1.2 Defining New Functions
In this section, we explore how commands work in inside a function, so we will be able to defined and design our own user-defined functions.
Definition 1.1 A function is a reusable block of code that performs a specific task. Functions take inputs (known as arguments or parameters), process them, and may return an output.
We start by examining how to express the idea of squaring. We might say, “To square something, multiply it by itself.” Of course, R already has this operation:
## [1] 9Now what if we do squaring just by using multiplication—a more primitive function that is included in most programming languages?
This defines a new function that has been given the name square.
Function definitions consist of the following:
a function statement that indicates a
<name>a comma-separated list of named
<formal parameters>.the
<function body>
<name> <- function(<formal parameters>){
<function body>
return(<return expression>)
}Definition 1.2 The values that will serve as function input are called formal parameters, or argument.
Definition 1.3 The function body is everything inside the curly braces {<function body>}. It contains a sequence of expressions and commands to be evaluated whenever the function is applied.
In the example, the return statement is part of the function body.
We can also write our function this way:
<name> <- function(<formal parameters>){return(<return expression>)}But for readability, we usually write the function body as a new line of code.
After running the lines of code defining our new function square, nothing will be displayed in your console YET. We still need to apply it with a call expression.
Definition 1.4 A call expression in R (or in programming in general) refers to an expression that invokes a function by specifying the function name and providing arguments within parentheses. It is how functions are executed.
212
## [1] 441(2+4)2
## [1] 49(32)2
## [1] 811.2.1 Environments
Questions:
What if a function assigns a value to a variable inside its function body that is bound to a different value already?
## [1] 40## [1] 5Can two functions share names without confusion?
stats::filter() dplyr::filter()
This is where environments will come in handy.
Definition 1.5 An environment can be thought of as a collection of objects (functions, variables, parent environments, etc.). Its job is to associate, or bind, a set of names to a set of values.
a <- FALSE
b <- "hello"
c <- 2.45
d <- function(){ # Function d takes in no arguments
return(0) # When called, function d returns the value 0
}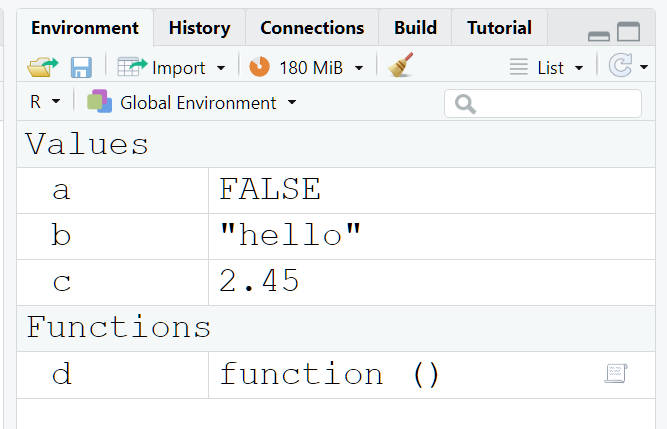
The global environment is the top-level environment available to us at the R command prompt.
What you see in the environment pane are the name-value bindings that are in the global frame. Take note: the global frame is the frame of the global environment.
We can use the environment() function to get the current environment.
## <environment: R_GlobalEnv>We can use the ls() function to show the bindings that are defined in the current environment.
## [1] "a" "ackermann"
## [3] "add_three" "age"
## [5] "age1" "age2"
## [7] "age3" "Anscombe"
## [9] "apply_function" "apply_trimmed"
## [11] "apply_twice" "arr"
## [13] "assignedSex1" "assignedSex2"
## [15] "assignedSex3" "b"
## [17] "B" "bank"
## [19] "beta" "betahat"
## [21] "bias_mean" "bias_median"
## [23] "binary_search" "boot.mpg"
## [25] "boot.reg" "boot_reg"
## [27] "boot2" "c"
## [29] "cards" "class_data"
## [31] "color" "con"
## [33] "customer_data" "customers"
## [35] "d" "dat1"
## [37] "dat2" "dat3"
## [39] "dat4" "data"
## [41] "data.star" "differentiate"
## [43] "double" "eval"
## [45] "exp_b" "f"
## [47] "factorial_loop" "fit"
## [49] "fit2" "fold"
## [51] "folds" "func"
## [53] "g" "generate_sequence"
## [55] "get_element" "gpa"
## [57] "grad_rosenbrock" "grade"
## [59] "h" "hess_rosenbrock"
## [61] "i" "index"
## [63] "j" "k"
## [65] "kfold" "kmeans_result"
## [67] "l" "lawschool"
## [69] "linear_search" "lm_fit"
## [71] "lm_validate" "loglik_lgstc"
## [73] "logreg_plot" "loocv"
## [75] "lsat" "M"
## [77] "mat" "mc_norm"
## [79] "mc_pois" "menu"
## [81] "method1" "method2"
## [83] "method3" "mod"
## [85] "mod1" "mod2"
## [87] "model" "mpg.boot"
## [89] "mtcars" "my_array"
## [91] "my_chr" "my_dbl"
## [93] "my_int" "my_list"
## [95] "my_log" "my_sorted_array"
## [97] "n" "N"
## [99] "names" "nested_loop"
## [101] "newton_optim" "newton_raphson_logistic"
## [103] "p" "P"
## [105] "pi_x" "pop"
## [107] "pred1" "predictions"
## [109] "prop" "Q"
## [111] "r.boot" "r.star"
## [113] "residual_bootstrap" "result"
## [115] "result_plot" "rmse"
## [117] "rmse_mod" "rmse_x"
## [119] "rosenbrock" "runtime"
## [121] "samp" "samp.star"
## [123] "sample_size" "simple_model"
## [125] "square" "square_d"
## [127] "square_loop" "start_time"
## [129] "SumOfDigits" "test"
## [131] "test_data" "time"
## [133] "tmp" "train"
## [135] "train_data" "train_indices"
## [137] "transactions" "update_x"
## [139] "validation" "var_mc"
## [141] "var_p" "var_phat"
## [143] "vari_mean" "vari_median"
## [145] "vec1" "vec2"
## [147] "vec3" "vec4"
## [149] "vec5" "vec6"
## [151] "x" "X"
## [153] "x0" "xbar.boot"
## [155] "xbar.star" "y"
## [157] "y1" "y2"
## [159] "y3" "years"
## [161] "z" "Z"
## [163] "Z_vec"
What if we call the function max? Recall that the only function in our current environment is d.
## function (..., na.rm = FALSE) .Primitive("max")Where is this function coming from?
Let’s check the parent environment.
## <environment: package:stats>
## attr(,"name")
## [1] "package:stats"
## attr(,"path")
## [1] "C:/Program Files/R/R-4.4.1/library/stats"## [1] "acf" "acf2AR" "add.scope"
## [4] "add1" "addmargins" "aggregate"
## [7] "aggregate.data.frame" "aggregate.ts" "AIC"
## [10] "alias" "anova" "ansari.test"
## [13] "aov" "approx" "approxfun"
## [16] "ar" "ar.burg" "ar.mle"
## [19] "ar.ols" "ar.yw" "arima"
## [22] "arima.sim" "arima0" "arima0.diag"
## [25] "ARMAacf" "ARMAtoMA" "as.dendrogram"
## [28] "as.dist" "as.formula" "as.hclust"
## [31] "as.stepfun" "as.ts" "asOneSidedFormula"
## [34] "ave" "bandwidth.kernel" "bartlett.test"
## [37] "BIC" "binom.test" "binomial"
## [40] "biplot" "Box.test" "bw.bcv"
## [43] "bw.nrd" "bw.nrd0" "bw.SJ"
## [46] "bw.ucv" "C" "cancor"
## [49] "case.names" "ccf" "chisq.test"
## [52] "cmdscale" "coef" "coefficients"
## [55] "complete.cases" "confint" "confint.default"
## [58] "confint.lm" "constrOptim" "contr.helmert"
## [61] "contr.poly" "contr.SAS" "contr.sum"
## [64] "contr.treatment" "contrasts" "contrasts<-"
## [67] "convolve" "cooks.distance" "cophenetic"
## [70] "cor" "cor.test" "cov"
## [73] "cov.wt" "cov2cor" "covratio"
## [76] "cpgram" "cutree" "cycle"
## [79] "D" "dbeta" "dbinom"
## [82] "dcauchy" "dchisq" "decompose"
## [85] "delete.response" "deltat" "dendrapply"
## [88] "density" "density.default" "deriv"
## [91] "deriv3" "deviance" "dexp"
## [94] "df" "df.kernel" "df.residual"
## [97] "DF2formula" "dfbeta" "dfbetas"
## [100] "dffits" "dgamma" "dgeom"
## [103] "dhyper" "diffinv" "dist"
## [106] "dlnorm" "dlogis" "dmultinom"
## [109] "dnbinom" "dnorm" "dpois"
## [112] "drop.scope" "drop.terms" "drop1"
## [115] "dsignrank" "dt" "dummy.coef"
## [118] "dummy.coef.lm" "dunif" "dweibull"
## [121] "dwilcox" "ecdf" "eff.aovlist"
## [124] "effects" "embed" "end"
## [127] "estVar" "expand.model.frame" "extractAIC"
## [130] "factanal" "factor.scope" "family"
## [133] "fft" "filter" "fisher.test"
## [136] "fitted" "fitted.values" "fivenum"
## [139] "fligner.test" "formula" "frequency"
## [142] "friedman.test" "ftable" "Gamma"
## [145] "gaussian" "get_all_vars" "getCall"
## [148] "getInitial" "glm" "glm.control"
## [151] "glm.fit" "hasTsp" "hat"
## [154] "hatvalues" "hclust" "heatmap"
## [157] "HoltWinters" "influence" "influence.measures"
## [160] "integrate" "interaction.plot" "inverse.gaussian"
## [163] "IQR" "is.empty.model" "is.leaf"
## [166] "is.mts" "is.stepfun" "is.ts"
## [169] "is.tskernel" "isoreg" "KalmanForecast"
## [172] "KalmanLike" "KalmanRun" "KalmanSmooth"
## [175] "kernapply" "kernel" "kmeans"
## [178] "knots" "kruskal.test" "ks.test"
## [181] "ksmooth" "lag" "lag.plot"
## [184] "line" "lm" "lm.fit"
## [187] "lm.influence" "lm.wfit" "loadings"
## [190] "loess" "loess.control" "loess.smooth"
## [193] "logLik" "loglin" "lowess"
## [196] "ls.diag" "ls.print" "lsfit"
## [199] "mad" "mahalanobis" "make.link"
## [202] "makeARIMA" "makepredictcall" "manova"
## [205] "mantelhaen.test" "mauchly.test" "mcnemar.test"
## [208] "median" "median.default" "medpolish"
## [211] "model.extract" "model.frame" "model.frame.default"
## [214] "model.matrix" "model.matrix.default" "model.matrix.lm"
## [217] "model.offset" "model.response" "model.tables"
## [220] "model.weights" "monthplot" "mood.test"
## [223] "mvfft" "na.action" "na.contiguous"
## [226] "na.exclude" "na.fail" "na.omit"
## [229] "na.pass" "napredict" "naprint"
## [232] "naresid" "nextn" "nlm"
## [235] "nlminb" "nls" "nls.control"
## [238] "NLSstAsymptotic" "NLSstClosestX" "NLSstLfAsymptote"
## [241] "NLSstRtAsymptote" "nobs" "numericDeriv"
## [244] "offset" "oneway.test" "optim"
## [247] "optimHess" "optimise" "optimize"
## [250] "order.dendrogram" "p.adjust" "p.adjust.methods"
## [253] "pacf" "Pair" "pairwise.prop.test"
## [256] "pairwise.t.test" "pairwise.table" "pairwise.wilcox.test"
## [259] "pbeta" "pbinom" "pbirthday"
## [262] "pcauchy" "pchisq" "pexp"
## [265] "pf" "pgamma" "pgeom"
## [268] "phyper" "plclust" "plnorm"
## [271] "plogis" "plot.ecdf" "plot.spec.coherency"
## [274] "plot.spec.phase" "plot.stepfun" "plot.ts"
## [277] "pnbinom" "pnorm" "poisson"
## [280] "poisson.test" "poly" "polym"
## [283] "power" "power.anova.test" "power.prop.test"
## [286] "power.t.test" "PP.test" "ppoints"
## [289] "ppois" "ppr" "prcomp"
## [292] "predict" "predict.glm" "predict.lm"
## [295] "preplot" "princomp" "printCoefmat"
## [298] "profile" "proj" "promax"
## [301] "prop.test" "prop.trend.test" "psignrank"
## [304] "psmirnov" "pt" "ptukey"
## [307] "punif" "pweibull" "pwilcox"
## [310] "qbeta" "qbinom" "qbirthday"
## [313] "qcauchy" "qchisq" "qexp"
## [316] "qf" "qgamma" "qgeom"
## [319] "qhyper" "qlnorm" "qlogis"
## [322] "qnbinom" "qnorm" "qpois"
## [325] "qqline" "qqnorm" "qqplot"
## [328] "qsignrank" "qsmirnov" "qt"
## [331] "qtukey" "quade.test" "quantile"
## [334] "quasi" "quasibinomial" "quasipoisson"
## [337] "qunif" "qweibull" "qwilcox"
## [340] "r2dtable" "rbeta" "rbinom"
## [343] "rcauchy" "rchisq" "read.ftable"
## [346] "rect.hclust" "reformulate" "relevel"
## [349] "reorder" "replications" "reshape"
## [352] "resid" "residuals" "residuals.glm"
## [355] "residuals.lm" "rexp" "rf"
## [358] "rgamma" "rgeom" "rhyper"
## [361] "rlnorm" "rlogis" "rmultinom"
## [364] "rnbinom" "rnorm" "rpois"
## [367] "rsignrank" "rsmirnov" "rstandard"
## [370] "rstudent" "rt" "runif"
## [373] "runmed" "rweibull" "rwilcox"
## [376] "rWishart" "scatter.smooth" "screeplot"
## [379] "sd" "se.contrast" "selfStart"
## [382] "setNames" "shapiro.test" "sigma"
## [385] "simulate" "smooth" "smooth.spline"
## [388] "smoothEnds" "sortedXyData" "spec.ar"
## [391] "spec.pgram" "spec.taper" "spectrum"
## [394] "spline" "splinefun" "splinefunH"
## [397] "SSasymp" "SSasympOff" "SSasympOrig"
## [400] "SSbiexp" "SSD" "SSfol"
## [403] "SSfpl" "SSgompertz" "SSlogis"
## [406] "SSmicmen" "SSweibull" "start"
## [409] "stat.anova" "step" "stepfun"
## [412] "stl" "StructTS" "summary.aov"
## [415] "summary.glm" "summary.lm" "summary.manova"
## [418] "summary.stepfun" "supsmu" "symnum"
## [421] "t.test" "termplot" "terms"
## [424] "terms.formula" "time" "toeplitz"
## [427] "toeplitz2" "ts" "ts.intersect"
## [430] "ts.plot" "ts.union" "tsdiag"
## [433] "tsp" "tsp<-" "tsSmooth"
## [436] "TukeyHSD" "uniroot" "update"
## [439] "update.default" "update.formula" "var"
## [442] "var.test" "variable.names" "varimax"
## [445] "vcov" "weighted.mean" "weighted.residuals"
## [448] "weights" "wilcox.test" "window"
## [451] "window<-" "write.ftable" "xtabs"Unfortunately, the max function is still not in the parent environment.
Now, let us check in the grandparent environment.
## <environment: package:graphics>
## attr(,"name")
## [1] "package:graphics"
## attr(,"path")
## [1] "C:/Program Files/R/R-4.4.1/library/graphics"Let’s see the name bindings in the graphics package.
## [1] "abline" "arrows" "assocplot" "axis"
## [5] "Axis" "axis.Date" "axis.POSIXct" "axTicks"
## [9] "barplot" "barplot.default" "box" "boxplot"
## [13] "boxplot.default" "boxplot.matrix" "bxp" "cdplot"
## [17] "clip" "close.screen" "co.intervals" "contour"
## [21] "contour.default" "coplot" "curve" "dotchart"
## [25] "erase.screen" "filled.contour" "fourfoldplot" "frame"
## [29] "grconvertX" "grconvertY" "grid" "hist"
## [33] "hist.default" "identify" "image" "image.default"
## [37] "layout" "layout.show" "lcm" "legend"
## [41] "lines" "lines.default" "locator" "matlines"
## [45] "matplot" "matpoints" "mosaicplot" "mtext"
## [49] "pairs" "pairs.default" "panel.smooth" "par"
## [53] "persp" "pie" "plot" "plot.default"
## [57] "plot.design" "plot.function" "plot.new" "plot.window"
## [61] "plot.xy" "points" "points.default" "polygon"
## [65] "polypath" "rasterImage" "rect" "rug"
## [69] "screen" "segments" "smoothScatter" "spineplot"
## [73] "split.screen" "stars" "stem" "strheight"
## [77] "stripchart" "strwidth" "sunflowerplot" "symbols"
## [81] "text" "text.default" "title" "xinch"
## [85] "xspline" "xyinch" "yinch"max is still not here…
In fact, it is found in the base package.
environment() |> # current
parent.env() |> # stats
parent.env() |> # graphics
parent.env() |> # grDevices
parent.env() |> # utils
parent.env() |> # datasets
parent.env() |> # methods
parent.env() |> # Autoloads
parent.env() |> # base
ls() # check name bindings## [1] "-" "-.Date"
## [3] "-.POSIXt" "!"
## [5] "!.hexmode" "!.octmode"
## [7] "!=" "$"
## [9] "$.DLLInfo" "$.package_version"
## [11] "$<-" "$<-.data.frame"
## [13] "$<-.POSIXlt" "%%"
## [15] "%*%" "%/%"
## [17] "%||%" "%in%"
## [19] "%o%" "%x%"
## [21] "&" "&&"
## [23] "&.hexmode" "&.octmode"
## [25] "(" "*"
## [27] "*.difftime" "/"
## [29] "/.difftime" ":"
## [31] "::" ":::"
## [33] "@" "@<-"
## [35] "[" "[.AsIs"
## [37] "[.data.frame" "[.Date"
## [39] "[.difftime" "[.Dlist"
## [41] "[.DLLInfoList" "[.factor"
## [43] "[.hexmode" "[.listof"
## [45] "[.noquote" "[.numeric_version"
## [47] "[.octmode" "[.POSIXct"
## [49] "[.POSIXlt" "[.simple.list"
## [51] "[.table" "[.warnings"
## [53] "[[" "[[.data.frame"
## [55] "[[.Date" "[[.factor"
## [57] "[[.numeric_version" "[[.POSIXct"
## [59] "[[.POSIXlt" "[[<-"
## [61] "[[<-.data.frame" "[[<-.factor"
## [63] "[[<-.numeric_version" "[[<-.POSIXlt"
## [65] "[<-" "[<-.data.frame"
## [67] "[<-.Date" "[<-.difftime"
## [69] "[<-.factor" "[<-.numeric_version"
## [71] "[<-.POSIXct" "[<-.POSIXlt"
## [73] "^" "{"
## [75] "|" "|.hexmode"
## [77] "|.octmode" "||"
## [79] "~" "+"
## [81] "+.Date" "+.POSIXt"
## [83] "<" "<-"
## [85] "<<-" "<="
## [87] "=" "=="
## [89] ">" ">="
## [91] "abbreviate" "abs"
## [93] "acos" "acosh"
## [95] "activeBindingFunction" "addNA"
## [97] "addTaskCallback" "agrep"
## [99] "agrepl" "alist"
## [101] "all" "all.equal"
## [103] "all.equal.character" "all.equal.default"
## [105] "all.equal.environment" "all.equal.envRefClass"
## [107] "all.equal.factor" "all.equal.formula"
## [109] "all.equal.function" "all.equal.language"
## [111] "all.equal.list" "all.equal.numeric"
## [113] "all.equal.POSIXt" "all.equal.raw"
## [115] "all.names" "all.vars"
## [117] "allowInterrupts" "any"
## [119] "anyDuplicated" "anyDuplicated.array"
## [121] "anyDuplicated.data.frame" "anyDuplicated.default"
## [123] "anyDuplicated.matrix" "anyNA"
## [125] "anyNA.data.frame" "anyNA.numeric_version"
## [127] "anyNA.POSIXlt" "aperm"
## [129] "aperm.default" "aperm.table"
## [131] "append" "apply"
## [133] "Arg" "args"
## [135] "array" "array2DF"
## [137] "arrayInd" "as.array"
## [139] "as.array.default" "as.call"
## [141] "as.character" "as.character.condition"
## [143] "as.character.Date" "as.character.default"
## [145] "as.character.error" "as.character.factor"
## [147] "as.character.hexmode" "as.character.numeric_version"
## [149] "as.character.octmode" "as.character.POSIXt"
## [151] "as.character.srcref" "as.complex"
## [153] "as.data.frame" "as.data.frame.array"
## [155] "as.data.frame.AsIs" "as.data.frame.character"
## [157] "as.data.frame.complex" "as.data.frame.data.frame"
## [159] "as.data.frame.Date" "as.data.frame.default"
## [161] "as.data.frame.difftime" "as.data.frame.factor"
## [163] "as.data.frame.integer" "as.data.frame.list"
## [165] "as.data.frame.logical" "as.data.frame.matrix"
## [167] "as.data.frame.model.matrix" "as.data.frame.noquote"
## [169] "as.data.frame.numeric" "as.data.frame.numeric_version"
## [171] "as.data.frame.ordered" "as.data.frame.POSIXct"
## [173] "as.data.frame.POSIXlt" "as.data.frame.raw"
## [175] "as.data.frame.table" "as.data.frame.ts"
## [177] "as.data.frame.vector" "as.Date"
## [179] "as.Date.character" "as.Date.default"
## [181] "as.Date.factor" "as.Date.numeric"
## [183] "as.Date.POSIXct" "as.Date.POSIXlt"
## [185] "as.difftime" "as.double"
## [187] "as.double.difftime" "as.double.POSIXlt"
## [189] "as.environment" "as.expression"
## [191] "as.expression.default" "as.factor"
## [193] "as.function" "as.function.default"
## [195] "as.hexmode" "as.integer"
## [197] "as.list" "as.list.data.frame"
## [199] "as.list.Date" "as.list.default"
## [201] "as.list.difftime" "as.list.environment"
## [203] "as.list.factor" "as.list.function"
## [205] "as.list.numeric_version" "as.list.POSIXct"
## [207] "as.list.POSIXlt" "as.logical"
## [209] "as.logical.factor" "as.matrix"
## [211] "as.matrix.data.frame" "as.matrix.default"
## [213] "as.matrix.noquote" "as.matrix.POSIXlt"
## [215] "as.name" "as.null"
## [217] "as.null.default" "as.numeric"
## [219] "as.numeric_version" "as.octmode"
## [221] "as.ordered" "as.package_version"
## [223] "as.pairlist" "as.POSIXct"
## [225] "as.POSIXct.Date" "as.POSIXct.default"
## [227] "as.POSIXct.numeric" "as.POSIXct.POSIXlt"
## [229] "as.POSIXlt" "as.POSIXlt.character"
## [231] "as.POSIXlt.Date" "as.POSIXlt.default"
## [233] "as.POSIXlt.factor" "as.POSIXlt.numeric"
## [235] "as.POSIXlt.POSIXct" "as.qr"
## [237] "as.raw" "as.single"
## [239] "as.single.default" "as.symbol"
## [241] "as.table" "as.table.default"
## [243] "as.vector" "as.vector.data.frame"
## [245] "as.vector.factor" "as.vector.POSIXlt"
## [247] "asin" "asinh"
## [249] "asNamespace" "asplit"
## [251] "asS3" "asS4"
## [253] "assign" "atan"
## [255] "atan2" "atanh"
## [257] "attach" "attachNamespace"
## [259] "attr" "attr.all.equal"
## [261] "attr<-" "attributes"
## [263] "attributes<-" "autoload"
## [265] "autoloader" "backsolve"
## [267] "balancePOSIXlt" "baseenv"
## [269] "basename" "besselI"
## [271] "besselJ" "besselK"
## [273] "besselY" "beta"
## [275] "bindingIsActive" "bindingIsLocked"
## [277] "bindtextdomain" "bitwAnd"
## [279] "bitwNot" "bitwOr"
## [281] "bitwShiftL" "bitwShiftR"
## [283] "bitwXor" "body"
## [285] "body<-" "bquote"
## [287] "break" "browser"
## [289] "browserCondition" "browserSetDebug"
## [291] "browserText" "builtins"
## [293] "by" "by.data.frame"
## [295] "by.default" "bzfile"
## [297] "c" "c.Date"
## [299] "c.difftime" "c.factor"
## [301] "c.noquote" "c.numeric_version"
## [303] "c.POSIXct" "c.POSIXlt"
## [305] "c.warnings" "call"
## [307] "callCC" "capabilities"
## [309] "casefold" "cat"
## [311] "cbind" "cbind.data.frame"
## [313] "ceiling" "char.expand"
## [315] "character" "charmatch"
## [317] "charToRaw" "chartr"
## [319] "chkDots" "chol"
## [321] "chol.default" "chol2inv"
## [323] "choose" "chooseOpsMethod"
## [325] "chooseOpsMethod.default" "class"
## [327] "class<-" "clearPushBack"
## [329] "close" "close.connection"
## [331] "close.srcfile" "close.srcfilealias"
## [333] "closeAllConnections" "col"
## [335] "colMeans" "colnames"
## [337] "colnames<-" "colSums"
## [339] "commandArgs" "comment"
## [341] "comment<-" "complex"
## [343] "computeRestarts" "conditionCall"
## [345] "conditionCall.condition" "conditionMessage"
## [347] "conditionMessage.condition" "conflictRules"
## [349] "conflicts" "Conj"
## [351] "contributors" "cos"
## [353] "cosh" "cospi"
## [355] "crossprod" "Cstack_info"
## [357] "cummax" "cummin"
## [359] "cumprod" "cumsum"
## [361] "curlGetHeaders" "cut"
## [363] "cut.Date" "cut.default"
## [365] "cut.POSIXt" "data.class"
## [367] "data.frame" "data.matrix"
## [369] "date" "debug"
## [371] "debuggingState" "debugonce"
## [373] "declare" "default.stringsAsFactors"
## [375] "delayedAssign" "deparse"
## [377] "deparse1" "det"
## [379] "detach" "determinant"
## [381] "determinant.matrix" "dget"
## [383] "diag" "diag<-"
## [385] "diff" "diff.Date"
## [387] "diff.default" "diff.difftime"
## [389] "diff.POSIXt" "difftime"
## [391] "digamma" "dim"
## [393] "dim.data.frame" "dim<-"
## [395] "dimnames" "dimnames.data.frame"
## [397] "dimnames<-" "dimnames<-.data.frame"
## [399] "dir" "dir.create"
## [401] "dir.exists" "dirname"
## [403] "do.call" "dontCheck"
## [405] "double" "dput"
## [407] "dQuote" "drop"
## [409] "droplevels" "droplevels.data.frame"
## [411] "droplevels.factor" "dump"
## [413] "duplicated" "duplicated.array"
## [415] "duplicated.data.frame" "duplicated.default"
## [417] "duplicated.matrix" "duplicated.numeric_version"
## [419] "duplicated.POSIXlt" "duplicated.warnings"
## [421] "dyn.load" "dyn.unload"
## [423] "dynGet" "eapply"
## [425] "eigen" "emptyenv"
## [427] "enc2native" "enc2utf8"
## [429] "encodeString" "Encoding"
## [431] "Encoding<-" "endsWith"
## [433] "enquote" "env.profile"
## [435] "environment" "environment<-"
## [437] "environmentIsLocked" "environmentName"
## [439] "errorCondition" "eval"
## [441] "eval.parent" "evalq"
## [443] "Exec" "exists"
## [445] "exp" "expand.grid"
## [447] "expm1" "expression"
## [449] "extSoftVersion" "F"
## [451] "factor" "factorial"
## [453] "fifo" "file"
## [455] "file.access" "file.append"
## [457] "file.choose" "file.copy"
## [459] "file.create" "file.exists"
## [461] "file.info" "file.link"
## [463] "file.mode" "file.mtime"
## [465] "file.path" "file.remove"
## [467] "file.rename" "file.show"
## [469] "file.size" "file.symlink"
## [471] "Filter" "Find"
## [473] "find.package" "findInterval"
## [475] "findPackageEnv" "findRestart"
## [477] "floor" "flush"
## [479] "flush.connection" "for"
## [481] "force" "forceAndCall"
## [483] "formals" "formals<-"
## [485] "format" "format.AsIs"
## [487] "format.data.frame" "format.Date"
## [489] "format.default" "format.difftime"
## [491] "format.factor" "format.hexmode"
## [493] "format.info" "format.libraryIQR"
## [495] "format.numeric_version" "format.octmode"
## [497] "format.packageInfo" "format.POSIXct"
## [499] "format.POSIXlt" "format.pval"
## [501] "format.summaryDefault" "formatC"
## [503] "formatDL" "forwardsolve"
## [505] "function" "gamma"
## [507] "gc" "gc.time"
## [509] "gcinfo" "gctorture"
## [511] "gctorture2" "get"
## [513] "get0" "getAllConnections"
## [515] "getCallingDLL" "getCallingDLLe"
## [517] "getConnection" "getDLLRegisteredRoutines"
## [519] "getDLLRegisteredRoutines.character" "getDLLRegisteredRoutines.DLLInfo"
## [521] "getElement" "geterrmessage"
## [523] "getExportedValue" "getHook"
## [525] "getLoadedDLLs" "getNamespace"
## [527] "getNamespaceExports" "getNamespaceImports"
## [529] "getNamespaceInfo" "getNamespaceName"
## [531] "getNamespaceUsers" "getNamespaceVersion"
## [533] "getNativeSymbolInfo" "getOption"
## [535] "getRversion" "getSrcLines"
## [537] "getTaskCallbackNames" "gettext"
## [539] "gettextf" "getwd"
## [541] "gl" "globalCallingHandlers"
## [543] "globalenv" "gregexec"
## [545] "gregexpr" "grep"
## [547] "grepl" "grepRaw"
## [549] "grouping" "gsub"
## [551] "gzcon" "gzfile"
## [553] "I" "iconv"
## [555] "iconvlist" "icuGetCollate"
## [557] "icuSetCollate" "identical"
## [559] "identity" "if"
## [561] "ifelse" "Im"
## [563] "importIntoEnv" "infoRDS"
## [565] "inherits" "integer"
## [567] "interaction" "interactive"
## [569] "intersect" "intToBits"
## [571] "intToUtf8" "inverse.rle"
## [573] "invisible" "invokeRestart"
## [575] "invokeRestartInteractively" "is.array"
## [577] "is.atomic" "is.call"
## [579] "is.character" "is.complex"
## [581] "is.data.frame" "is.double"
## [583] "is.element" "is.environment"
## [585] "is.expression" "is.factor"
## [587] "is.finite" "is.finite.POSIXlt"
## [589] "is.function" "is.infinite"
## [591] "is.infinite.POSIXlt" "is.integer"
## [593] "is.language" "is.list"
## [595] "is.loaded" "is.logical"
## [597] "is.matrix" "is.na"
## [599] "is.na.data.frame" "is.na.numeric_version"
## [601] "is.na.POSIXlt" "is.na<-"
## [603] "is.na<-.default" "is.na<-.factor"
## [605] "is.na<-.numeric_version" "is.name"
## [607] "is.nan" "is.nan.POSIXlt"
## [609] "is.null" "is.numeric"
## [611] "is.numeric.Date" "is.numeric.difftime"
## [613] "is.numeric.POSIXt" "is.numeric_version"
## [615] "is.object" "is.ordered"
## [617] "is.package_version" "is.pairlist"
## [619] "is.primitive" "is.qr"
## [621] "is.R" "is.raw"
## [623] "is.recursive" "is.single"
## [625] "is.symbol" "is.table"
## [627] "is.unsorted" "is.vector"
## [629] "isa" "isatty"
## [631] "isBaseNamespace" "isdebugged"
## [633] "isFALSE" "isIncomplete"
## [635] "isNamespace" "isNamespaceLoaded"
## [637] "ISOdate" "ISOdatetime"
## [639] "isOpen" "isRestart"
## [641] "isS4" "isSeekable"
## [643] "isSymmetric" "isSymmetric.matrix"
## [645] "isTRUE" "jitter"
## [647] "julian" "julian.Date"
## [649] "julian.POSIXt" "kappa"
## [651] "kappa.default" "kappa.lm"
## [653] "kappa.qr" "kronecker"
## [655] "l10n_info" "La.svd"
## [657] "La_library" "La_version"
## [659] "labels" "labels.default"
## [661] "lapply" "lazyLoad"
## [663] "lazyLoadDBexec" "lazyLoadDBfetch"
## [665] "lbeta" "lchoose"
## [667] "length" "length.POSIXlt"
## [669] "length<-" "length<-.Date"
## [671] "length<-.difftime" "length<-.factor"
## [673] "length<-.POSIXct" "length<-.POSIXlt"
## [675] "lengths" "letters"
## [677] "LETTERS" "levels"
## [679] "levels.default" "levels<-"
## [681] "levels<-.factor" "lfactorial"
## [683] "lgamma" "libcurlVersion"
## [685] "library" "library.dynam"
## [687] "library.dynam.unload" "licence"
## [689] "license" "list"
## [691] "list.dirs" "list.files"
## [693] "list2DF" "list2env"
## [695] "load" "loadedNamespaces"
## [697] "loadingNamespaceInfo" "loadNamespace"
## [699] "local" "lockBinding"
## [701] "lockEnvironment" "log"
## [703] "log10" "log1p"
## [705] "log2" "logb"
## [707] "logical" "lower.tri"
## [709] "ls" "make.names"
## [711] "make.unique" "makeActiveBinding"
## [713] "Map" "mapply"
## [715] "margin.table" "marginSums"
## [717] "mat.or.vec" "match"
## [719] "match.arg" "match.call"
## [721] "match.fun" "Math.data.frame"
## [723] "Math.Date" "Math.difftime"
## [725] "Math.factor" "Math.POSIXt"
## [727] "matrix" "max"
## [729] "max.col" "mean"
## [731] "mean.Date" "mean.default"
## [733] "mean.difftime" "mean.POSIXct"
## [735] "mean.POSIXlt" "mem.maxNSize"
## [737] "mem.maxVSize" "memCompress"
## [739] "memDecompress" "memory.profile"
## [741] "merge" "merge.data.frame"
## [743] "merge.default" "message"
## [745] "mget" "min"
## [747] "missing" "Mod"
## [749] "mode" "mode<-"
## [751] "month.abb" "month.name"
## [753] "months" "months.Date"
## [755] "months.POSIXt" "mostattributes<-"
## [757] "mtfrm" "mtfrm.default"
## [759] "mtfrm.POSIXct" "mtfrm.POSIXlt"
## [761] "nameOfClass" "nameOfClass.default"
## [763] "names" "names.POSIXlt"
## [765] "names<-" "names<-.POSIXlt"
## [767] "namespaceExport" "namespaceImport"
## [769] "namespaceImportClasses" "namespaceImportFrom"
## [771] "namespaceImportMethods" "nargs"
## [773] "nchar" "ncol"
## [775] "NCOL" "Negate"
## [777] "new.env" "next"
## [779] "NextMethod" "ngettext"
## [781] "nlevels" "noquote"
## [783] "norm" "normalizePath"
## [785] "nrow" "NROW"
## [787] "nullfile" "numeric"
## [789] "numeric_version" "numToBits"
## [791] "numToInts" "nzchar"
## [793] "objects" "oldClass"
## [795] "oldClass<-" "OlsonNames"
## [797] "on.exit" "open"
## [799] "open.connection" "open.srcfile"
## [801] "open.srcfilealias" "open.srcfilecopy"
## [803] "Ops.data.frame" "Ops.Date"
## [805] "Ops.difftime" "Ops.factor"
## [807] "Ops.numeric_version" "Ops.ordered"
## [809] "Ops.POSIXt" "options"
## [811] "order" "ordered"
## [813] "outer" "package_version"
## [815] "packageEvent" "packageHasNamespace"
## [817] "packageNotFoundError" "packageStartupMessage"
## [819] "packBits" "pairlist"
## [821] "parent.env" "parent.env<-"
## [823] "parent.frame" "parse"
## [825] "parseNamespaceFile" "paste"
## [827] "paste0" "path.expand"
## [829] "path.package" "pcre_config"
## [831] "pi" "pipe"
## [833] "plot" "pmatch"
## [835] "pmax" "pmax.int"
## [837] "pmin" "pmin.int"
## [839] "polyroot" "pos.to.env"
## [841] "Position" "pretty"
## [843] "pretty.default" "prettyNum"
## [845] "print" "print.AsIs"
## [847] "print.by" "print.condition"
## [849] "print.connection" "print.data.frame"
## [851] "print.Date" "print.default"
## [853] "print.difftime" "print.Dlist"
## [855] "print.DLLInfo" "print.DLLInfoList"
## [857] "print.DLLRegisteredRoutines" "print.eigen"
## [859] "print.factor" "print.function"
## [861] "print.hexmode" "print.libraryIQR"
## [863] "print.listof" "print.NativeRoutineList"
## [865] "print.noquote" "print.numeric_version"
## [867] "print.octmode" "print.packageInfo"
## [869] "print.POSIXct" "print.POSIXlt"
## [871] "print.proc_time" "print.restart"
## [873] "print.rle" "print.simple.list"
## [875] "print.srcfile" "print.srcref"
## [877] "print.summary.table" "print.summary.warnings"
## [879] "print.summaryDefault" "print.table"
## [881] "print.warnings" "prmatrix"
## [883] "proc.time" "prod"
## [885] "prop.table" "proportions"
## [887] "provideDimnames" "psigamma"
## [889] "pushBack" "pushBackLength"
## [891] "q" "qr"
## [893] "qr.coef" "qr.default"
## [895] "qr.fitted" "qr.Q"
## [897] "qr.qty" "qr.qy"
## [899] "qr.R" "qr.resid"
## [901] "qr.solve" "qr.X"
## [903] "quarters" "quarters.Date"
## [905] "quarters.POSIXt" "quit"
## [907] "quote" "R.home"
## [909] "R.version" "R.Version"
## [911] "R.version.string" "R_compiled_by"
## [913] "R_system_version" "range"
## [915] "range.Date" "range.default"
## [917] "range.POSIXct" "rank"
## [919] "rapply" "raw"
## [921] "rawConnection" "rawConnectionValue"
## [923] "rawShift" "rawToBits"
## [925] "rawToChar" "rbind"
## [927] "rbind.data.frame" "rcond"
## [929] "Re" "read.dcf"
## [931] "readBin" "readChar"
## [933] "readline" "readLines"
## [935] "readRDS" "readRenviron"
## [937] "Recall" "Reduce"
## [939] "reg.finalizer" "regexec"
## [941] "regexpr" "registerS3method"
## [943] "registerS3methods" "regmatches"
## [945] "regmatches<-" "remove"
## [947] "removeTaskCallback" "rep"
## [949] "rep.Date" "rep.difftime"
## [951] "rep.factor" "rep.int"
## [953] "rep.numeric_version" "rep.POSIXct"
## [955] "rep.POSIXlt" "rep_len"
## [957] "repeat" "replace"
## [959] "replicate" "require"
## [961] "requireNamespace" "restartDescription"
## [963] "restartFormals" "retracemem"
## [965] "return" "returnValue"
## [967] "rev" "rev.default"
## [969] "rle" "rm"
## [971] "RNGkind" "RNGversion"
## [973] "round" "round.Date"
## [975] "round.POSIXt" "row"
## [977] "row.names" "row.names.data.frame"
## [979] "row.names.default" "row.names<-"
## [981] "row.names<-.data.frame" "row.names<-.default"
## [983] "rowMeans" "rownames"
## [985] "rownames<-" "rowsum"
## [987] "rowsum.data.frame" "rowsum.default"
## [989] "rowSums" "sample"
## [991] "sample.int" "sapply"
## [993] "save" "save.image"
## [995] "saveRDS" "scale"
## [997] "scale.default" "scan"
## [999] "search" "searchpaths"
## [1001] "seek" "seek.connection"
## [1003] "seq" "seq.Date"
## [1005] "seq.default" "seq.int"
## [1007] "seq.POSIXt" "seq_along"
## [1009] "seq_len" "sequence"
## [1011] "sequence.default" "serialize"
## [1013] "serverSocket" "set.seed"
## [1015] "setdiff" "setequal"
## [1017] "setHook" "setNamespaceInfo"
## [1019] "setSessionTimeLimit" "setTimeLimit"
## [1021] "setwd" "shell"
## [1023] "shell.exec" "showConnections"
## [1025] "shQuote" "sign"
## [1027] "signalCondition" "signif"
## [1029] "simpleCondition" "simpleError"
## [1031] "simpleMessage" "simpleWarning"
## [1033] "simplify2array" "sin"
## [1035] "single" "sinh"
## [1037] "sink" "sink.number"
## [1039] "sinpi" "slice.index"
## [1041] "socketAccept" "socketConnection"
## [1043] "socketSelect" "socketTimeout"
## [1045] "solve" "solve.default"
## [1047] "solve.qr" "sort"
## [1049] "sort.default" "sort.int"
## [1051] "sort.list" "sort.POSIXlt"
## [1053] "sort_by" "sort_by.data.frame"
## [1055] "sort_by.default" "source"
## [1057] "split" "split.data.frame"
## [1059] "split.Date" "split.default"
## [1061] "split.POSIXct" "split<-"
## [1063] "split<-.data.frame" "split<-.default"
## [1065] "sprintf" "sqrt"
## [1067] "sQuote" "srcfile"
## [1069] "srcfilealias" "srcfilecopy"
## [1071] "srcref" "standardGeneric"
## [1073] "startsWith" "stderr"
## [1075] "stdin" "stdout"
## [1077] "stop" "stopifnot"
## [1079] "storage.mode" "storage.mode<-"
## [1081] "str2expression" "str2lang"
## [1083] "strftime" "strptime"
## [1085] "strrep" "strsplit"
## [1087] "strtoi" "strtrim"
## [1089] "structure" "strwrap"
## [1091] "sub" "subset"
## [1093] "subset.data.frame" "subset.default"
## [1095] "subset.matrix" "substitute"
## [1097] "substr" "substr<-"
## [1099] "substring" "substring<-"
## [1101] "sum" "summary"
## [1103] "summary.connection" "summary.data.frame"
## [1105] "Summary.data.frame" "summary.Date"
## [1107] "Summary.Date" "summary.default"
## [1109] "Summary.difftime" "summary.factor"
## [1111] "Summary.factor" "summary.matrix"
## [1113] "Summary.numeric_version" "Summary.ordered"
## [1115] "summary.POSIXct" "Summary.POSIXct"
## [1117] "summary.POSIXlt" "Summary.POSIXlt"
## [1119] "summary.proc_time" "summary.srcfile"
## [1121] "summary.srcref" "summary.table"
## [1123] "summary.warnings" "suppressMessages"
## [1125] "suppressPackageStartupMessages" "suppressWarnings"
## [1127] "suspendInterrupts" "svd"
## [1129] "sweep" "switch"
## [1131] "sys.call" "sys.calls"
## [1133] "Sys.chmod" "Sys.Date"
## [1135] "sys.frame" "sys.frames"
## [1137] "sys.function" "Sys.getenv"
## [1139] "Sys.getlocale" "Sys.getpid"
## [1141] "Sys.glob" "Sys.info"
## [1143] "Sys.junction" "sys.load.image"
## [1145] "Sys.localeconv" "sys.nframe"
## [1147] "sys.on.exit" "sys.parent"
## [1149] "sys.parents" "Sys.readlink"
## [1151] "sys.save.image" "Sys.setenv"
## [1153] "Sys.setFileTime" "Sys.setLanguage"
## [1155] "Sys.setlocale" "Sys.sleep"
## [1157] "sys.source" "sys.status"
## [1159] "Sys.time" "Sys.timezone"
## [1161] "Sys.umask" "Sys.unsetenv"
## [1163] "Sys.which" "system"
## [1165] "system.file" "system.time"
## [1167] "system2" "t"
## [1169] "T" "t.data.frame"
## [1171] "t.default" "table"
## [1173] "tabulate" "Tailcall"
## [1175] "tan" "tanh"
## [1177] "tanpi" "tapply"
## [1179] "taskCallbackManager" "tcrossprod"
## [1181] "tempdir" "tempfile"
## [1183] "textConnection" "textConnectionValue"
## [1185] "tolower" "topenv"
## [1187] "toString" "toString.default"
## [1189] "toupper" "trace"
## [1191] "traceback" "tracemem"
## [1193] "tracingState" "transform"
## [1195] "transform.data.frame" "transform.default"
## [1197] "trigamma" "trimws"
## [1199] "trunc" "trunc.Date"
## [1201] "trunc.POSIXt" "truncate"
## [1203] "truncate.connection" "try"
## [1205] "tryCatch" "tryInvokeRestart"
## [1207] "typeof" "unCfillPOSIXlt"
## [1209] "unclass" "undebug"
## [1211] "union" "unique"
## [1213] "unique.array" "unique.data.frame"
## [1215] "unique.default" "unique.matrix"
## [1217] "unique.numeric_version" "unique.POSIXlt"
## [1219] "unique.warnings" "units"
## [1221] "units.difftime" "units<-"
## [1223] "units<-.difftime" "unix.time"
## [1225] "unlink" "unlist"
## [1227] "unloadNamespace" "unlockBinding"
## [1229] "unname" "unserialize"
## [1231] "unsplit" "untrace"
## [1233] "untracemem" "unz"
## [1235] "upper.tri" "url"
## [1237] "use" "UseMethod"
## [1239] "utf8ToInt" "validEnc"
## [1241] "validUTF8" "vapply"
## [1243] "vector" "Vectorize"
## [1245] "version" "warning"
## [1247] "warningCondition" "warnings"
## [1249] "weekdays" "weekdays.Date"
## [1251] "weekdays.POSIXt" "which"
## [1253] "which.max" "which.min"
## [1255] "while" "with"
## [1257] "with.default" "withAutoprint"
## [1259] "withCallingHandlers" "within"
## [1261] "within.data.frame" "within.list"
## [1263] "withRestarts" "withVisible"
## [1265] "write" "write.dcf"
## [1267] "writeBin" "writeChar"
## [1269] "writeLines" "xor"
## [1271] "xpdrows.data.frame" "xtfrm"
## [1273] "xtfrm.AsIs" "xtfrm.data.frame"
## [1275] "xtfrm.Date" "xtfrm.default"
## [1277] "xtfrm.difftime" "xtfrm.factor"
## [1279] "xtfrm.numeric_version" "xtfrm.POSIXct"
## [1281] "xtfrm.POSIXlt" "xzfile"
## [1283] "zapsmall"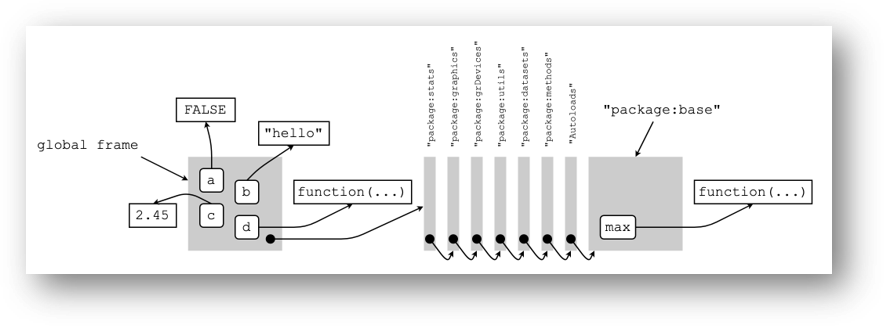
Special Environments in R
In R, there are four special environments:
The
globalenv(), or global environment, is the interactive workspace. This is the environment in which you normally work.The
baseenv(), or base environment, is the environment of the base package. Its parent is the empty environment.The
emptyenv(), or empty environment, is the ultimate ancestor of all environments, and the only environment without a parent.The
environment()is the current environment.
