values, which may not represent the exact values they want to represent, which then propagates after the matrix multiplication with X
As an alternative, you may use the following:
X−ˉJX
## x1 x2 x3 ## [1,] -2 -4 -5 ## [2,] -1 -2 -3 ## [3,] 0 0 0 ## [4,] 1 2 3 ## [5,] 2 4 5With this,
ˉJX is computed first, which is just a matrix of column means.
Subtracting this from X only involves basic subtraction.
Fewer floating-point operations = less accumulation of error.
2.3.5 Doing as little as possible
The easiest way to make a function faster is to let it do less work. One way to do that is use a function tailored to a more specific type of input or output, or to a more specific problem.
For example:
rowSums(),colSums(),rowMeans(), andcolMeans()are faster than equivalent invocations that useapply()because they are vectorised.Example
Suppose we have the matrix Y
Let’s try getting the sum of each column of
Yin two ways:Using
sumwithapply## [1] 5050 15050 25050 35050 45050The
colSumsfunction## [1] 5050 15050 25050 35050 45050
Benchmarking the two functions, we have:
expression<bnch_xpr>min<bench_tm>median<bench_tm>itr/sec<dbl>mem_alloc<bnch_byt>gc/sec<dbl><bnch_xpr> 34.9µs 38.5µs 23709.04 6.375KB 0 <bnch_xpr> 4.5µs 5µs 190439.92 0B 0 The performance of vector operations, matrix operations, and certain specialized functions may vary. When developing a program, it is advisable to experiment to determine the most efficient approach for your specific case.
vapply()is faster thansapply()because it pre-specifies the output type through its parameterFUN.VALUE.Example
## a b c ## 500500 1500500 2500500## a b c ## 500500 1500500 2500500expression<bnch_xpr>min<bench_tm>median<bench_tm>itr/sec<dbl>mem_alloc<bnch_byt>gc/sec<dbl><bnch_xpr> 20.5µs 21.9µs 42603.91 0B 8.522486 <bnch_xpr> 8.8µs 9.7µs 93825.83 0B 9.383521 Explanation:
sapply():Returns a vector or matrix, depending on the result of the applied function. It does not require you to specify the output type.vapply(): Requires you to specify the output type, which makes it more efficient since it avoids guessing the result structure.
Some functions coerce their inputs into a specific type. If your input is not the right type, the function has to do extra work. Instead, look for a function that works with your data as it is, or consider changing the way you store your data.
Other functions will do less work if you give them more information about the problem.
read.csv(): specify known column types withcolClasses. (Also consider switching toreadr::read_csv()ordata.table::fread()which are considerably faster thanread.csv().)factor(): specify known levels withlevels.cut(): don’t generate labels withlabels = FALSEif you don’t need them, or, even better, usefindInterval()as mentioned in the “see also” section of the documentation.unlist(x, use.names = FALSE)is much faster thanunlist(x).interaction(): if you only need combinations that exist in the data, usedrop = TRUE.
The best way to solve this problem is to read widely so that you can build up your own vocabulary over time.
Record all solutions that you find, not just those that immediately appear to be faster. Some solutions might be slower initially, but end up being faster because they’re easier to optimize.
2.3.6 Parallel Computing
Parallel computing is an essential tool for statisticians working with large datasets or complex models, offering a way to split computations into smaller tasks that can be executed simultaneously.
By leveraging multiple processors or cores, parallel computing can significantly reduce computation time and improve efficiency.
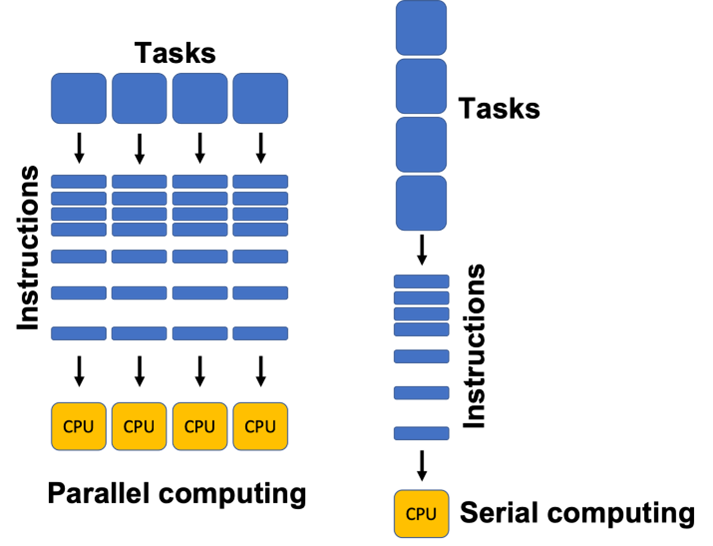
Definition 2.4
Parallel computing uses multiple computer cores to attack several operations at once.
