Chapter 5 Cross-Validation
In creating statistical models, it is important to evaluate the performance of your model when deployed on new data.
Definition 5.1 Cross-validation is a statistical technique used to assess the performance and generalizability of a predictive model. It involves partitioning the dataset into multiple subsets, training the model on some of these subsets, and validating it on the remaining ones.
The goal is to evaluate how well the model will perform on unseen data and to reduce the risk of overfitting.
In this section, we will discuss some cross-validation methods to assess how good the model is in predicting a set of new observations.
There will be three cross-validation methods that will be discussed here:
- Validation Set Approach
- Leave-One-Out Cross-Validation (LOOCV)
- K-Fold Cross-Validation
All of these approaches involve calculating some evaluation metrics or cross-validation estimate. For an overview, here are some of them:
Regression
Let yi be the actual observed value and let ˆyi be the predicted value computed using ˆf(xi). The following are possible evaluation metrics in regression. Lower values are better.
- MSE=1n∑ni=1(yi−ˆyi)2
- RMSE=√1n∑ni=1(yi−ˆyi)2
- MAE=1n∑ni=1|yi−ˆyi|
- BIAS=1n∑ni=1(yi−ˆyi)
Classification
Suppose we are classifying y into two values (positive or negative). After modelling and predicting the classification of y, we have the following confusion matrix.
| Predicted Positive | Predicted Negative | |
|---|---|---|
| Actual Positive | TP | FN |
| Actual Negative | FP | TN |
The following are some evaluation metrics for a binary classification problem. Higher values are better.
Accuracy: TP+TNTP+TN+FP+FN
Precision (Positive Predictive Value): TPTP+FP
Recall (Sensitivity or True Positive Rate): TPTP+FN
Specificity (True Negative Rate): TNTN+FP
F1 Score: 2⋅Precision⋅RecallPrecision+Recall
The choice of an evaluation metric will be based on the context of your data. All of these have functions in the Metrics package.
In this Chapter, we will focus on cross-validation of models that predict numeric variables, specifically with applications on multiple linear regression only.
Validation Set Approach
The train-and-test-set validation, or simply validation set approach, is a a very simple strategy for cross-validation.
The idea is to randomly split the set of observations into two parts, a training set and a test set (also called validation set, or hold-out set).
The model is fit on the training set, and the fitted model is used to predict the responses for the observations in the test set.
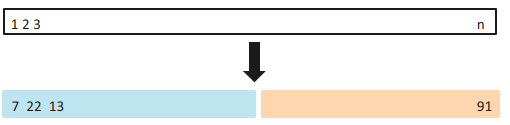
It is common to split the dataframe to 70-30: 70% of the dataset will be used for training the model, and the rest will be used for testing the model.
The resulting test set error rate is typically assessed using MSE or RMSE for quantitative response variables.
education <int> | income <int> | young <dbl> | urban <int> | |
|---|---|---|---|---|
| ME | 189 | 2824 | 350.7 | 508 |
| NH | 169 | 3259 | 345.9 | 564 |
| VT | 230 | 3072 | 348.5 | 322 |
| MA | 168 | 3835 | 335.3 | 846 |
| RI | 180 | 3549 | 327.1 | 871 |
| CT | 193 | 4256 | 341.0 | 774 |
| NY | 261 | 4151 | 326.2 | 856 |
| NJ | 214 | 3954 | 333.5 | 889 |
| PA | 201 | 3419 | 326.2 | 715 |
| OH | 172 | 3509 | 354.5 | 753 |
validation <- function(formula, data, size = 0.7, criterion){
train_indices <- sample(1:nrow(data),
size = 0.7 * nrow(data))
train_data <- data[train_indices, ]
test_data <- data[-train_indices, ]
# Fit model on training data
lm_fit <- lm(formula, data = train_data)
# Predict on test data
pred <- predict(lm_fit, newdata = test_data)
# Extract the vector of observed y from test data
obs <- test_data[[all.vars(formula)[1]]]
# compute and return evaluation metric
eval <- criterion(obs, pred)
return(eval)
}validation(income ~ urban, Anscombe, size = 0.7, Metrics::rmse)
validation(income ~ urban + education, Anscombe, size = 0.7, Metrics::rmse)
validation(income ~ urban + education + young, Anscombe, size = 0.7, Metrics::rmse)## [1] 552.6391
## [1] 368.039
## [1] 280.6608Leave-One-Out Cross Validation
Leave-one-out cross-validation (LOOCV) is closely related to the validation set approach, but it attempts to address that method’s drawbacks.
Like the validation set approach, LOOCV involves splitting the set of observations into two parts. However, instead of creating two subsets of comparable size, a single observation (x1,y1) is used for the validation set, and the remaining observations (x2,y2),...,(xn,yn) make up the training set.