Hierarchy of Environments
Names are evaluated using the hierarchy of environments
Remember this operator: <-? This is called the assignment operator.
What it does is create a bind between the value (or more generally the object) on the right of it to the name of the left.
<NAME> <- <OBJECT>Hence, the name is associated with the object (which exists virtually in our computer’s memory).
Consequently, an object can be associated with many different names.
Here, the object 2 is only a single entity, i.e., we are not creating multiple copies of the entity 2.
Rather, we are associating 2 with different names—x, y, and z. In short, 2 can be called using x, y, and z.
But note that the assignment operator is unidirectional—it doesn’t go both ways.
It means that, while an object can be associated with many different names, a name can only be associated with a single object.
That’s why, in the lines of code above, the second line rebinds the name x to the new value 3, and when x is called after running the lines of code, 3 will be returned by the computer and not 2.
But, if names have to be unique, does this mean you cannot repeat a name when you want to refer to different objects?
- While an object can be associated with many different names, a name can only be associated with a single object in a particular environment.
So what is an environment?
- An environment is a collection of name-object bindings (which are located in a frame) + the environment’s parent environment.
Consider the following bindings below done in the global environment (our default working environment).
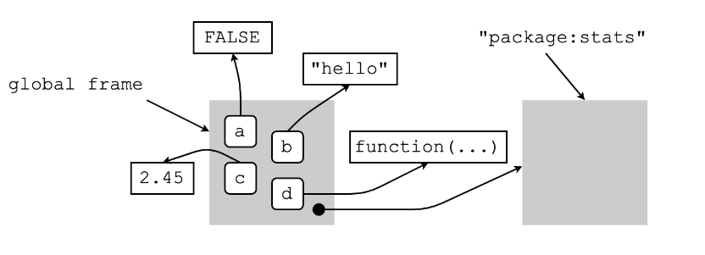
What is this telling us?
the four name-object bindings we made above are found in the global frame (i.e., the frame of the global environment), and
global frame + the global environment’s parent environment (which in this case is package:stats) are what make up the global environment.
What do we mean by “a name can only be associated with a single object in a particular environment”?
Well, it means that, in a particular environment—say, the global environment—the name
arefers toFALSEonly,bto"hello"only, and so on.But, in a different environment, these names can be bound to different values or objects.
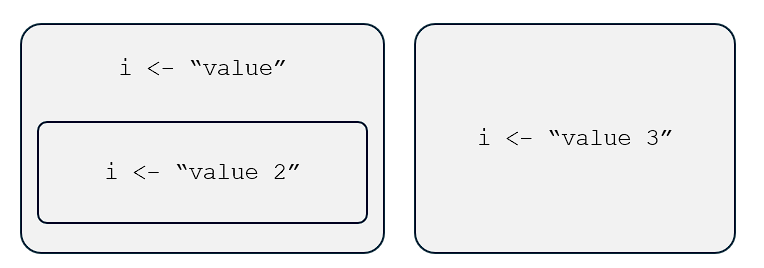
Why is this relevant?
First, we can reuse names without our computers getting confused. Depending on the context—the environment—in which the computer is operating on, a particular name will refer to a particular object relevant to that context.
Second, our names don’t have to be convoluted. We can keep them simple like
counter,n,i, etc., and use them for a particular context we are working on.Third, it gives us the ability to simplify complex tasks.
If we call a name in a particular environment, our computer will:
Look for a binding to that name in the frame of the environment.
- If a binding was found, our computer will return the object associated with that name.
- If not, our computer will go to the parent environment of the particular environment.
- In the parent environment of the particular environment, our computer will look for a binding to that name.
- If a binding was found, our computer will return the object associated with that name.
- If not, our computer will go to the grandparent environment of the particular environment. And so on….
- In the parent environment of the particular environment, our computer will look for a binding to that name.
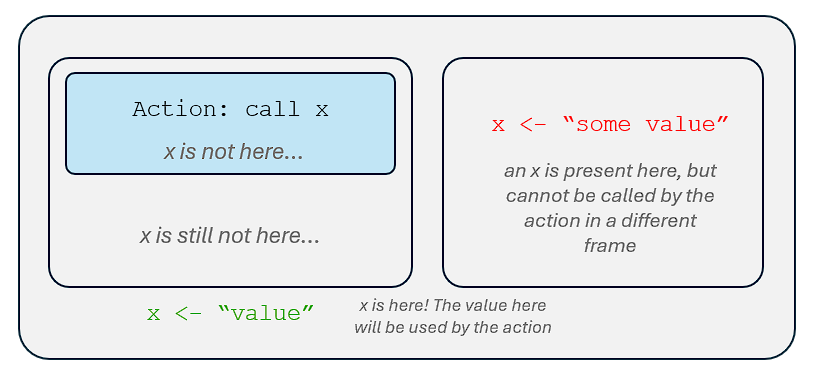
Why are environments important?
They give context to the names that we are calling in our computer, so that the name we can use to bind to our objects will not necessarily be unique.
Hierarchy

Now that we already know what are environments, we now explore how calling a function creates their own environment.
1.2.2 Calling User-Defined Functions
To evaluate a call expression whose operator names a user-defined function, the R interpreter follows a computational process. As with any call expression, the interpreter evaluates the operator and operand expressions, and then applies the named function to the resulting arguments.
Applying a user-defined function introduces a local frame, which is only accessible to that function. To apply a user-defined function to some arguments,
first, bind the arguments to the names of the function’s formal parameters in a new local frame,
then, execute the body of the function in the environment that starts with this frame.
The environment in which the body is evaluated consists of two frames:
first the local frame that contains formal parameter bindings,
then the global frame that contains everything else.
Each instance of a function application has its own independent local frame.
Example 1:
Let’s define a new function (let’s call it square) that has a formal parameter (we call x). Our function is simple: we want the function to get a square of a number.
The function square should now appear in the R_GlobalEnv.
## [1] "a" "ackermann"
## [3] "add_three" "age"
## [5] "age1" "age2"
## [7] "age3" "Anscombe"
## [9] "apply_function" "apply_trimmed"
## [11] "apply_twice" "arr"
## [13] "assignedSex1" "assignedSex2"
## [15] "assignedSex3" "b"
## [17] "B" "bank"
## [19] "beta" "betahat"
## [21] "bias_mean" "bias_median"
## [23] "binary_search" "boot.mpg"
## [25] "boot.reg" "boot_reg"
## [27] "boot2" "c"
## [29] "cards" "class_data"
## [31] "color" "con"
## [33] "customer_data" "customers"
## [35] "d" "dat1"
## [37] "dat2" "dat3"
## [39] "dat4" "data"
## [41] "data.star" "differentiate"
## [43] "double" "eval"
## [45] "exp_b" "f"
## [47] "factorial_loop" "fit"
## [49] "fit2" "fold"
## [51] "folds" "func"
## [53] "g" "generate_sequence"
## [55] "get_element" "gpa"
## [57] "grad_rosenbrock" "grade"
## [59] "h" "hess_rosenbrock"
## [61] "i" "index"
## [63] "j" "k"
## [65] "kfold" "kmeans_result"
## [67] "l" "lawschool"
## [69] "linear_search" "lm_fit"
## [71] "lm_validate" "loglik_lgstc"
## [73] "logreg_plot" "loocv"
## [75] "lsat" "M"
## [77] "mat" "mc_norm"
## [79] "mc_pois" "menu"
## [81] "method1" "method2"
## [83] "method3" "mod"
## [85] "mod1" "mod2"
## [87] "model" "mpg.boot"
## [89] "mtcars" "my_array"
## [91] "my_chr" "my_dbl"
## [93] "my_int" "my_list"
## [95] "my_log" "my_sorted_array"
## [97] "n" "N"
## [99] "names" "nested_loop"
## [101] "newton_optim" "newton_raphson_logistic"
## [103] "p" "P"
## [105] "pi_x" "pop"
## [107] "pred1" "predictions"
## [109] "prop" "Q"
## [111] "r.boot" "r.star"
## [113] "residual_bootstrap" "result"
## [115] "result_plot" "rmse"
## [117] "rmse_mod" "rmse_x"
## [119] "rosenbrock" "runtime"
## [121] "samp" "samp.star"
## [123] "sample_size" "simple_model"
## [125] "square" "square_d"
## [127] "square_loop" "start_time"
## [129] "SumOfDigits" "test"
## [131] "test_data" "time"
## [133] "tmp" "train"
## [135] "train_data" "train_indices"
## [137] "transactions" "update_x"
## [139] "validation" "var_mc"
## [141] "var_p" "var_phat"
## [143] "vari_mean" "vari_median"
## [145] "vec1" "vec2"
## [147] "vec3" "vec4"
## [149] "vec5" "vec6"
## [151] "x" "X"
## [153] "x0" "xbar.boot"
## [155] "xbar.star" "y"
## [157] "y1" "y2"
## [159] "y3" "years"
## [161] "z" "Z"
## [163] "Z_vec"BUT, the body of the this function will not be executed yet, until the function is called (and not when it is defined).
When we call this function with argument equal to -2, a local frame will be created.
## [1] 4In this local frame, x will be bound to the value -2, and result will be bound to the value of the expression x*x.
Let’s modify the function to visualize the statements above.
square <- function(x){
result <- x*x
print(ls()) # show the bindings in the local frame
return(result)
}
square(-2)## [1] "result" "x"## [1] 4The “Return value” in the local frame of function square is not a name binding. Instead it indicates the value returned by the function call that created the frame.
Even in this simple example, 2 environments were used:
1. The local environment of the function call.
2. The global environment, where the top level expression square(-2) was evaluated.
All about context
Each time a function is called, bindings are created in a separate environment.
| Name | Global Environment | New Environment
|
New Environment
|
|---|---|---|---|
x |
3 |
2+5=7 |
3+5=8 |
a |
5 |
5 |
5 |
b |
2 |
x=3 |
|
func |
<definition> |
Name evaluation
A name evaluates to the value bound to that name in the earliest frame of the current environment in which that name is found.
Names are bound to values, which are distributed across many independent local frames, along with a single global frame that contains shared names. A new local frame is introduced every time a function is called, even if the same function is called twice.
Example:
To visualize further, let’s have the following:
y <- 10
square <- function(x){
return(x*x)
}
d <- function(x){
z <- square(x) + square(y)
print(ls())
return(z)
}If we call it using d(5), we can see the object that are found in the function’s local frame.
## [1] "x" "z"## [1] 125x and z are the only bindings found in the function call’s local frame.
y was called (used in an expression), but not bound to any value inside function’s local frame. Instead y can be found in the local frame’s parent environment, which is the global frame.
Note that even if there was no binding to x in the function body, there was still a binding to it since x is an argument.
When d(5) was called, there was a binding x <- 5 that occurred in the function call.
The function square was also called twice within the function d. When it was called, local frames were created temporarily.
All three local frames contain a binding for the name x, but that name is bound to different values in different frames. Local frames keep these names separate.
Local Names
An argument in a function is not related with the argument of another function, even if they have the same name.
One detail of a function’s implementation that should not affect the function’s behavior is the implementer’s choice of names for the function’s formal parameters.
Example 1: Same structure, different parameters
Even if we use different parameter names in the 2 functions, the result should be the same.
Example 2: Different structure, same parameters
Even if the parameter names are the same in the two functions, they should not affect each other.
We say that the scope of a local name is limited to the body of the user-defined function that defines it. When a name is no longer accessible, it is out of scope.
Lexical Scoping
Locally defined functions also have access to the name bindings in the scope in which they are defined.
This discipline of sharing names among nested definitions is called lexical scoping. Critically, the inner functions have access to the names in the environment where they are defined (not where they are called).
The scoping rules of a language determine how a value is associated with a free variable in a function.
Consider the function below:
Free variables are not formal arguments and are not local variables (assigned inside the function body). In this example, x is a free variable.
R uses lexical scoping or static scoping.
An alternative to lexical scoping is dynamic scoping which is implemented by some languages.
We require two extensions to our environment model to enable lexical scoping.
Each user-defined function has a parent environment: the environment in which it was defined.
When a user-defined function is called, its local frame extends its parent environment.
To further illustrate, we have another example
The y in f()
The y in the definition of f() is not a formal parameter, but defined inside the body. This is a local variable.
The y in g()
On the other hand, in the new function g(), y is still not a formal argument and also not locally defined. This is a free variable.
Its value will be based on the parent environment where it was defined, not on the local frame of g().
Questions:
What is the value of
f(3)?What is the value of
g(3)?
1.2.3 Function Parameters
So far, in our examples, parameters take single numeric values. Functions in different programming languages make take different data types as input parameters, provided that the commands in the function body can be applied on that object type.
For example, we can have a vector as an input in the square function.
## [1] 1 4 9The output will be consistent with the corresponding result of the vector operation.
## [1] 1 4 9We can also create a function that may take character objects.
## [1] "Hello, UserName!"Practice Exercises
Scope of a function
Before doing this exercise, make sure that your environment is empty.
Create a function called
modify_value()that takes an argumentxand adds10to it.Inside the function, define local variable
y <- 5.Print the values of
xandyinside the function.Without calling the function, try printing
youtside the function. Did it print?
Trimmed mean
A trimmed mean is a measure much like the mean that discards the smallest and largest values. Using only basic arithmetic operations (
+,-,*, and/) and the functionsmaxandmin, write a simple function that computes for the trimmed mean of five numbers.The function should be named
trimmed_mean_5that takes in five arguments and discards the smallest value and largest value. Then use the functiontrimmed_mean_5to find the trimmed mean of2,4,20,0, and-3that discards the smallest and largest values.