Unlike serial computing, parallel architecture can break down a job into its component parts and multi-task them. Parallel computer systems are well suited to modeling and simulating real-world phenomena.
Key Concepts
Parallelism: The simultaneous execution of multiple tasks.
Shared Memory vs. Distributed Memory: Shared memory systems allow processors to access the same memory, while distributed memory systems use separate memories and communicate over a network.
Multithreading: Running multiple threads within a single process concurrently.
Parallel Computing in R
R offers several tools for parallel computing, including the parallel package.
## [1] 16Note that the value returned by detectCores() is dependent on the operating system and hardware of your machine.
The most used parallel applications are parallelized replacements of lapply(), sapply() and apply() (i.e., parLapply , parApply, and parSapply)
We will have examples of this in Section 3.3.
To those interested about learning more about parallel computing in R, I suggest you read Parallel Processing in R by Josh Errickson.
We will not be discussing this in detail since this would be better to be discussed in another course.
2.3.7 Compiling Code
Compilation is the process of translating R code into byte-code that can be executed more efficiently.
The compiler package, introduced in R 2.13.0, compiles R functions into byte-code for faster execution by reducing interpretation overhead.
Since R 2.14.0, base R functions like mean() are pre-compiled by default.
## function (x, ...)
## UseMethod("mean")
## <bytecode: 0x0000023530f3f7e0>
## <environment: namespace:base>The third line contains the bytecode of the function. This means that the compiler package has translated the R function into another language that can be interpreted by a very fast interpreter. Amazingly the compiler package is almost entirely pure R, with just a few C support routines.
Compiling your own function
The compiler package, introduced in R 2.13.0, enables this by reducing interpretation overhead. Since R 2.14.0, base R functions like mean() are pre-compiled by default.
The compiler package comes with R, so we just need to load the package in the usual way.
To demonstrate, we create an inefficient function for calculating the mean. This function takes in a vector, calculates the length and then updates the m variable.
This is clearly a bad function and we should just use the mean() function, but it’s a useful comparison.
Compiling the function is straightforward, using the cmpfun function.
## [1] 0.4937434
## [1] 0.4937434However, in some cases, using bench::mark() may show that an uncompiled function runs faster. This can happen because compiling introduces additional overhead, especially for very short functions where the compilation time outweighs the performance gains.
The benefits of compilation are more noticeable in complex functions that involve many computations or loops.
Exercises
Consider this piece of R code:
log_sum1 <- function(x){ log_sum <- 0 for (i in 1:length(x)){ log_sum <- log_sum + log(x[i]) } return(log_sum) }Create another function
log_sum2that shows the same outputs, but uses vectorization.Compare results by running this chunk:
Also, check its median relative performance by computing
relative performance of log_sum1=median run time of log_sum1 median run time of log_sum2
The following codes create a growing dataframe per iteration. It has two columns,
a, which contains integers from 1 to n, andbwhich contains the square of the integers from 1 to n.no_prealloc_df <- function(n) { df <- data.frame() for (i in 1:n) { df <- rbind(df, data.frame(a = i, b = i^2)) } return(df) }a<int>b<dbl>1 1 2 4 3 9 4 16 5 25 Create a new function that creates the same output above, but with pre-allocation.
You may initialize a dataframe with zeroes using the following:
data.frame(a = numeric(n), b = numeric(n))Note that when indexing dataframes or matrices, there are two parameters.
Supose
datais a dataframe object. To get the rth row and cth column, we usedata[r,c].