The processes is repeated for all observation to obtain n evaluation metrics, and their average will be obtained for an overall evaluation metric.
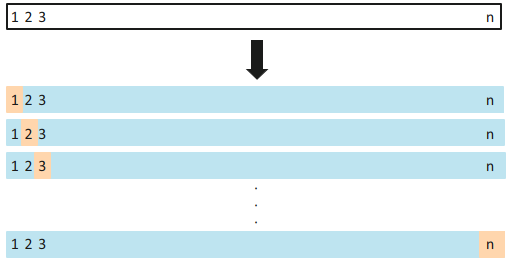
Most functions in R already show statistics from LOOCV approach (such as the PRESS), but we will demonstrate the LOOCV approach manually here.
loocv <- function(formula, data, criterion){
n <- nrow(data)
eval <- numeric(n)
for(i in 1:n){
train_i <- data[-i,]
test_i <- data[i,]
mod_i <- lm(formula,train_i)
pred_i <- predict(mod_i, test_i)
y <- all.vars(formula)[1]
eval[i] <- criterion(test_i[[y]], pred_i)
}
return(mean(eval))
}loocv(income ~ urban, Anscombe, Metrics::rmse)
loocv(income ~ urban + education, Anscombe, Metrics::rmse)## [1] 324.9626
## [1] 232.3712k-Fold Cross-validation
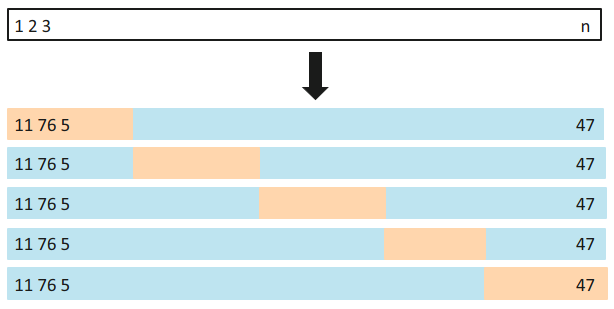
An alternative to LOOCV is k-fold CV.
This approach involves randomly dividing the set of observations into k groups, or folds, of approximately equal size.
The first fold is treated as a validation set, and the method is fit on the remaining k−1 folds. This process results in k estimates of the evaluation metric. The k-fold cross validation estimate is computed as the average of the k metrics.
LOOCV is a special case of k-Fold CV, where k=n.
kfold <- function(formula, data, k = 10, criterion){
n <- nrow(data)
# Assigning folds
folds <- sample(rep(1:k, length.out = n))
# Loop
eval <- numeric(k)
for (i in 1:k){
#splitting the dataset
train <- data[folds!=i,]
test <- data[folds==i,]
# modelling
mod <- lm(formula, data = train)
pred <- predict(mod, test)
y <- all.vars(formula)[1]
eval[i] <- criterion(test[[y]], pred)
}
return(mean(eval))
}kfold(income ~ urban, data = Anscombe, k = 10, Metrics::rmse)
kfold(income ~ urban + young,data = Anscombe, k = 10, Metrics::rmse)## [1] 389.2178
## [1] 432.4814Example/Exercise: Model performance through the years
In this exercise, we explore if a model created in 1952 is still useful for prediction as years go by using some exploratory data analysis. Install and load the package gapminder for this exercise.
Create a new function
lm_validatethat inputs the following:formula: the formula to be usedtrain: training set to be usedtest: test set to be usedcriterion: the evaluation metric function to be used
This should perform the Validation Set Approach for linear regression, but you will have more control which data will be used for training and testing.
Load the dataset
gapminderincluded in the packagegapminder.country<fct>continent<fct>year<int>lifeExp<dbl>pop<int>gdpPercap<dbl>Afghanistan Asia 1952 28.80100 8425333 779.4453 Afghanistan Asia 1957 30.33200 9240934 820.8530 Afghanistan Asia 1962 31.99700 10267083 853.1007 Afghanistan Asia 1967 34.02000 11537966 836.1971 Afghanistan Asia 1972 36.08800 13079460 739.9811 Afghanistan Asia 1977 38.43800 14880372 786.1134 Afghanistan Asia 1982 39.85400 12881816 978.0114 Afghanistan Asia 1987 40.82200 13867957 852.3959 Afghanistan Asia 1992 41.67400 16317921 649.3414 Afghanistan Asia 1997 41.76300 22227415 635.3414 Filter the dataset to show only values in 1952. Fit a linear regression model that predicts
lifeExpusinglog(gdpPercap)andpop.Using the model in (3) and the function in (1), predict the
lifeExpof each country per year, and show how the RMSE changes through the years. It should look something like this: